Identification, Phylogeny and Expressional Profiles of Peptidoglycan Recognition Protein (PGRP) Gene Family in Sinonovacula constricta
MENG Yiping, Lü Liyuan, DAI Wenfang, ZHANG Weiwei, LIN Zhihua2), , *,and DONG Yinghui
Identification, Phylogeny and Expressional Profiles of Peptidoglycan Recognition Protein () Gene Family in
MENG Yiping1), 2), Lü Liyuan3), DAI Wenfang3), ZHANG Weiwei1), LIN Zhihua2), 3), *,and DONG Yinghui2), *
1),,315010,2),,,315100,3),,315604,
Peptidoglycan recognition protein (PGRP) plays a vital role in invertebrate innate immunity system as a specific pattern recognition receptor for peptidoglycan. Bivalves possess various PGRP systems for self-defense; however, it has not been characterized in razor clam. In this study, eightcoding sequences were identified and analyzed fromgenome, which are designated as,,,,,,,The results of molecular evolutionary analyses showed that all eightgenes were highly conserved and exhi- bited a typical PGRP/amidase_2 domain asgenes in other mollusks. Moreover, the presence of signal peptides was predicted in,and, while a transmembrane structure only existed in. Notably, a tertiary struc- ture analysis indicated that no disulfide bond was observed inand. The mRNA transcripts analysis ofrevealed that the high expression patterns ofandwere found in mantle, adductor muscle and foot, while those of,andwere observed in hepatopancreas. Furthermore, the temporal expression profiles ofin the hepatopancreas were analyzed by qPCRafter Gram-negativeand Gram-positivechallenges. The mRNA expressions of,andcould be induced byand. Overall, our findings indicated thatwere involved in the immune defense against invaders, which constituted a comprehensive understanding of the potential role ofgenes in.
; peptidoglycan recognition protein; gene family; gene expression
1 Introduction
Invertebrates merely rely on innate immunity to efficient ly defend themselves against pathogens invasion. Among the innate immunity responses, it is a premier and crucial step by discriminating infectious nonself from noninfection self (Janeway Jr., 1992). Organism recognizes the patho- genic invaders through a limited number of germ-line-en- coded receptors,which are termed as pattern recognitionreceptors (PRRs) (Medzhitov and Janeway Jr., 1997, 2000). The major targets of innate immune recognition are pa- thogen-associated molecular patterns (PAMPs), whichare highly conserved in microorganisms (Medzhitov andJaneway Jr., 1997, 2000). Specially, in invertebrates the peptidoglycan recognition proteins (PGRPs) can specifical- ly recognize and bind the peptidoglycan (PGN), an impor-tant cell wall component of bacterium, thereby triggering downstream host immuneresponse.
PGRP, a 19kDa protein, was first identified in the he- molymph and cuticle of(Yoshida., 1996). It has been demonstrated that this protein has affinity for peptidoglycan and enables to activate the phenol-oxidase cascade(Yoshida., 1996). Additionally, a number of studies have showed that PGRPs are highly conserved frominsects to mammals (Kang., 1998; Kiselev., 1998;Werner., 2000). Typically, based on the length of mRNA transcripts, PGRPs can be divided into three cate- gories: short extracellular PGRPs (20–25kDa, PGRP-S), intermediate PGRPs with two predicted transmembrane domains (40–45kDa, PGRP-I) and intracellular or trans- membrane long PGRPs (>90kDa, PGRP-L) (Dziarski, 2004). Among them, PGRP-S and PGRP-L mainly exist in vertebrates and some invertebrates, while PGRP-I is only identified in mammals (Dziarski and Gupta, 2006). The di-versity of PGRPs in both category and structure implies thatPGRPs perform multifunction in innate immunity. General- ly, in insects, the major functions of PGRPs are categorized intofour classes, including activation of the prophenol-oxi- dase cascade, amidase activity, activation of Toll or Imd sig- nal transduction pathway, and induction of phagocytosis (Yoshida., 1996; Werner., 2000; Michel., 2001; Gottar., 2002; Bischoff., 2004; Takehana., 2004; Garver., 2006; Park, 2006). For example, the PGRP-S inandis able to bind bacteria and Lys- and DAP-peptidoglycan, and then activate the prophenol-oxidase cascade that generates antimicrobial products (such as melanin and reactive oxygen species) to surround the infection site (Yoshida., 1996; Park., 2006). It has been proved that thePGRP-SA, PGRP-SD and PGRP-SC1 enable to activate Toll pathway. The mechanism is summarized as follows: after recogniz- ing bacterial PGN, the protease that can cleave Spaetzle is activated, and then the cleavage product acts as an endo- genous activator to activate Dorsal and Dif transcription fac- tors, which can enter the nucleus and initiate the transcrip- tion of drosomycin and other antimicrobial peptides (Mi- chel., 2001; Bischoff., 2004; Gaver., 2006). Activation of the Imd pathway by Gram-negative bacteria and Gram-positiveinis mediated through PGRP-LC and PGRP-LE (Michel., 2001; Gottar., 2002; Bischoff., 2004; Takehana., 2004; Garver., 2006). Specifically, the binding of Dap-type pepti- doglycan to Drosophila PGRP-LC induces oligomerization of the model receptor and recruits and activates the Imd protein, which further activates Relish transcription Factors, and induce the transcription of dipterin and other antimi- crobial peptides that are active against gram-negative bac- teria. In mammals, the symbol of PGRP is also denoted to PGLYRP (Liu., 2001). Notably, the PGLYRPs in mam- mals have two vital functions, amidase activity and bacte- ricidal activity (Dziarski., 2003; Gelius., 2003; Wang., 2003; Lu., 2006). It has been reported thatPGLYRP-1, PGLYRP-3, PGLYRP-4 and heterodi- mers formed by PGLYRP-3 and PGLYRP-4 have antibac- terial effects on many pathogenic and non-pathogenic bac- teria,and their activity depends on the presence of Ca2+(Dziarski., 2003; Lu., 2006). These findings sug- gest thatgenes play an important role in host im- munity responses.
Currently, several PGRPs have been identified in scal- lops (Ni., 2007; Su., 2007; Yang., 2010), oysters (Itoh and Takahashi, 2009; Zhang and Yu, 2013; Iizuka., 2014; Yang., 2016), mussels (Yang., 2013; Tao., 2014; Huang., 2019),(Wei., 2012, 2018), abalone (Premachandra., 2014) and(Yang., 2019). It has been demonstrated that many molluscan PGRPs can function as PRRs, scavengers and bactericidal amidase in innate immunity. For example, the mRNA expressions ofare significantly up-regulated after bacteria chal-lengein(Su., 2007),(Itoh and Takahashi, 2009; Yang., 2016),(Yang., 2013),(Wei., 2012) and(Yang., 2019).The recombinant CfPGRP-S1 in the presence of Zn2+can work as a bactericidal amidase to degrade the PGN and strongly inhibit the growth ofand(Yang., 2010). Similarly, the recom- binant HcPGRPS1 in the presence of Zn2+display a strong antibacterial activity to both Gram-negative bacteria includ-ing,,and Gram- positive bacteria such as(Yang., 2013). How- ever, compared with the remarkable progress in arthropods and mammals, the information on the prominent roles of PGRPs in innate immunity defense in mollusk is still li- mited.
, an economically important mol-lusk, is widely distributed in intertidal zones along the coasts of southeastern China and western Paci?c Ocean. Nowa- days, its farming industries are being threatened by increas- ing bacterial diseases due to high-density culture and con- tinuous water deterioration of farming environment. There- fore, in order to develop effective strategies to deal with bacterial diseases, it is of great significance to explore the innate immune system of. In this study, bio- informatics analysis was performed on 8CDS se- quences from thegenome. Moreover, their ex- pression patterns in different tissues and time-course expres-sion profiles in hepatopancreas challenged by Gram-nega- tiveand Gram-positivewere investigated. Our study enriches the theoretical know- ledge of innate immunity in mollusks and lays a founda- tion for the further functional research of.
2 Materials and Methods
2.1 Experimental Animals and Microbes
The razor clamswere collected from a com- mercial shellfish farming in Ningbo, Zhejiang Province (121?78?N, 29?65?E), and then were maintained in aerated seawater (temperature 19℃±1.5℃, salinity 18±1.5) for 3 days.andused for bacte- rial challenge were respectively cultured in 2216E medium in 28℃ and LB medium in 37℃ incubators.andsuspensions for infection were pre- pared by centrifuging the cultures at 5000×for 5min, re-moving the corresponding media, and re-suspending the ma- terials in filtered sterile seawater, and diluting the concen- tration of 1×108CFUmL?1and 1×109CFUmL?1, respec- tively.
2.2 Identification and Sequence Analysis
To identify thegenes in thege- nome(NCBI under accession numbers WSYO00000000.1), the known amino acid sequences of(BAH66 800.1) and(AHK22768.1) were selected as the query sequence, and then the BLASTP retrieval was performed with an E value threshold of 1e-10. Furthermore,the candidate sequences ofwere screened by searching the conserved domain using NCBI CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Pfam (http://pfam.xfam.org/search#tabview=tab1). Next, these sequences werecompared with other known proteins through the BLAST program. The basic physico- chemical properties ofproteins, including pro- tein length, isoelectric point (pI), molecular weight (MW) and grand average of hydropathicity (GRAVY), were de-termined in ProtParam (https://web.expasy.org/protparam/). Their signal peptides and transmembrane structures were further predicted using the online software SingalP 5.0(http://www.cbs.dtu.dk/services/SignalP/) and TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/), respectively.
2.3 Multiple Sequence Alignment and Phylogenetic Analysis
The multiple sequence alignments of the conserved do- main sequences of all predictedproteins andprotein were accomplished through the Clus- talX 2.1 software. A phylogenetic tree of 64 full-length PGRP protein sequences from amphibia, mammalia, in- secta, mollusca and fish was established to compare the evolutionary relationships using the MEGA X with neigh- bor-joining method (1000 bootstrap replicates). The con- structed phylogenetic tree was visualized by the iTOL v5 tool (https://itol.embl.de/).
2.4 Motif Patterns, Conserved Domains and Chromosome Location Analysis
The Deduced protein sequences ofgenes were submitted to MEME (Multiple Expectation Maximization for Motif Elicitation) (http://alternate.meme-suite.org/) for identification of the conserved motifs. The combination of phylogenetic tree, motifs and domains was then generated using TBtools. The distributions ofgenes onchromosomes were visualized using TBtools.
2.5 Prediction of Tertiary Structure of ScPGRP Proteins
The protein structures ofgenes were construct- ed on the SWISS-MODEL (https://swissmodel.expasy.org/) and then were visualized by the Swiss-pdb viewer 4.1.
2.6 Tissue Distribution Expression of ScPGRP Genes
Six tissues samples including mantle, gill, foot, hemo- cytes, hepatopancreas and adductor muscle were dissect- ed from stocked. The total RNA of these tis- sues was extracted with Trizol (Sangon, Shanghai) and thencDNAwas synthesized with Prime-ScriptTMRT reagentKit (TaKaRa, Japan) according to the manufacturer’s in- struction.
The tissue distribution pattern oftranscripts was examined by qPCR using iTaq?universal SYBR?Green Supermix (Bio-Rad, USA) in LightCycler?480II (Roche, USA). The primers used in the experiment were shown in Table 1. The 18S rRNA gene was selected as internal re- ference gene. The PCR amplification system was as fol- lows: 8μL of cDNA (100-fold dilution), 1μL forward pri- mer, 1μL reverse primer and 10μL of Mix. The reactionprocedure was 95℃ for 30s, followed by 40 cycles of de- naturation at 95℃ for 15s and annealing at 60℃ for 1min. All samples were performed with three biological pa- rallels and three technical replicates. The expression levelsofgenes were calculated using the method of 2???CT, and then was processed in GraphPad Prism 8.0.

Table 1 Sequences of the primers used in the study
2.7 Temporal Expression Patterns of ScPGRP Genes in the Hepatopancreas After Bacteria Challenge
The 180 clams were equally divided into two groups. One group was injectedwith, while the other one was injectedwith. 50μL of bacte- rial suspension was injected into the foot muscle. After in- jection, the hepatopancreases were collected from six ran- domly chosen clams at 0h, 6h, 12h, 24h, 48h and 72h, re- spectively. The qPCR reaction conditions and the calcula- tion of relativemRNA expression levels were the same as described above. Statistical analysis of the dif- ferences between groups was analyzed by one-way analy- sis of variance (one-way ANOVA) using SPSS 21.0 soft- ware (*<0.05; **<0.01).
3 Results
3.1 Sequence Analysis of ScPGRPs Genes
Totally, eightgenes’ sequences were obtained from the genome of. The identity analysis showed that thefamily genes had high identity with thein other mollusks(Table 2), which pro- vided a good guidance for studying the function offamily genes. Moreover, it was found that all the 8proteins possessed one PGRP and amidase-2 conserved do- main.
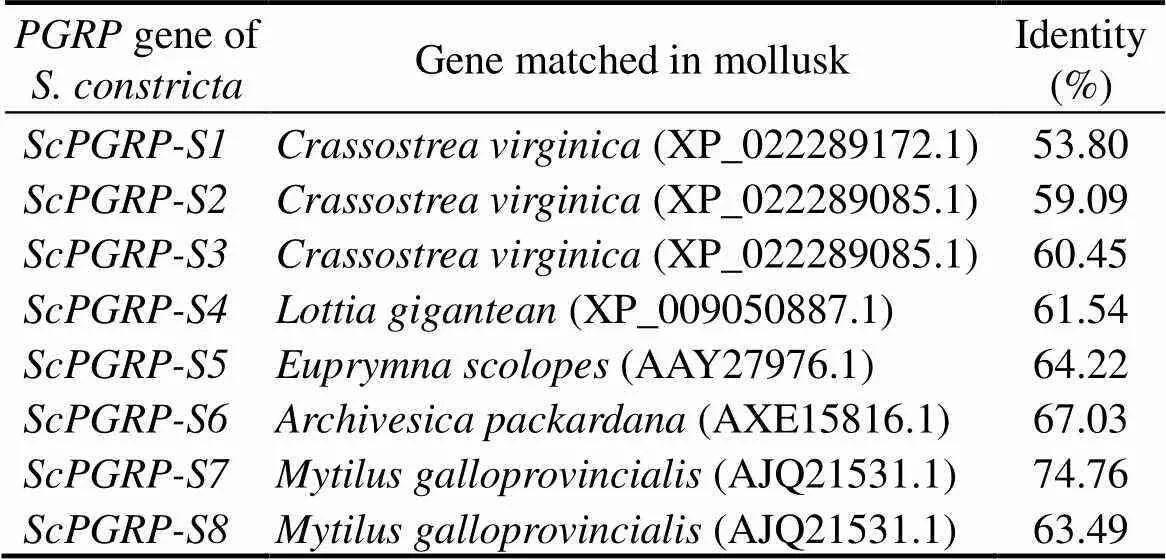
Table 2 Identity of ScPGRPs with corresponding genes in mollusks
The basic physicochemical properties ofgenes inwere shown in Table 3. Briefly, the lengths ofproteins are from 128 to 233 amino acids, with the putative molecular weights from 14.2kDa to 25.0kDa, and the theoretical pI values from 5.90 to 9.06. With the grand average of hydropathicity (GRAVY) values are <0, all thegenes are hydrophilic. The results of sig- nal peptide analysis indicated that,andall contain an N-terminal signal peptide. Notably, onlyexited the transmembrane struc- ture, which implied thatmight be a transmem- brane protein.
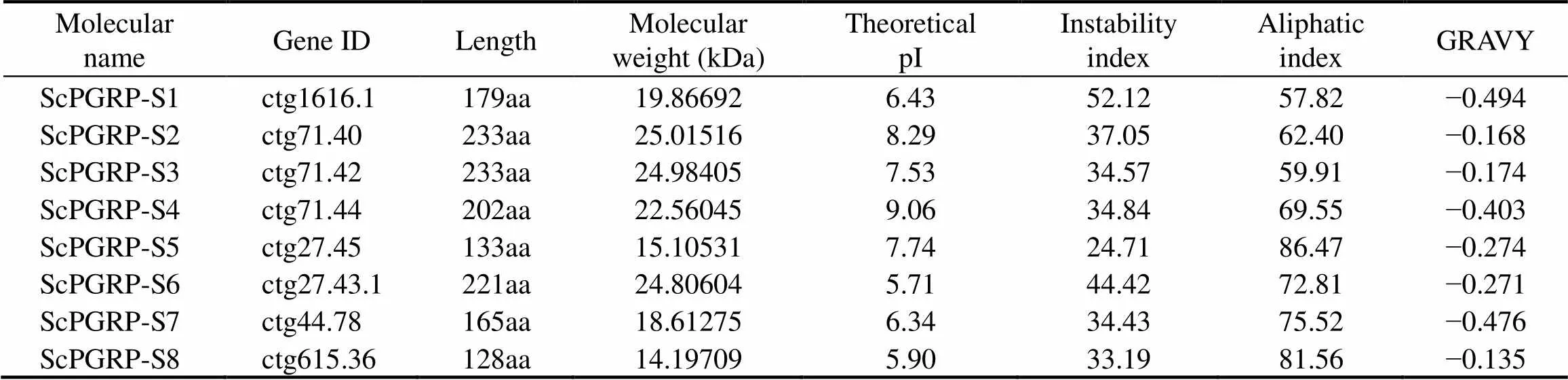
Table 3 ScPGRP family genes in S. constricta and their basic physicochemical properties
3.2 Multiple Sequence Alignment and Phylogenetic Tree of ScPGRPs
The multi-sequence alignment showed that the four key catalytic residues responsible for amidase activity (His42, Tyr158, His152 and Cys160 in) were well con- served in(H42-H151-T157-C159),(H96-H205-T211-C213),(H64-H172-T178- C180) and(H68-H177-T183-C185), whereasandhad no amidase activity sites (Fig.1). Notably, the(H96-Q205-T211-C213) detected a substitution, in which the 205th amino acid Q replaced the residue H, whereas the(H142- T148-C150) lacked the conserved residue H.

Fig.1 Multiple alignment of PGRP domain sequences. The red-shaded boxes represent the same amino acids and the blue-shaded boxes represent similar amino acids. The triangle (△) indicates the amidase catalytic site, and the circle (○) indicates Zn2+ binding site. GenBank accession number of the DmPGRP sequence used in the analysis is AAG23731.1.
The phylogenetic tree showed that all 8genes had high homology with other mollusks (Fig.2). For in-stance,,,andwere clustered together with the PGRPs of other mol- lusks such asand, whereas,,andwere cluster- ed in another branch of the mollusks. Among them,was clustered together withand, whilewas closely related with the.andwere clustered in the same branch.
3.3 Motif Patterns, Conserved Domain and Chromosome Location of ScPGRPs
The eightprotein sequences were analyzed and 10 conserved motifs were identified, ranging in length from6 to 50 amino acid residues. Most ofproteins con- tained motif 1, 2, 3, with the exception of,and. Overall, PGRPs are quite con- served during the evolution (Fig.3).

Fig.2 Phylogenetic tree of the amino acid sequences of ScPGRPs. The tree is constructed by the neighbor-joining (NJ) algorithm using the MEGA X software based on the multiple sequence alignment by ClustalW. The reliability of the branching was tested by bootstrap re-sampling (1000 pseudo-replicates).

Fig.3 Phylogenetic tree (a), motif patterns (b) and domain (c) of ScPGRPs.
Thegenes were mapped on thechromosomes. As shown in Fig.4, these 8genes were distributed across 4 chromosomes with a maximumof 3 genes on chromosome 8. Of the rest, chromosomes 13 and 17 contain the same number ofgenes (=2), and only one gene was on chromosome 7. In addition, some gene clusters were observed in thegenome:,andclustered on the48Mb region of chromosome 8;andclustered on 39Mb of chromosome 13, indicating thatgenes may exist tandem duplication events.

Fig.4 Chromosome distribution of ScPGRP genes in S. constricta genome. The gene numbers of each chromosome are showed at left and the scale is in megabases (Mb).
3.4 Potential Tertiary Structure of ScPGRP Proteins
The potential protein tertiary structures ofgeneswere shown in Fig.5. Almost allhad a typical PGRP structure that was composed of three α-helices and β-strands, with the exception of. In detail, the PGRP domains ofandwere unable to form disulfide bonds, while the other sixproteins had one or more pairs of disulfide bonds.
3.5 Tissue Expression Pattern of ScPGRP Genes
The tissue distribution analysis ofrevealed that the mRNA transcripts of fivegenes were express- ed in tested six tissues with the lowest expressions in the hemocytes (Fig.6). Specifically,andexhibited higher expression levels in the adductor mus- cle, mantle, and foot compared to other tissues.,andexhibited highly expres- sionlevels in the hepatopancreas, and their relative abun- dances correspondingly increased to 749 folds, 316 folds and 3267 folds of those in the hemocytes, respectively.

Fig.5 The tertiary structure of ScPGRPs. The α helix and β sheet is shown in red and yellow respectively. The disulfide bond is marked in blue and indicated by green arrows.

Fig.6 The mRNA expression levelsof ScPGRPgenes in different tissues ofS. constricta. The transcript levels in mantle, gill, foot, hemocytes, hepatopancreas and adductor muscle of six adult razor clams is normalized to that of hemocytes. Vertical bar represents mean±SD (n=3).
3.6 Temporal Expression of ScPGRP Genes Post Bacterial Challenge
Due to the higher expression levels ofgenes in the hepatopancreas, we further monitored the temporal ex- pression of mRNA transcripts ofin the hepato- pancreas stimulated by two bacteriaand. As shown in Fig.7, afterchallenge, compared with the mRNA expression level ofat 0h, a significant up-regulation expression was found in,and, and thepeak appeared at 24h, 24h, and 12h, respectively (<0.01). There was no significant difference in the expression le-vel ofat each time point (>0.05). The ex- pression level ofdecreased significantly (<0.01). Intriguingly, an inconsistent expression pattern was found in the experimental group challenged byrelative to(Fig.7). For example, the expression level of,andwere increased at 6h. Only the mRNA temporal expres- sion ofandshowed significant increase compared to 0 h, corresponding increased to 1.27 and 1.57 folds. Furthermore, the expression pattern ofexhibited down-regulation post-stimulation (<0.01). The expression ofshowed a trend of increasing firstly and then decreasing.

Fig.7 Time-dependent expression patterns of ScPGRPs in the hepatopancreas ofS. constricta after bacterial challenge. The blue and pink bars represent V. parahaemolyticus and S. aureus stimulation, respectively. The difference was statistically significant (* P<0.05; ** P<0.01).
4 Discussion
PGRPs are one of PRRs family members, which play an important role in innate immunity. Studies have been well characterized in insects and mammals, showing that PGRPs exert amidase activity and participate in Toll or Imd path- way (Gottar., 2002; Takehana., 2004; Garver., 2006). In this study, eightgenes were iden- tified from the genome and the number is similar to that ofand. All eightproteins po-ssess the PGRP/amidase_2 domain and evolve closely with the PGRPs in other mollusks, suggesting that the detected 8genes belong tofamily. A gene family that consists of two or more copies from gene duplication or doubling exhibits the similar structures and functions (Dayhoff, 1976; Demuth and Hahn, 2009). Consistently, the,andgenes were close in evolution, as demonstrated in the chromosomal lo- cation that the three genes congregate at certain positionsof the chromosome. Theandwere adjacent on chromosome 13; however, a phylogenetic tree showed that they were classified into two branches. Nota- bly, the structures of the two proteins are quite different, implying that they may have undergone functional diffe- rentiation. PGRPs are divided into three types: small ex- tracellular PGRP (PGRP-S, 20–25kDa), long PGRP (PGRP-L, >90kDa) and intermediate PGRP (PGRP-I, 40–45kDa) (Dziarski, 2004). Intriguingly, all eightproteins here were short-type PGRPs with their molecular weight between 14kDa and 25kDa. It is worth noting that most of the reported molluscan PGRPs are short-type PGRPs (Ni., 2007; Wei., 2012; Premachandra., 2014; Yang., 2019), indicating that the immunerecognition and response mechanisms of PGRPs in mol- lusk are different from those in higher organisms.
In general, the transmembrane-PGRPs are involved in the activation of Imd signaling pathway or induction of phagocytosis by recognizing meso-diaminopimelic acid con- taining PGN (DAP-type) (Hoffmann, 2003). In this study, the amino acid sequence analysis manifested that thepossessed a transmembrane domain. Addition- ally,was orthologous tofrommusselswith a transmembrane domain. It is predicted thatmay have an affinity for trachealcytotoxin (TCT) derived from DAP-type PGN (Ikuta., 2019). In this regard,may have the same pro- perties as BsPGRP-L. Nevertheless, on account of lack- ing the key Imd adaptor molecule and no identified func- tionally homologous component in bivalves (Gerdol., 2018), it is still unknown whether the Imd pathway in- volved in the response to Gram-negative bacteria existing in bivalves. Thus, further investigations are required to con- firm the role ofScPGRP-S2 in triggering an immune cas- cade. In fact, all PGRPs with amidase activity have con- served Zn binding sites in their activate centers that are composed of two histidines, one tyrosine and one cysteine, while the cysteine is replaced by a serine in non-amidase PGRPs (Gelius et al., 2003; Mellroth., 2003; Wang., 2003). For instance, two short PGRPs (ApPGRP-1 and -2) fromhave both conservedZn2+binding sites (H-H-C) and amidase catalytic sites (H- Y-H-T-C), and exhibit amidase activity with selective zinc ion dependence (Kong., 2018). Similarly, the,,andproteins po-ssessed complete active sites, speculating that they mayhave amidase activity. In contrast, theandproteins lacking all active sites may lose the abi- lity to hydrolyze peptidoglycan. In addition, the mutations of conserved sites inandcannot prove that the two proteins have no amidase activity. Thus, further analyses are needed to explore the structure and amidase activity ofandproteins. Moreover, almost all PGRPs have two closely spaced con-served cysteines in the PGRP domain formed by a disul- fide bond, which plays an important role in the structure and function of PGRPs (Dziarski, 2004). Indeed, a muta- tion in one of these cysteines inPGRP-SA(Cys80Tyr) abolishes the ability of PGRP-SA to activate the Toll pathway (Michel., 2001). Furthermore, a mu- tation in one of these cysteines in human PGRP-L (Cys- 419Ala) abolishes its amidase activity (Wang., 2003). For the tertiary structure, it was speculated thatandproteins did not function in eliminat- ing the threat of various exogenous microorganisms. In con- trast, the other sixpossessed one or two disul- fide bonds, but whether those genes function in innate im- mune system remains to be studied.
Unveiling the mRNA distribution pattern ofin different tissues would be bene?cial for exploring their potential functions in innate immunity. Most of the insectare expressed in immune organs, consistent with their role in insect immunity (Dziarski, 2004). Humanfamily members are selectively expressed in various organs and tissues (Liu., 2001). On the basis of existing re- ports,in mollusks display highly variable expres- sion in various tissues (Ni., 2007; Itoh and Takahashi, 2009; Wei., 2012; Yang., 2013; Premachandra., 2014; Huang., 2019). For instance,are highly expressed in the hemocytes of(Ni., 2007), whileandmainly present in the adductor muscle and hepatopan- creas of, respectively (Yang., 2016).Theis predominantly expressed in hepatopancreas, gonad, and intestine (Wei., 2012). Here,had a ubiquitously distribution in the tested tissues of. Considering that the razor clams were completely exposed to mudflats with large number of pathogenic microorganism, an analysis on the wide tis- sues distribution ofgenes may contribute to un- derstanding the immune mechanism. In this study, fivegenes exhibited different tissue expression patterns, indicating that these genes might contribute important roles in the immune response of. Specially,andshowed high expression levels in the mantle, suggesting that the proteins coded by the two genes may be related to the early identification of invasive pa- thogenic microorganisms, while the high expression le- vels of,andamidase activity.
After exposure to bacteria, the expression patterns ofconsiderably varied in different species (Ni., 2007; Su., 2007; Takahashi, 2009; Hu., 2020). For instance, the expression level ofgene inwas upregulated significantly after the stimula- tion of bacteriaand, and the expression ofwas increased by injection of the bacteriaand(Su., 2007; Itoh and Takahashi, 2009). Similar patterns were observed in,3 andandchallenges, reflecting the importance of thesegenes in immune defense in. Notably, Gram-nega- tiveinduced higher expression com- pared with Gram-positive, suggesting that a thin- ner PGN layer of Gram-negative bacteria may trigger stronger PGRP expression in hepatopancreas than the thick- er PGN layer of Gram-positive bacteria. Finally, it was spe- culated thatandmay be negative regulators whose expression will be inhibited when the host needs a strong immune response to resist the invasion of microorganisms. For example, thegene, a specific negative regulator of the Drosophila Imd pathway, binds to PGRP-LCx in the absence of tracheal cy- totoxin (TCT) and prevents the formation of PGRP-LCa/ -LCx active heterodimerization, thereby inhibiting the ac- tivation of the Imd pathway (Basbous., 2011).
5 Conclusions
In summary, eightgenes with typical PGRP/ami- dase_2 domain were identified fromgenome. All eightproteins were classified to PGRP-S and had high homology with the PGRPs of mollusks. The,,,andgenes exhibited constitutive and wide tissues dis- tribution. Specially, the expression levels of,andwere significantly up-regu- lated in the hepatopancreas after bacterial stimulation, sug-gesting their important roles in the innate immune defenseagainst bacterial infections in. These findingsnot only expand our understanding of the role ofgene family in mollusks, but also build a good basis for fur- ther research on the functions of.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2018YFD0901 405), the Zhejiang Major Program of Science and Tech- nology (No. 2016C02055-9), the Ningbo Major Project of Science and Technology (No. 2019B10005), and the China Agriculture Research System of MOF and MARA, National Marine Genetic Resource Center Program.
Basbous, N., Coste, F., Leone, P., Vincentelli, R., Royet, J., Kel- lenberger, C.,., 2011. Thepeptidoglycan-re- cognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway., 12(4): 327-333.
Bischoff, V., Vignal, C., Boneca, I. G., Michel, T., Hoffmann, J. A., and Royet, J., 2004. Function of thepattern-recognition receptor PGRP-SD in the detection of Gram-po- sitive bacteria., 5(11): 1175-1180.
Dayhoff, M. O., 1976. The origin and evolution of protein su- perfamilies., 35(10): 2132-2138.
Demuth, J. P., and Hahn, M. W., 2009. The life and death of gene families., 31(1): 29-39.
Dziarski, R., 2004. The peptidoglycan recognition proteins (PGRPs)., 40(12): 877-886.
Dziarski, R., and Gupta, D., 2006. The peptidoglycan recogni-tion proteins (PGRPs)., 7(8): 232.
Dziarski, R., Platt, K. A., Gelius, E., Steiner, H. K., and Gupta, D., 2003. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in pep-tidoglycan recognition protein-S (PGRP-S)-deficient mice., 102(2): 689-697.
Garver, L. S., Wu, J., and Wu, L. P., 2006. The peptidoglycan re- cognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis ofin., 103(3): 660-665.
Gelius, E., Persson, C., Karlsson, J., and Steiner, H. K., 2003. A mammalian peptidoglycan recognition protein with N-acetyl- muramoyl-L-alanine amidase activity., 306(4): 988-994.
Gerdol, M., Gomez-Chiarri, M., Castillo, M. G., Figueras, A., Fio-rito, G., Moreira, R.,., 2018. Immunity in molluscs: Re- cognition and effector mechanisms, with a focus on bivalvia. In:. Cooper, E., ed., Springer, Cham, 225-341.
Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoff- mann, J. A.,., 2002. Theimmune response against Gram-negative bacteria is mediated by a peptidogly- can recognition protein., 416(6881): 640-644.
Hoffmann, J. A., 2003. The immune response of., 426(6962): 33-38.
Hu, Z., Cao, X., Guo, M., and Li, C., 2020. Identification and characterization of a novel short-type peptidoglycan recogni- tion protein in., 99: 257-266.
Huang, Y., Pan, J., Li, X., Ren, Q., and Zhao, Z., 2019. Molecu- lar cloning and functional characterization of a short pepti- doglycan recognition protein from triangle-shell pearl mussel ()., 86: 571-580.
Iizuka, M., Nagasaki, T., Takahashi, K. G., Osada, M., and Itoh, N., 2014. Involvement of Pacific oyster CgPGRP-S1S in bac- terial recognition, agglutination and granulocyte degranulation., 43(1): 30-34.
Ikuta, T., Tame, A., Saito, M., Aoki, Y., Nagai, Y., Sugimura, M.,., 2019. Identification of cells expressing two peptidogly- can recognition proteins in the gill of the vent mussel,., 93: 815-822.
Itoh, N., and Takahashi, K. G., 2009. A novel peptidoglycan re- cognition protein containing a goose-type lysozyme domain from the Pacific oyster,., 46(8-9): 1768-1774.
Janeway Jr., C. A., 1992. The immune system evolved to discri- minate infectious nonself from noninfectious self., 13(1): 11-16.
Kang, D., Liu, G., Lundstrom, A., and Steiner, G. H., 1998. A pep- tidoglycan recognition protein in innate immunity conserved from insects to humans., 95(17): 10078-10082.
Kiselev, S. L., Kustikova, O. S., Korobko, E. V., Prokhortchouk,E. B., Kabishev, A. A., Lukanidin, E. M.,., 1998. Molecu- lar cloning and characterization of the mouse tag7 gene en- coding a novel cytokine., 273(29): 18633-18639.
Kong, X., Liu, H., Li, Y., and Zhang, H., 2018. Two novel short peptidoglycan recognition proteins (PGRPs) from the deep sea Vesicomyidae clam: Identification, re- combinant expression and bioactivity., 9: 1476.
Liu, C., Xu, Z. J., Gupta, D., and Dziarski, R., 2001. Peptidogly- can recognition proteins–A novel family of four human innate immunity pattern recognition molecules., 276(37): 34686-34694.
Lu, X., Wang, M., Qi, J., Wang, H., Li, X., Gupta, D.,., 2006. Peptidoglycan recognition proteins are a new class of human bactericidal proteins., 281(9): 5895-5907.
Medzhitov, R., and Janeway Jr., C. A., 1997. Innate immunity: Impact on the adaptive immune response., 9(1): 4-9.
Medzhitov, R., and JanewayJr., C. A., 2000. Innate immunity., 343(5): 338-344.
Mellroth, P., Karlsson, J., and Steiner, H., 2003. A scavenger func- tion for a Drosophila peptidoglycan recognition protein., 278(9): 7059-7064.
Michel, T., Reichhart, J. M., Hoffmann, J. A., and Royet, J., 2001.Toll is activated by Gram-positive bacteria througha circulating peptidoglycan recognition protein., 414(6865): 756-759.
Ni, D., Song, L., Wu, L., Chang, Y., Yu, Y., Qiu, L.,., 2007. Molecular cloning and mRNA expression of peptidoglycan re-cognition protein (PGRP) gene in bay scallop (, Lamarck 1819)., 31(6): 548-558.
Park, J. W., Je, B. R., Piao, S., Inamura, S., Fujimoto, Y., Fukase, K.,., 2006. A synthetic peptidoglycan fragment as a com- petitive inhibitor of the melanization cascade., 281(12): 7747-7755.
Premachandra, H. K. A., Elvitigala, D. A. S., Whang, I., and Lee, J., 2014. Identification of a novel molluscan short-type pep- tidoglycan recognition protein in disk abalone () involved in host antibacterial defense., 39(1): 99-107.
Su, J., Ni, D., Song, L., Zhao, H., and Qiu, L., 2007. Molecular cloning and characterization of a short type peptidoglycan re- cognition protein (CfPGRP-S1) cDNA from Zhikong scallop., 23(3): 646-656.
Takehana, A., Yano, T., Mita, S., Kotani, A., Oshima, Y., and Ku-rata, S., 2004. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically inimmunity., 23(23): 4690-4700.
Tao, Y., Yang, Z., Zhang, X., and Wu, H., 2014. Molecular cloning and mRNA expression of the peptidoglycan recognition pro- tein gene HcPGRP1 and its isoform HcPGRP1a from the fresh-water mussel., 37(3): 508-517.
Wang, Z. M., Li, X., Cocklin, R. R., Wang, M., Wang, M., Fu-kase, K.,., 2003. Human peptidoglycan recognition pro- tein-L is an N-acetylmuramoyl-L-alanine amidase., 278(49): 49044-49052.
Wei, X., Yang, D., Li, H., Zhao, T., Jiang, H., Liu, X.,., 2018.Peptidoglycan recognition protein of(SgPGRP-S1) mediates immune recognition and bacteria clearance., 73: 30-36.
Wei, X., Yang, J., Yang, D., Xu, J., Liu, X., Yang, J.,., 2012. Molecular cloning and mRNA expression of two peptidogly- can recognition protein (PGRP) genes from mollusk., 32(1): 178-185.
Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H., and Hult- mark, D., 2000. A family of peptidoglycan recognition proteins in the fruit fly., 97(25): 13772-13777.
Yang, C., Wang, L., Jia, Z., Yi, Q., and Song, L., 2016. Two shortpeptidoglycan recognition proteins fromwith similar structure exhibited different PAMP binding activity., 70: 9-18.
Yang, D., Han, Y., Liu, Y., Cao, R., Wang, Q., Dong, Z.,.,2019. A peptidoglycan recognition protein involved in immune recognition and immune defenses in., 88: 441-448.
Yang, J., Wang, W., Wei, X., Qiu, L., Wang, L., Zhang, H.,., 2010. Peptidoglycan recognition protein of(CfPGRP-S1) mediates immune defenses against bacterial in- fection., 34(12): 1300-1307.
Yang, Z., Li, J., Li, Y., Wu, H., and Wang, X., 2013. Molecular cloning and functional characterization of a short peptidogly- can recognition protein (HcPGRPS1) from the freshwater mus- sel,., 56(4): 729-738.
Yoshida, H., Kinoshita, K., and Ashida, M., 1996. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm,., 271(23): 13854-13860.
Zaidman-Rémy, A., Poidevin, M., Hervé, M., Welchman, D. P., Paredes, J. C., Fahlander, C.,., 2011.immunity: Analysis of PGRP-SB1 expression, enzymatic activity and func-tion., 6(2): e17231.
Zhang, Y., and Yu, Z., 2013. The first evidence of positive selec- tion in peptidoglycan recognition protein (PGRP) genes of., 34(5): 1352-1355.
January4, 2021;
February25, 2021;
July20, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
. E-mail: zhihua9988@126.com
E-mail: dongyinghui118@126.com
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- Complete Mitochondrial Genome of Myra affinis (Decapoda:Brachyura: Leucosiidae) and Its Phylogenetic Implications for Brachyura
- Multisource Target Classification Based on Underwater Channel Cepstral Features
- Joint Model of Wind Speed and Corresponding Direction Based on Wind Rose for Wind Energy Exploitation
- Elastic-Wave Reverse Time Migration Random Boundary-Noise Suppression Based on CycleGAN
- Molecular Characterization,Expression Pattern and Transcriptional Regulation of Figla During Gonad Development in Japanese Founder(Paralichthys olivaceus)
- Expressions of Toll Like Receptor (TLR) Genes in Paralichthys olivaceus After Induced by Different Extracts of Edwardsiella tarda
