Expressions of Toll Like Receptor (TLR) Genes in Paralichthys olivaceus After Induced by Different Extracts of Edwardsiella tarda
SHAN Yanan, ZHENG Jinhui, GAO Hong, and SUN Jinsheng
Expressions of Toll Like Receptor (TLR) Genes inAfter Induced by Different Extracts of
SHAN Yanan, ZHENG Jinhui, GAO Hong*, and SUN Jinsheng*
,,,300387,
Toll like receptors (TLRs) are the main innate immune ‘pattern recognition receptors’ of animals,which play a central role in host cell recognition and responses to invasive pathogens, particularly common structures of microbial pathogens. In this study, the gene expression profiles of TLRs in the spleen, head kidney,gill, small intestine, liver, muscle, and heart of healthywere detected by real-time quantitative PCR (qPCR).The TLR family members were widely expressed in different tissues with different basic expression profiles. The highest expressions of TLR1, 5m, 7, 8, 9, 14, and 21 were found in the spleen; the highest expressions of TLR3 and TLR21 were found in the gill; the highest expressions of TLR2 and 5swere found in the small intestine. The second highest expressions of TLR3, 7, and 8 were found in small intestine.The gene expression profiles of TLRs stimulated withDNA, RNA, and lipopolysaccharide (LPS)were also detected inspleen, head kidney and gill. TLR9 and TLR21 were sensitive toDNA; TLR 8 and TLR21 were sensitive toRNA; and TLR1 and TLR14 were sensitive toLPS. The expressions of the other TLR genes showed no significant changes. The results imply that the expressions of these TLR genes inare differently regulated in the whole body and play important roles in the immune response againstinfection.
;;TLRs; expression; real-time fluorescence quantitative PCR
1 Introduction
belongs to the flounder family and is also called flounder or partial mouth in China.It is delicious with high nutritional value. It is mainly distri-buted in the east and west coasts of South America and North America as well as in Bohai Sea, Yellow Sea, East China Sea, South China Sea and far east coastal areas of Korea, Japan, and Russia in Asia.
Toll-like receptors (TLRs)are a class of important pro- tein molecules involved in pattern recognition receptors(PRRs) and have recognition function (Zhou., 2018). TLR is a single transmembrane protein that can recognize microorganisms. When they break through the physical bar-riers of the body, such as skin and mucous membrane, TLRscan recognize such barriers and activate immune cell re-sponse. The TLR family includes many members. At pre- sent, 17 TLR members have been found in fish, including TLRs 1, 2, 3, 4, 5, 5s, 7, 8, 9, 13, 14, 18, 19, 20, 21, 22, and 23. In this study, we mainly investigate the expression and distribution of 10 TLR genes (1, 2, 3, 5m, 5s, 7, 8, 9, 14, and 21) in important immune tissues of.
Although fish is a relatively lower vertebrate, it has in-nate immune response and acquired immune response (Whyte, 2007). Fish is in a special evolution position be- tween higher vertebrates and lower invertebrates, and its acquired immune mechanism is not perfect (Wilson, 2017). For the two kinds of immune responses, innate immune re- sponse plays a very important role.
is a pathogenic bacterium that is harm-ful to freshwater and marine fishes.It can cause diseases in more than 20 kinds of fishes, such as flounder,, carp, and so on.is a common pathogen forboth human and fish, and can pose a threat to human health(Gao., 2013; Zhou and Sun, 2016). Therefore, research ofhas attracted extensive attention. This bacterium mainly invades fish through the digestive tract, gill, or in- jured epidermis, causing great losses to aquaculture (Qin., 2017). A comprehensive and in-depth study on the molecular mechanism of the immune response ofto the pathogen will provide a theoretical basis for the establishment of an effective immune control measure to prevent and controlinfection.
TLRs are widely distributed in tissues and cells of the body, and the types and numbers of TLRs are different in different tissues or cells. On the other hand, a pathogen of- ten has a variety of pathogen related molecular patterns PAMP that can be recognized by animal bodies. PAMP is recognized by different TLRs members at different time periods of infection, How these TLRs family members work together to activate the innate immune system and activate the specific immune response is still unknown. Therefore, it is not enough to study the immunoregulation mechanismof TLRs only from the perspective of single tissue or singleTLR member. The aim of this study is to investigate the immune responses of TLRs in various tissues of, and to obtain the temporal and spatial expression pro- files of TLRs family members duringinfection, so as to reveal the natural law and immunobiological sig- nificance of the interaction between TLRs and.
2 Materials and Methods
2.1 Materials
2.1.1 Experimental fish
Healthywas purchased from a fish farm in Tianjin. Flounders with normal morphology and a length of 10–12cmwere selected. Theywere raised at 25℃ in artificial sea water in shaded aquarium for a week to adapt to the environment and fed once a day.
2.1.2 Pathogen
was provided by Tianjin Aquaculture Disease Control Center and preserved in our laboratory.
2.2 Methods
2.2.1 Pathogen culture
was inoculated into LB medium (pH=7) on the ultra-clean stage, and cultured at 37℃ with shaking until OD600value was 0.6. The bacteria were fully washed with sterile PBS buffer three times and stored in PBS buffer so- lution. The concentration was adjusted to 1×107cellsmL?1.
2.2.2 Extraction of DNA from E. tarda
Phenol-chloroform extraction method was used. The bac- teria were placed into 500μL of 20×SSC solution and mix- ed overnight with 1% SDS. The next day, the same volumes of phenol, chloroform, and isoamyl alcohol were added. After mixing, the bacteria were centrifuged at 4℃ and 9590.4for 20min. The suspension was added with two volumes of anhydrous ethanol, placed at ?20℃ for 2h, and centrifuged. The precipitate was placed in double SSC so- lution, added with RNase, placed at 37℃ for 1h, added with protease K, and maintained for 1h. The DNA solution in the aqueous phase was mixed evenly with sodium ace- tate (10%, pH=5.2) and 2 volumes of pre-cooled absolute ethanol. DNA was precipitated in an ice bath for 2h. The supernatant was centrifuged at 4℃ with 9590.4for 10min. The supernatant was precipitated twice with 70% ethanol, centrifuged again, dissolved in TE buffer solution, and stored at 4℃ for standby or at ?20℃ for further analyses.
2.2.3 Extraction of RNA from E. tarda
The frozen tissue (fresh or preserved at ?70℃ or in li- quid nitrogen) was placed in a centrifuge tube, added with 1mL of Trizol, homogenized, and allowed to stand at room temperature for 5min. The solution was then added with 0.2mL of chloroform, shaken for 15s, and allowed to standfor 2min. The supernatant was centrifuged at 4℃ with9590.4for 15min, added with 0.5mL of isopropanol, mix-ed gently, and allowed to stand at room temperature for 10min. The supernatant was centrifuged at 4℃ with 9590.4for 10min and added with 1mL of 75% ethanol. The pre- cipitate was washed gently. The supernatant was discarded after the pellet was centrifuged at 4℃ with 3746.25for 5min. The supernatant was dried in the air and added with appropriate amount of DEPC H2O to dissolve (promote the dissolution at 65℃ for 10–15min). The extracted RNA was placed in a refrigerator at ?80℃ prior to use.
2.2.4 Extraction of Lipopolysaccharides (LPS) from the cell wall of E. tarda
About 10mL of the bacterial suspension (107piecemL?1) and 90% phenol solution were preheated for 10min in a wa-ter bath at 66℃. The suspension was added with the same volume of phenol (preheated), stirred for 25min, cooled at room temperature, and refrigerated overnight at 4℃. The next day, centrifugation was performed with 2775for 15min, and the supernatant was removed. The residue and phe- nol layer were added with 10mL of water, stirred in a wa- ter bath at 66℃ for 30min, cooled, and centrifuged to ob- tain the supernatant. The supernatant extracted twice was dialyzed in 0.85% normal saline for 48h, during which the normal saline was changed, and the crude lipopolysaccha- ride was obtained by measuring phenol free (purple free) with ferric chloride. The crude LPS was concentrated with polyethylene glycol 6000 and added with 50μgmL?1DNaseand RNase. Enzymolysis was carried out at 37℃ for 4h. The solution was heated in 66℃water bath for 10min, cooled, and centrifuged at 4℃ with 249.75for 30min. The super-natant was collected and added with two volumes of ace- tone to obtain the purified LPS.
2.2.5 Immunity of P. olivaceus
Each tail ofwas intraperitoneally inject- ed with 100μL of pathogenic stimulants (DNA, RNA, orLPS). The control group was injected with the samevolume of PBS. At each time point after injection, three fish were randomly chosen to be executed for analyses. The tailswere taken, and the other tissues were dissected under ste- rile conditions and then were frozen in liquid nitrogen. The next day, the tissues were transferred to ?80℃for storage.
2.2.6 Quantitative analysis of gene expression
The cDNA kit purchased from Sangon Biotech? Inc. was used to synthesize the first strand of TLR genes in this ex- perimentaccording to its recommended method.
Transcriptome sequencing kit was provided by Beijing Nuohe Zhiyuan Bioinformation Technology Co., Ltd. The sequencing platform was Illumina hiseqTM2500.
The relative quantitative expression data of 10 genes in this experiment were obtained using ABI 7500 fluorescent PCR instrument through the method recommended by Pro- mega’s GoTaq? qPCR Master Mix kit with reverse tran- scriptional cDNA as template.The primer sequences are listed in Table 1. The reaction system (24μL) consisted of 4μL of the cDNA template, 0.4μL of upstream primer (10μmolL?1), 0.4μL of downstream primer (10μmolL?1), 10μL of Promega’s GoTaq? qPCR master mix, and 9.2μL of nuclear free water. The relative quantitative analysis was performed by 2?ΔΔCTmethod, and histogram analysis was performed by Origin 8 software.

Table 1 Gene specific primers used in real time RT-PCR in this project
3 Results
3.1 Expression Profile of TLRs Genes in Normal Tissues of Healthy P. olivaceus
All members of the TLRs family ofwere widely expressed in various tissues, but their basic expres- sion profiles were different (Fig.1). Many TLR members were expressed with a high level in the spleen, gill, and small intestine. The highest expression levels of TLR1, 5m, 7, 8, 9, 14, and 21 were found in the spleen; and the highest ex- pression of TLR3 was found in the gill tissue. TLR21 was also highly expressed in the gill tissue. The highest expres- sions of TLR2 and 5S were found in the small intestine, where TLR 3, 7, and 8 were also expressed with a high le- vel. The spleen and gill of fish are important immune organs,and TLRs have a high basic expression level in immune or- gans, consistent with their defense function(Rebl., 2010). The small intestine is an organ that comes in contact with foreign food and is exposed to a large number of mi- croorganisms for a long time. TLR members exhibited ba- sic expression distribution in this organ, which is also re- lated to the environment of small intestine (Jault., 2004; Meijer., 2004). In contrast to expectations, as another important immune organ, the head kidney showed low ex- pression of TLR genes,though TLR1, 2, 3, and 21 could be detected in head kidney. Under the health status, the spleen functions as a main preventive immune organ, and the head kidney can keep the lowest energy consumption of the body. Their function are different. However, the real reason re- mains to be further investigated. Our subsequent immune stimulation experiments showed that the gene expression of TLRs in the head kidney ofwas significantly up-regulated in response to pathogen invasion upon stimu- lation by pathogenic microorganisms. This expression cha-racteristic may be used as an indicator for health monitor- ing of.
To better understand the difference in the basal expres- sion levels of TLR genes in the same healthy tissue, we re- arranged the data and presented the basal expression pro- file of TLRs in the same tissue (Fig.2). TLR2 and TLR9 genes generally had higher basal expression levels in each tissue than the other TLR members. The overall basal ex- pression levels of TLR family genes were relatively higher in the spleen than in the other tissues.
TLRs were widely expressed in all the tissues of healthy, but the basic expression pattern of TLRs va- ried among different tissues. There are many TLRs in the spleen of healthy, and their basic expression values are high, which indicates that the spleen plays an im-portant role in the prevention of infection. However, the expression of TLRs in the head kidney was low, and the expression of some TLRs increased after immune stimula- tion, suggesting that the head kidney was more focused on the immune response during acute infection.This may sug- gest that the spleen is the main preventive immune organ in healthy. Various TLR genes were also expressed in the heart, liver, and muscle that are non-immune organs, but the expression level was low, which may be beneficial to maintain the overall health of the body. Among the 10 TLR genes, TLR2 and TLR9 had the high- est basal expression levels.
3.2 TLR Gene Expression Profile of P. olivaceus Immunized with E. tarda DNA Extract
The gene expression profiles of TLR 1, 2, 3, 5m, and 5s after immunization withDNAare shown in Fig.3. The expressions of TLR1 and TLR2 were up-regulated. The TLR3 gene expression in the gill and spleen was down-regulated at less than 1h and was also down-regulated and up-regulated in the head kidney and reached the peak at 6 h. The expressions of TLR5m and TLR5s were up-regulated and reached the highest levels in the gill and spleen at 6h and in the head kidney at 3h. The expression profiles of TLR7, 8, 9, 14, and 21 genes are shown in Fig.4. Only TLR9 and TLR21 genes were significantly up-regulated, among which TLR9 was the most up-regulated in the spleen and presented two peaks at 3–6h and at 1d. TLR21 showed the largest up-regulation in the head kidney and reached the peak in the three immune organs at 6h. Few significant changes were observed in other genes. Hence, TLR9 and TLR21 were sensitive to the DNA of.

Fig.2 Basic expression profile of TLR genes in the same tissues of healthy P. olivaceus. Data are expressed as mean±SD (n=3) (P<0.05).
3.3 TLR Gene Expression Profile of P. olivaceus Immunized with E. tarda RNA Extract
The expression profiles of TLR1, 2, 3, 5m, and 5s inRNA-immunizedare shown in Fig.5. The expression of all five genes changed, but the amplitude was not very significant. The gene expression profiles of TLR7, 8, 9, 14, and 21 are shown in Fig.6. The expression of TLR8 and 21 genes was significantly up-regulated. TLR8 show- ed two peaks in the head kidney and was up-regulated in different degrees in the gill and spleen. The TLR21 expres- sion increased at a lower rate and showed the largest in- crease in the gills. TLR7 was only weakly up-regulated in the gills. The expression of the other genes had no signi-ficant change in the three immune organs. Hence, TLR8 and TLR21 were sensitive toRNA.
3.4 TLR Gene Expression Profile in P. olivaceus Immunized with LPS Extracted from E. tarda Cell Wall
The expression profiles of TLR1, 2, 3, 5m, and 5s genes inimmunized with LPS ofare shown in Fig.7, and those of TLR7, 8, 9, 14, and 21 genes are shown in Fig.8. The expressions of TLR2 and 14 genes were significantly up-regulated in all the three immune organs. The TLR1 gene was up-regulated in the head kidney only at 3h, whereas the other genes did not change significantly. Hence, TLR2 and 14 were sensitive to the LPS of.

Fig.3 Gene expression profiles of TLR 1, 2, 3, 5m, and 5s in P. olivaceus immunized with E. tarda DNA. Data are expressed as mean±SD (n=3) (P<0.05).
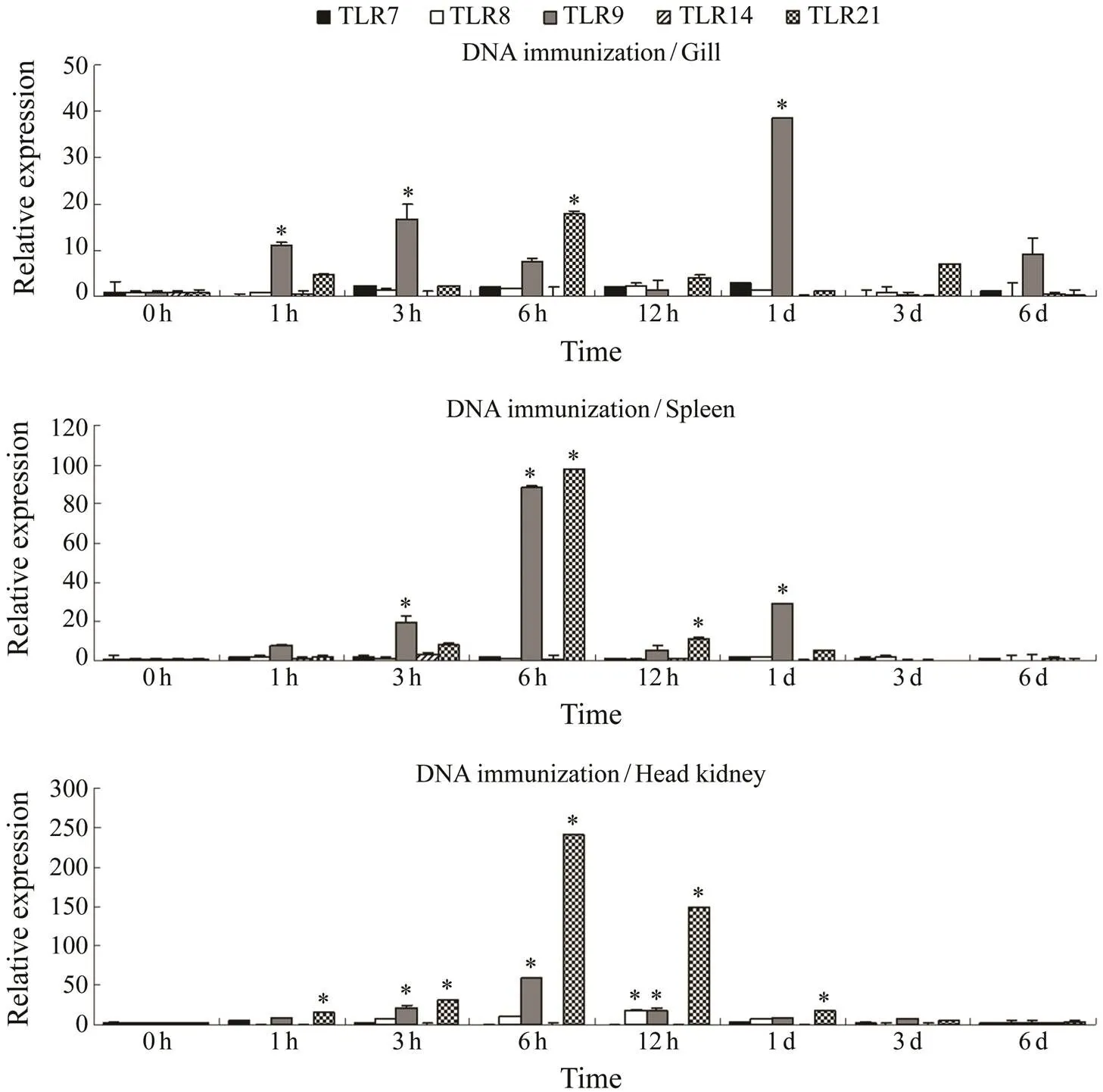
Fig.4 Gene expression profiles of TLR 7, 8, 9, 14, and 21 in P. olivaceus immunized with E. tarda DNA. Data are expressed as mean±SD (n=3) (P<0.05).

Fig.5 Gene expression profiles of TLR1, 2, 3, 5m, and 5s in P. olivaceus immunized with E. tarda RNA. Data are expressed as mean±SD (n=3) (P<0.05).

Fig.6 Gene expression profiles of TLR7, 8, 9, 14, and 21 in P. olivaceus immunized with E. tarda RNA. Data are expressed as mean±SD (n=3) (P<0.05).

Fig.7 Gene expression profiles of TLR1, 2, 3, 5m, and 5s in P. olivaceus immunized with E. tarda LPS. Data are expressed as mean±SD (n=3) (P<0.05).

Fig.8 Gene expression profiles of TLR7, 8, 9, 14, and 21 in P. olivaceus immunized with E. tarda LPS. Data are expressed as mean±SD (n=3) (P<0.05).
4 Discussion
Up to now, there are many studies on the identification and expression of immunity-related genes in fish. As an im- portant immunity-related gene, the immune regulatory me-chanism of TLR is still unclear in teleost fish. Some studies have shown that stimulation with bacterial pathogens can up-regulate TLR gene expression in fish. For example, TLR20 and TLR21 have been identified in several teleost fishes, such as,, large yel-low croaker, zebrafish(Li., 2012; Gao., 2013;Yeh., 2013; Sun., 2016). TLR21, TLR22 andTLR25 are also expressed in(Qi.,2018). Fan isolated and identified full-length cDNA ofTLR2 in giant yellow crocker and determined gene expres- sion after immune stimulation (Fan., 2015). In catfish, the changes of TLR genes spectrum were detected after pa- rasite ciliate infection at different time points, which con- firmed that some TLRs may play an important role in pro- tecting the fish from infection (Zhao., 2013). High expressions of TLR3, TLR4, TLR9 and TLR22 genes can be induced when yellow catfish is infected with(Zhang., 2017). The expression of TLR5s and 5m was up-regulated ininfected byand(Hwang., 2010). In addition to the original innate immunity, Rauta. (2014) investigated the additional role of acquired immunity. By supplement- ing TLR activator to stimulate dendritic cells, they can pre- pare effective vaccines against major diseases in many aqua- tic animals.
Although many TLRs have been cloned and identified in different fishes, the function and mechanism of TLRs are still unclear. Previous studies mainly analyzed the immune expression regulation of single TLR from a specific tissue of fish, and then determine their biological function and immune role. However, TLRs are widely distributed in various tissuesof the body, and the types and numbers of TLRs are different in different tissues. On the other hand, a pathogen often has a variety of molecular patterns that can be recognized by immune system. In different periods of in-fection, PAMP is recognized by different members of TLRs.How these family members work accurately in various tis-sues to activate the innate immune system and activate the specific immune response is still unknown. Therefore, it is not enough to study the immune regulation mechanism of TLRs only from the perspective of single tissue or single TLR gene. It is necessary to investigate the full range of immune responses of various TLR members in a variety of important tissues of fish, and to analyze the interaction and relationship between TLR members and pathogenic micro- organisms, so as to obtain a more systematic and in-depth understanding.
In this study, ten TLR (1,2,3,5m,5s,7,8,9,14,21) genes were detected by real-time quantitative PCR in dif- ferent tissues ofinfected withto find the expression patterns of ten TLR genes after pathogen sti- mulation.
The results of real-time PCR detection in healthyshowed that all members of the TLRs family were widely expressed in the tested tissues, but the basic expres- sion profiles of each member varied among different tissues. As immune organs, the spleen and gill had variety mem- bers of TLR with the highest or high expression. The in- testine also had variety members of TLR with the highest or higher expression. In the head kidney, the expression le- vels of TLR genes were very low, and some expression le- vels were nearly zero. The result may suggest the spleen is the main preventive immune organ in healthy. Ten TLRs were also detected in non-immune organs such as heart, liver, and muscle, but their expressionswere verylow. The low expression may be helpful to maintain the overall health of the body. Among the ten TLRs, TLR2 and TLR9 showed the highest basal expression, while TLR1 showed the highest expression in spleen. Previous studies also showed that the relative expression of TLR1 in spleen was the highest in grouper tissues (Wei., 2011).
According to the ligands and the subcellular location, TLRs can be divided into two subgroups. TLR1, TLR2, TLR4, TLR5, TLR6, and TLR11 are located primarily on the cell surface and recognize mainly microbial membrane components such as lipids, lipoproteins, and lipopolysac- charide(LPS). On the other hand, TLR3, TLR7, TLR8, and TLR9 reside on the membranes of intracellular compart- ments, such as endosomes, lysosomes, endolysosomes and endoplasmic reticulum, and are responsible for the recog- nition of microbial nucleic acids (Palti, 2011). Eight mem- bers of TLRs (TLR5S, 14, 18, 19, 20, 21, 22, 23) were spe- cific in teleost, which have not been identified in mam- mals. A unique feature of teleost TLRs is the presence of a soluble TLR5 molecule (TLR5S). TLR5S ligands have notbeen identified in fish, and mammalian TLR5 are thought to recognize flagellin. Teleost TLR14 shares sequence and structural similarity with TLR1 and 2 (Palti, 2011), which was divided into subgroups of TLR1 in fish and recognize gram-negative bacteria, gram-positive bacteria, polyI:C and LPS (Fan.,2015).
In the present study, most of the immune stimulation stud- ies of fish are based on the complete pathogen. In order to explore which component of the pathogen can effectively activate the TLRs response, and further understand the in- teraction betweenand TLRs family of,we examined the effects of LPS, DNA and RNA extracted fromon TLRs family genes expression in immune organs (spleen, head kidney, and gill) of.
The results showed that only TLR9 and TLR21 gene ex- pression were significantly up-regulated afterDNAimmunization in. This implied that both TLR9 and TLR21 were sensitive toDNA.The expres- sions of TLR8 and TLR21 genes were significantly up-re- gulated afterRNA immunization. The results im- plied that TLR8 and TLR21 were sensitive toRNA. The expressions of TLR2 and TLR14 genes were signifi- cantly up-regulatedafterLPS immunization, and no significant changes were found in other genes. The re- sults implied that TLR2 and TLR14 were sensitive toLPS.
In our experiments, TLR1 and TLR5s were up-regulated unexpectedly after the immunization withDNA, which may be caused by the stimulation of inflammatory factors produced byimmune system in re- sponse toDNA. Another interesting phenomenon is that there were two peaks of up regulation in TLR8 af- ter immunization withRNA and in TLR14 after immunization withLPS. The maximum expres- sion peak values of TLR8 and TLR14 were much higher than those of other TLR genes, which implied that TLR8 and TLR14 were more sensitive to the stimulation over- expression. However, over-expression may be suppressed by negative feedback of immune system. The expressions of TLR8 and TLR14 were restored and reached the se- cond peak after the negative regulation stopped.
From all of the gene expression profiling in this study, we can get lots of information about the dynamic changes in the gene expression of the immune organs and non-im- mune organs under different stresses or stimulations. The results will be helpful to clarify the interaction betweeninfection and host recognition, and provide a theo- retical basis forvaccine development for.
Acknowledgements
This research was supported by the Applied Basic Re- search Programs of the Science and Technology Commis- sion Foundation of Tianjin, China (Nos. 19JCZDJC34300, 14JCZDJC34200 and 18JCYBJC96100).
Fan, Z. J., Jia, Q. J.,and Yao, C. L., 2015. Characterization and expression analysis of Toll-like receptor 2 gene in large yellow croaker,., 44: 129-137, DOI:10.1016/j.fsi.2015.01.037.
Fan, Z. J., Zou, P. F., and Yao, C. L., 2015. Toll-like receptors(TLR) and its signaling pathway in teleost., 39: 173-184(in Chinese with English abstract).
Gao, H., Wu, L., Sun, J. S., Geng, X. Y.,and Pan, B. P., 2013. Molecular characterization and expression analysis of Toll-like receptor 21 cDNA from., 35: 1138-1145, DOI:10.1016/j.fsi.2013.07.027.
Hwang, S. D., Asahi, T., Kondo, H., Hirono, I.,and Aoki, T., 2010. Molecular cloning and expression study on Toll-like re- ceptor 5 paralogs in Japanese flounder,., 29: 630-638, DOI: 10.1016/j.fsi. 2010.06.011.
Jault, C., Pichon, L.,and Chluba, J., 2004. Toll-like receptor gene family and TIR-domain adapters in., 40(11): 759-771, DOI: 10.1016/j.molimm.2003.10.001.
Li, Y. W., Luo, X. C., Dan, X. M., Qiao, W., Huang, X. Z.,and Li, A. X., 2012. Molecular cloning of orange-spotted grouper () TLR21 and expression analysis postinfection., 32: 476-481, DOI: 10.1016/j.fsi.2011.11.021.
Meijer, A. H., Gabby Krens, S. F., Medina Rodriguez, I. A., He, S., Bitter, W., Ewa Snaar-Jagalska, B.,., 2004. Expression analysis of the Toll-like receptor and TIR domain adaptor fa- milies of zebrafish., 40(11): 773-783, DOI: 10.1016/j.molimm.2003.10.003.
Palti, Y., 2011. Toll-like receptors in bony fish: From genomics to function., 35: 1263-1272, DOI:10.1016/j.dci.2011.03.006.
Qi, Z. T., Wang, S. S., Zhu, X. Z., Yang, Y. Y., Han, P. P., Zhang, Q. H.,., 2018. Molecular characterization of three Toll-like receptors (TLR21, TLR22, and TLR25) from a primitive ray-finned fish Dabry’s sturgeon ()., 82: 200-211, DOI: 10.1016/j.fsi.2018.08. 033.
Qin, L., Li, F. H., Wang, X. Q., Sun, Y. Y., Bi, K. R.,and Gao, Y. L., 2017. Proteomic analysis of macrophage in response to-infection., 111: 86-93, DOI: 10.1016/j.micpath.2017.08.028.
Rauta, P. R., Samanta, M., Dash, H. R., Nayak, B.,and Das, S., 2014. Toll-like receptors (TLRs) in aquatic animals: Signaling pathways, expressions and immune responses.,158: 14-24, DOI: 10.1016/j.imlet.2013.11.013.
Rebl, A., Goldammer, T.,and Seyfert, H. M., 2010. Toll-like re- ceptor signaling in bony fish., 134(3-4): 139-150, DOI:10.1016/j.vetimm.2009.09.021.
Sun, M., Mu, Y. N., Ding, Y., Ao, J. Q.,and Chen, X. H., 2016. Molecular and functional characterization of Toll-like receptor 21 in large yellow croaker ()., 59: 179-188, DOI: 10.1016/j.fsi.2016.10.024.
Wei, Y. C., Pan, T. S., Chang, M. X., Huang, B., Xu, Z., Lou, T. R.,., 2011. Cloning and expression of Toll-like receptors 1 and 2 from a teleost fish, the orange-spotted grouper., 141: 173-182, DOI: 10.1016/j.vetimm.2011.02.016.
Whyte, S. K., 2007. The innate immune response of finfish–A review of current knowledge.,23: 1127-1151, DOI:10.1016/j.fsi.2007.06.005.
Wilson, A. B., 2017. MHC and adaptive immunity in teleost fishes.,69: 521-528, DOI:10.1007/s00251-017-1009-3.
Yeh, D. W., Liu, Y. L., Lo, Y. C., Yuh, C. H., Yu, G. Y., Lo, J. F.,., 2013.Toll-like receptor 9 and 21 have different ligand re-cognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish., 110(51): 20711- 20716, DOI: 10.1073/pnas.1305273110.
Zhang, X. T., Zhang, G. R., Shi, Z. C., Yuan, Y. J., Zheng, H., Lin, L.,., 2017. Expression analysis of nine Toll-like receptors in yellow catfish () responding tochallenge., 63: 384-393, DOI: 10.1016/j.fsi.2017.02.021.
Zhao, F., Li, Y. W., Pan, H. J., Shi, C. B., Luo, X. C., Li, A. X.,., 2013. Expression profiles of toll-like receptors in chan- nel catfish () after infection with., 35: 993-997, DOI:10.1016/j.fsi.2013.05.023.
Zhou, Z. J.,and Sun, L., 2016.-induced inhi- bition of apoptosis: A strategy for intracellular survival,6: 76, DOI:10.3389/fcimb.2016.00076.
Zhou, Z.X., Lin, Z.J., Peng, X., Shan, P.P.,and Wang, J.X., 2018.MicroRNA regulation of Toll-like receptor signaling pathways in teleost fish., 75: 32-40, DOI: 10.1016/j.fsi.2018.01.036.
December 28, 2020;
March 12, 2021;
June 29, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
.E-mail: skygh@tjnu.edu.cn
E-mail: skysjs@tjnu.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- Complete Mitochondrial Genome of Myra affinis (Decapoda:Brachyura: Leucosiidae) and Its Phylogenetic Implications for Brachyura
- Multisource Target Classification Based on Underwater Channel Cepstral Features
- Joint Model of Wind Speed and Corresponding Direction Based on Wind Rose for Wind Energy Exploitation
- Elastic-Wave Reverse Time Migration Random Boundary-Noise Suppression Based on CycleGAN
- Identification, Phylogeny and Expressional Profiles of Peptidoglycan Recognition Protein (PGRP) Gene Family in Sinonovacula constricta
- Molecular Characterization,Expression Pattern and Transcriptional Regulation of Figla During Gonad Development in Japanese Founder(Paralichthys olivaceus)
