Molecular Characterization,Expression Pattern and Transcriptional Regulation of Figla During Gonad Development in Japanese Founder(Paralichthys olivaceus)
QU Jiangbo, LI Rui, LIU Yuxiang, SUN Minmin, YAN Weijie, LIU Jinxiang, 2),WANG Xubo, and ZHANG Quanqi, 2), *
Molecular Characterization,Expression Pattern and Transcriptional Regulation ofDuring Gonad Development in Japanese Founder()
QU Jiangbo1), LI Rui1), LIU Yuxiang1), SUN Minmin1), YAN Weijie1), LIU Jinxiang1), 2),WANG Xubo1), and ZHANG Quanqi1), 2), *
1),,,266003,2),,266237,
The factor in the germline alpha (), as a member of the basic helix-loop-helix family, has been reported to be in- volved in ovary development in mammals and teleosts. However, the regulatory mechanisms ofin teleosts remain unclear. Here,in() was characterized with encoding a 202 amino acid protein that contains a conserved basic region and helix-loop-helix (HLH) domain. Amino acids alignment and synteny analysis revealed thatwas conserved with the ortholo- gous gene sequences in other vertebrates. The results of qRT-PCR showedwas maternally inherited during embryonic de- velopment. For tissue distribution,showed a sexually dimorphic gene expression in the gonad of different genders, with a higher expression in ovary than in testis.hybridization (ISH) results demonstratedwas specifically expressed in germ cells including oocytes, spermatogonia and spermatocytes. By screening and analyzing two proximal regions (?2966/?2126 and ?772/?444) with high promoter activity, we found SOX5, LEF1, FOXP1 and GATA1 may play important roles in the transcriptional regulation of. Furthermore, we observed the co-localization between Figla and LEF1 in HEK 293T cells. And the significant up-regulation effect of the canonical Wnt signaling pathway on the expression ofwas found in cultured ovarian cells. This study provided the first evidence thatnot only has an important function in ovary development, but also plays some potential roles in testis development and/or male germ cell differentiation during early testis development in. The results provide valuable reference in exploring the regulatory network ofin teleost.
; transcriptional regulation; Wnt signaling pathway; gonad development;
1 Introduction
bHLH gene family is a very diverse transcription fac- tors superfamily with basic helix-loop-helix structural do- main (Liu., 2014; Li., 2016; Tarczewska and Greb-Markiewicz, 2019). This domain is conserved with a approximate length of 60 amino acids, which may form homodimeric or heterodimeric complexes with each other to bind to specific DNA sequence (Zhang., 2017). Thefamily members are involved in the development of nu-merous tissues including gonad, spleen, lung and heart (Wang., 2017; Chiba., 2019;Tan., 2019). Given the complexity and diversity of the family, it has been studied in multiple developmental processes including neurogene- sis, mesoderm formation, cranial vault development and sex determination (Zhang., 2017; Blumel., 2019).
, as a transcription factor gene belonging to bHLH family, plays an important role in the formation of pri- mordial follicles in some vertebrates (Soyal., 2000;Bayne., 2004). Many researches have revealed some molecular mechanisms about the function ofin mam- mals. In mouse, primordial follicles were not formed at birth once mutagenesis happened in embryonic stem cells. Sub- sequently, massive depletion of oocytes resulted in shrun- ken ovaries and female sterility. In addition, zona pelluci- da genes (,,and) are not expressed innull female mouse (Soyal., 2000), which indicatescan regulate the expression of zona pellucida genes.also functions as a key regulatory molecule in coordinat- ing expression of the NALP family genes, as well as some genes related to oogenesis such as,and(Joshi., 2007). In human, the expression ofincreases across mid-gestation and reaches the peak by the time of primordial follicle formation. Further- more, Figla can heterodimerize with E12 protein and bind to the E-box of the humanpromoter to regulate its ex- pression level (Bayne., 2004).
In teleosts, the studies ongene were far less de- tailed than in mammals. Twohomologues with dif- ferent functions during sex differentiation were identified in tongue sole (Li., 2016). In Nile tilapia,was only expressed in ovary and over-expression ofinXY fish resulted in the disruption of spermatogenesis along with the depletion of meiotic spermatocytes and spermat- ids in testis (Qiu., 2015), which was consistent with impaired meiosis and germ cell apoptosis due to the ec- topicexpression in testis of mouse (Hu., 2010). Although there have been some studies aboutin te- leosts, studies examining its expression regulation involved signal pathways are limited. Japanese flounder () is an economically important cultured fish in China, Korea and Japan. Due to its remarkable sexual dimorphismduring growth (females grow faster and have superior body length than males) (Yamamoto, 1999), finding ways to in- crease the proportion of female during the breeding pro- cess has been a research hotspot. In order to achieve this goal, exploring and understanding the regulatory mecha- nisms of genes that participate in gonad development will be fundamental and meaningful. Recently, Liang(2020) have characterizedand explored its role in the pro- liferation of oocytes in.
Although there have been some researches aboutin flounder, studies examining its regulation of expression by upstream genes or signaling pathways are lacking. Interest- ingly, in our results, quantitative real-time polymerase chain reaction (qRT-PCR) andhybridization (ISH) indi- cated thatwas not only expressed in the ovary, butalso expressed in the testis in the early developmental stage, which was slightly different from the results of Liang(2020). In addition, we made deletion constructs and site- directed mutagenesis of the promoter regions, combined with dual luciferase assays to identify potential transcrip- tion factors that regulatetranscription, and explored the regulation of canonical Wnt signaling pathway on. This is the first attempt to study the potential regulator ofat molecular level in Japanese flounder, which could be helpful for understanding the gonad development, arti- ficial breeding and further gene regulation studies ofin other species.
2 Materials and Methods
2.1 Ethics Statement
All experimental protocols were performed in accordance with the Institutional Care and Use Committee of the Ocean University of China (Qingdao, China) and were conduct- ed to fit in the guidelines of Chinese Government’s Prin- ciples for the Utilization and Care of Vertebrate Animals Used in Testing, Research and Training (China, 1988).
2.2 Fish and Embryo Sampling
All healthy fish samples and embryos were reared in the recirculating aquaculture system in Yellow Sea Aquatic Pro-duct Co., Ltd., Yantai, Shandong, China. Three females and three males of Japanese flounder (1-year-old) were select- ed randomly. Tissue samples including heart, liver, spleen, kidney, brain, gill, muscle, intestine, testis and ovary were collected for tissue distribution analysis. Testes and ovaries of 8-months, 1-year and 1.5 years old individuals were col- lected, respectively. Each sample was collected in triplicate. Embryo samples at twelve different developmental stages, including 1 cell, 16 cells, morula, high blastula, low blas- tula, early gastrula, late gastrula, neurula, tailbud formation,hearting-beating, before hatching and hatching stages werecollected for gene expression analyses. Each sample, which contained 30 embryos, was collected in triplicate. All sam- ples were immediately frozen using liquid nitrogen and stored in ?80℃ for RNA extraction.
2.3 RNA Extraction and cDNA Synthesis
Total RNA was extracted from tissues and embryos us- ing TRIzol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions, then genomic DNA was removed with RNase-free DNase I (TaKaRa, Dalian, Chi- na). RNA quality and quantity were assessed by electro- phoresis detection and NanoDrop determination, respec- tively. The cDNA was synthesized with 1μg high-quality total RNA and 25μmolL?1random primer using the Re- verse Transcriptase M-MLV Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
2.4 Isolation, Sequence Analysis of Pofigla
The available sequences oforthologs were identi- fied in other teleosts from NCBI and Ensemble to obtainsequence ofby tBlastx. The core cDNA sequence of thewas amplified by PCR with de- generate primers-core-FW/RV (Table 1). PCR pro- ducts were separated by agarose gel electrophoresis, puri- fied using the Gel Extraction Kit (CWBIO, Beijing, Chi- na), and cloned and ligated into the pEASY-Blunt Simple Vector (TransGen Biotech, Beijing, China) for sequencing. Homologous protein sequences were confirmed through BLAST searches in NCBI and Ensemble (Table 2). Multi- ple sequence alignments were conducted using ClustalX 2.1. The amino acid Logos of alignments were generated with WebLogo (http://weblogo.berkeley.edu/logo.cgi). Structure of floundergenewas determined by the online Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn). Chromosome synteny was performed according to the lo- cation ofand its flanking genes obtained from NCBI online genome database (MPLB00000000.1).
2.5 Quantitative Real-Time PCR (qRT-PCR)
The primers (in Table 1) used for qRT-PCR were design- ed in the non-conserved regions by the online Integrated DNA Technologies (IDT) (http://sg.idtdna.com/Primerquest/ Home/Index). qRT-PCR was performed using a Light- Cycler 480 (Roche, Forrentrasse, Switzerland) with a total 20μL reaction volume, which contained 10ng template cDNA, 0.4μL of each primer (10μmolL?1) and 10μL 2×SYBR Green qPCR Master Mix (US Everbright Inc., Su- zhou, China). The specificity of primers was validated ac- cording to the single peak of melting curves generated af- ter amplification reaction.was selected as the re- ference gene according to the previous study (Zhang., 2013). All qRT-PCR assays were performed in triplicate. The relative expression levels of the target gene were cal- culated by the 2?ΔΔCtcomparative Ct method.
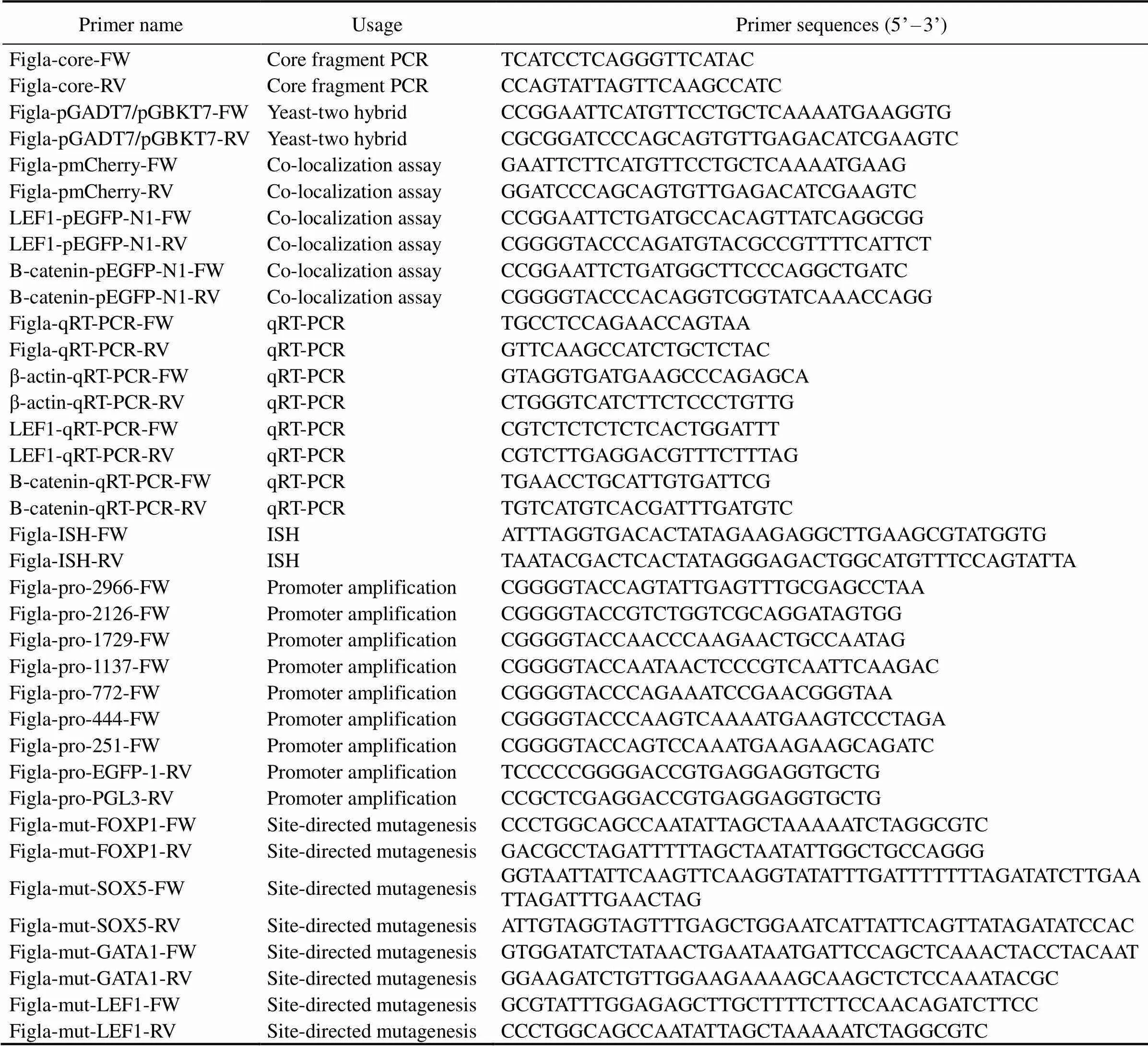
Table 1 Primers used in this study
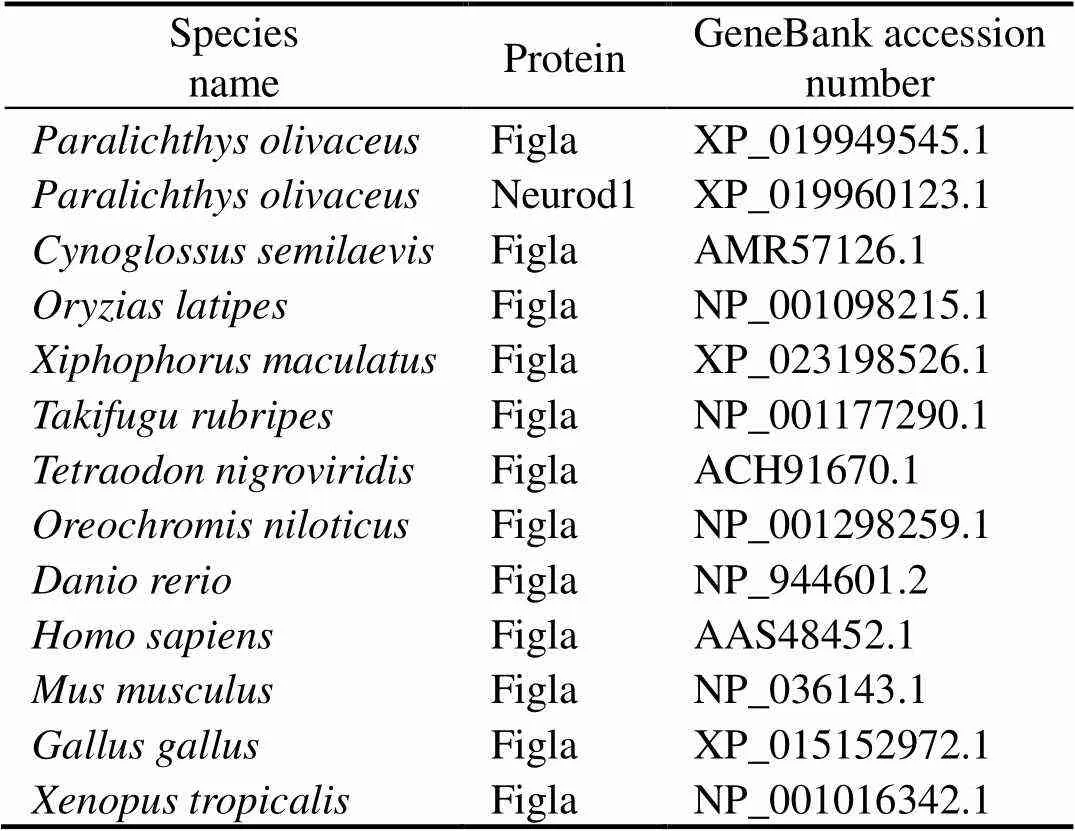
Table 2 Sequence used for alignments
2.6 In Situ Hybridization
ISH analyses in 8-month-old male and female gonads were performed using the probe spanning the non-con- served region of cDNA (Table 1). DIG-labeled RNA sense and antisense probes were synthesized using a DIG RNA Labeling Kit (SP6/T7) (Roche, Mannheim, Germany) ac- cording to the manufacturer’s protocol. The experiment of tissue ISH has been described in our previous research (Gao., 2013). Briefly, gonad tissues were dehydrated with gradient methanol, then transited to xylene and em- bedded by paraffin ultimately. The ovary and testis blocks were sectioned at 7 and 5μm thickness, respectively. And then all sections were fixed in the microscope (Solarbio, Shanghai, China) coated with polylysine for 12h at 37℃. Each sample sections were incubated with 20μgmL?1pro- tease K at 37℃ for 15min and fixed with 4% PFA for 20min. The hybridization was carried out at 60℃ for 15h (3h for pre-hybridization process and 12h for hybridization process). After hybridization, all sample sections were wash-ed twice in 2×SSC, 1×SSC and 0.2×SSC for 15min at 60℃, respectively. Then they were washed twice in maleic acid buffer at room temperature for 5min each time. Sam- ples were blocked in blocking buffer for 2h, then incubated with blocking buffer mixed with anti-DIG alkaline phos- phatase-conjugated F (ab)’ 2 fragments for 12h at 4℃. Thepositive signals were observed after reaction with NBT/BCIP substrate solution (Roche, Mannheim, Germany). Then the sample sections were stained with 1% neutral redfor 20–30min and covered with neutral resin. The sectionswere observed and photographed using a Nikon Eclipse Ti-U microscope (Nikon, Tokyo, Japan).
2.7 Plasmid Construction, Matinspector Analysis and Site-Directed Mutagenesis
For the Yeast two-hybrid assay, the coding sequence ofwas cloned and inserted into pGADT7 and pGBKT7 (Clontech, San Francisco, USA) withanddigestion method. For the analysis of promoter activity, the promoter regions with different lengths of(?251/ ?44, ?444/?44, ?772/?44, ?1137/?44, ?1729/?44, ?2126/ ?44, and ?2966/?44) were amplified from Japanese flound- er genome and cloned intoandrestriction en- zyme sites of pEGFP-1 (Clontech, San Francisco, USA), respectively. For the analysis of cis-acting elements regu- latingpromoter activity, the ?772/?44 and ?2966/?44regions were cloned intoandrestriction enzymesites of pGL3-basic vector (Clontech, San Francisco, USA).The potential transcription factor binding sites within thesetwo regions were forecasted using the online program Mat-Inspector (http://www.genomatix.de/matinspector.html) with the Matrix Family Library Version 9.3 (Cartharius., 2005). The transcription factor binding sites were deleted with the QuickChange Site-Directed Mutagenesis Kit (Agi- lent Technologies, Santa Clara, USA) using the wild-type constructs pGL3?2966/?44 and pGL3?772/?44 as the tem- plates following the manufacturer’s instructions. The cod- ing sequences ofandwere amplified and cloned intoandrestriction enzyme sites of thepEGFP-N1 for the co-transfection and dual-reporter assays. The coding sequence ofwas amplified and cloned intoandsites of pmCherry-N1 for the co- localization assay. All constructed recombinant plasmids were proofed by sequencing (BGI, Shenzhen, China). The primers used in this study were listed in Table 1.
2.8 Yeast Two-Hybrid Assay
The recombinant plasmids PoFigla-pGADT7 and Po-Figla-pGBKT7 were co-transformed into Y2HGold com- petent yeast cells (Weidi Biotechnology, Shanghai, China) according to the manufacturer’s protocol. The transform- ed yeast cells were cultured in leucine and tryptophan au- xotrophic media for 3d at 29℃. Then the yeast colonies were transferred into adenine, histidine, leucine, and trypto- phan auxotrophic media supplemented with aureobasidin A (AbA) and Alpha-galactosidase (X-α-Gal) for identifi- cation of the protein-protein interaction. pGADT7-T and pGBKT7-53 plasmids were co-transformed as positive con-trol. pGADT7-T and pGBKT7-lam co-transformation was used as negative control. The self-activation of yeast two- hybrid system was validated before experimental assay.
2.9 Transfection, Dual-Luciferase Reporter and Co-Localization Assay
About 2×105HEK 293T cells were seeded in 24-well plates, cultured in the Dulbecco modified Eagle medium (Biological Industries, Bet-Haemek, Israel) containing 10% fetal bovine serum (Gibco, Carlsbad, USA) and 1% peni- cillin-streptomycin (Gibco, Carlsbad, USA) for 12h under 5% CO2at 37℃, and transfected with the plasmids using Lipofectamine 3000 Reagent (Life Technologies, Carlsbad, USA) according to the manufacturer’s protocol. For dual- luciferase assay, the total 1000ng plasmids including over- expression plasmids B-catenin or LEF1 with different doses (0, 50, 200ng), 20ng pRL-TK Vector (Promega, Madison, USA) and the remaining empty pEGFP-N1 were transfect- ed into HEK 293T cells. At 36h after transfection, the lu- ciferase values were obtained with the Dual-Luciferase Re- porter Assay System (Promega, Mannheim, Germany) ac- cording to the manufacturer’s protocol. Each sample in thedual-luciferase reporter assays was performed in triplicates. For GFP reporter assay, the cells were transfected with thesame amount of recombinant pEGFP-1 vectors (500ng per well), the expression of GFP in transfected cells was ob- served with an inverted ?uorescence microscope at 48h after transfection. pEGFP-C1 was selected as a positive control for successful transfection. For co-localization as- say, the recombinant PoLEF1-pEGFP-N1 and PoFigla-pm- Cherry-N1 were constructed and the different combina- tions of plasmids were transfected into HEK 293T cells. After 36h of incubation, the cells were fixed with 4% pa- raformaldehyde for 20min and washed 3 times with PBS, stained with DAPI (Sangon Biotech, Shanghai, China) for 15min followed by washing 3 times. Lastly, the cell fluo- rescence photographs of each combination group were ac-quired through the laser confocal scanning microscope A1R (Nikon, Tokyo, Japan).
2.10 Expression of β-catenin, Lef1 and Figla in Ovarian Cells Treated with BIO and XAV-939
Healthy Japanese flounder adults were obtained from a commercial hatchery located in Qingdao City of China. Adult fish were cultured for one week in seawater tanks at 20℃±1℃ under laboratory conditions. The ovarian tis- sue was carefully dissected and cut into pieces (mean size =5mm3) in PBS (pH 7.4). To avoid the bacterial infections and remove the white ovarian walls, the ovary pieces werewashed 5 to 6 times in PBS added with 5% penicillin-streptomycin. Finally, ovary tissue pieces were culturedat 24℃ in DMEM/F12 medium (Thermo Fisher Sci- entifc Inc, Waltham, USA) containing 15% FBS (Gibco,Carlsbad, USA), 10mmolL?1HEPES, 10mmolL?1non-es- sential amino acids (NEAA), 2mmolL?1 L-glutamine (So- larbio, Beijing, China), 50μgmL?1streptomycin and 50UmL?1penicillin (Gibco, Carlsbad, USA).
In order to explore the influence of Wnt signaling path- way on the expression of, we selected the activator(BIO) (Cayman Chemical, Ann Arbor, USA) and inhibitor (XAV-939) (Cayman Chemical, Ann Arbor, USA) of cano-nical Wnt signaling pathway (Niu., 2018), to treat thecultured ovary tissue. The tissue was stimulated with 10μmolL?1BIO and 10μmolL?1XAV-939, respectively. The tissue treated with solvent DMSO (0.1%) was considered as a control. Total RNA was extracted and qRT-PCR of,andwas carried out at 24h after treat- ment.
2.11 Expression Analysis of Genes in Canonical Wnt Signaling Pathway Basing on Transcriptome Data
The transcriptome of testis and ovary in our laboratory (Zhang., 2016)were applied to analyze the expressionof genes,,,,,,,,and. The expression levels were evaluated bynormalized Fragments Per Kilobase per Million (FPKM) value.
2.12 Statistical Analysis
All experimental data were expressed as the mean±SEM (standard error of mean) of three independent experiments.The data were statistically analyzed using one-way ANOVA followed by Games-Howell or Independent-Samples-testusing SPSS 20.0 (IBM, New York, USA) with<0.05 in- dicating significant difference.
3 Results
3.1 Sequence Characteristics, Gene Structure and Evolution of Pofigla
The sequence ofwas identified fromtranscriptome by tBlastx and verified by PCRThecDNA sequence contains an open reading frame (ORF) of 609bp, which encodes a polypeptide of 202 amino acids with a predicted molecular weight of 23.126kDa. Genecontains two functional domains: basic region and helix-loop-helix (HLH) region (Fig.1A). In these regions, the percent identity of most amino acid in the alignments exceeded 60% (Fig.1A). Yeast two-hybrid assay showed that all groups could grow on 2Q deficient medium, which demonstrated plasmids were successfully transformed into yeast. In the positive control group (PGADT7-T and PGB-KT7-53), a few green colonies could grow on the 4Q/X/ Aba deficient medium. However, no yeast colonies could grow on the multiple defective medium in the experiment group (PGADT7-Figla and PGBKT7-Figla) and negative control group (PGADT7-T and PGBKT7-lam), which strongly suggested that PoFigla itself does not have the po- tential to interact and form homodimers (Fig.1B). So it may regulate downstream gene expression by combining other transcription factors to form heterodimers. Compari- son of cDNA and genomic sequences showed thatcontains six exons, which differs from other teleost spe- cies such as tongue sole, fugu, zebrafish and medaka (Fig.2). A conserved syntenic relationship was obtained forand its surrounding genes in vertebrates from fish to te- trapods although fragment betweenandis re- versed in(Fig.3). These results to- gether revealed thatis greatly conserved during the evolution.
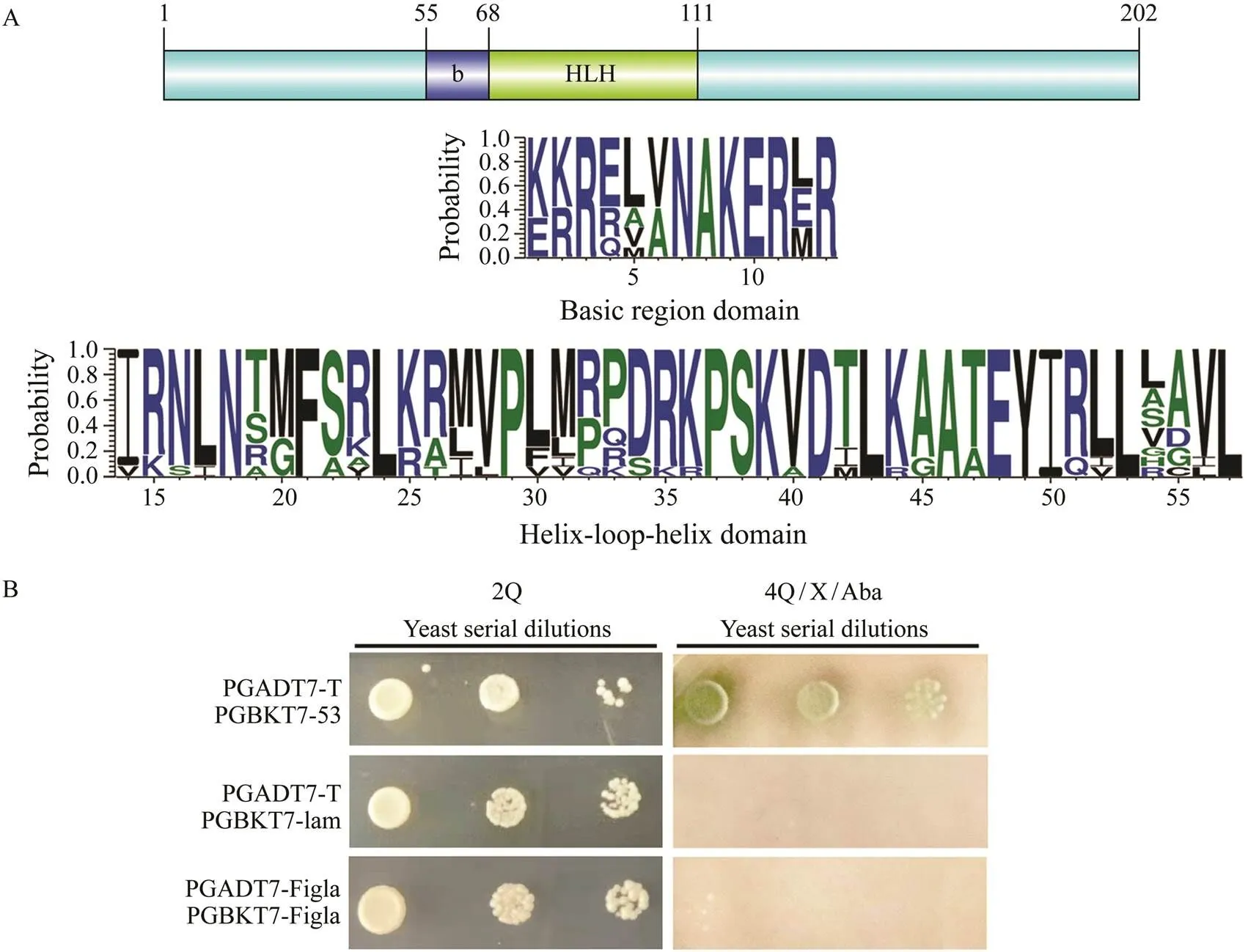
Fig.1 Sequence characterization and verification of non-homodimer for PoFigla. (A) Above: Location information of con- served domains of PoFigla. Below: The consensus logos of basic region and HLH domain. The height of each letter indi- cates the percentage of corresponding amino acid residues at that position in the multiple sequence alignments. Default color scheme was selected for different amino acids. Multiple sequence alignment of Figla protein sequences from P. oli- vaceus and other vertebrates was performed by Clustal X (2.0). The abbreviated species names and GenBank accession numbers are available in Table 2. (B) Yeast two-hybrid assay to confirm PoFigla could not form homodimers. 2Q: leucine and tryptophan auxotrophic medium; 4Q/X/Aba: adenine, histidine, leucine, and tryptophan auxotrophic media were supplemented with aureobasidin A (AbA) and Alpha-galactosidase (X-α-Gal).
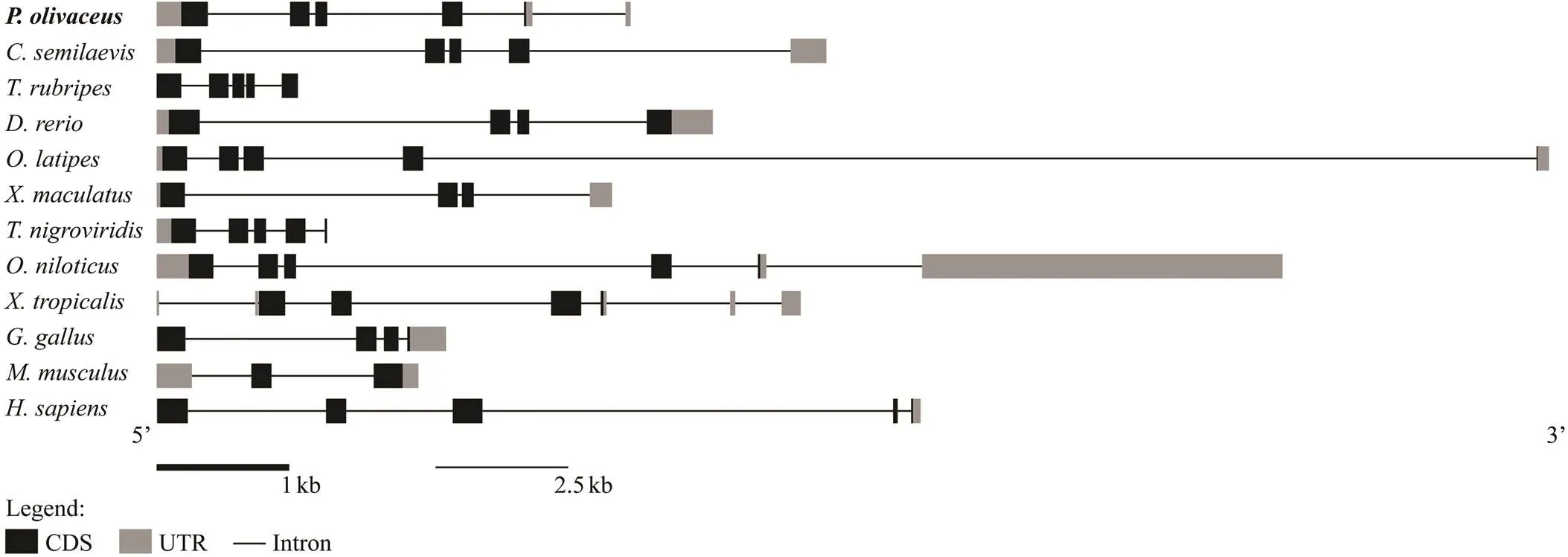
Fig.2 The genomic structure of figla in some teleosts and tetrapods. Coding sequence and UTRs are indicated by dark and gray boxes, respectively, whereas introns are represented by lines.
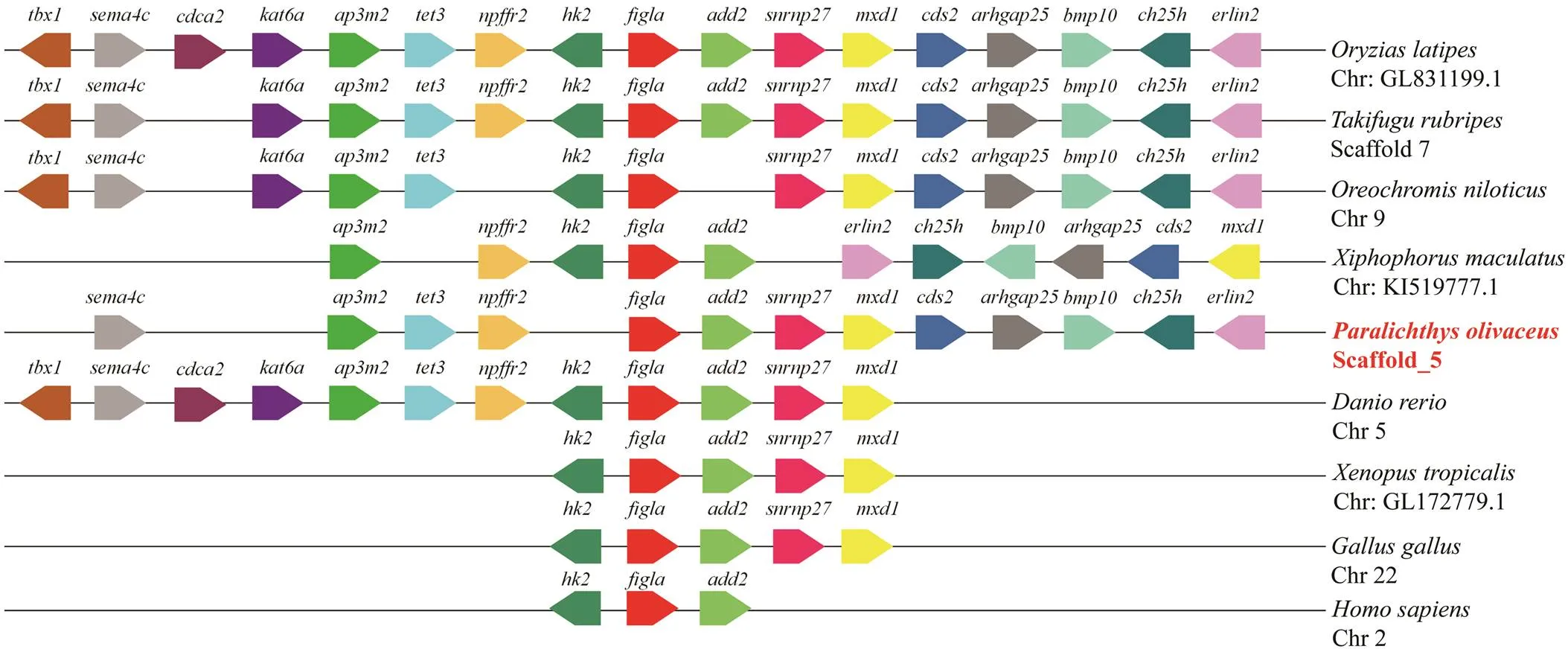
Fig.3 Chromosome synteny of Pofigla with those of other vertebrates. The pentagons with different colors represent va- rious genes and their respective directions. Gene order was determined according to the relative position of genes in the same chromosome (Chr) or scaffold.
3.2 Expression Pattern of Pofigla
The temporal expression pattern ofat the early stages of embryonic development was investigatedqRT-PCR.transcripts showed a maternal expression pattern in view of the result that transcripts maintained a high le- vel until blastula stage and then decreased significantly at gastrula stage and reached the undetectable level after neu- rula stage (Fig.4A).
The distribution pattern ofin different tissues was shown in Fig.4B.transcripts were specifical- ly distributed in gonads, with a sexually dimorphic expres- sion pattern observed. More specifically, a significantly higher expression level was found in the ovary. By con- trast,was moderately expressed in liver and intestine and not expressed in spleen and kidney (Fig.4B). To bet- ter understand the expression change ofduring go- nadal developmental stages, we performed qRT-PCR on go- nads at different stages, including 8 months (immature go- nads), 1 year (testis is typically mature) and 1.5 year (ovary is typically close to maturity).The expression rose in ovary from 8-month-old stage to 1-year-old and 1.5-year-old stage although there was no significant difference between 1 year and 1.5 year stages. Significant difference between ovary and testis sustained in these three phases with ovary show- ing higher expression than testis (Fig.4C).

Fig.4 The relative expression patterns of Pofigla during embryonic development stage from 1 cell stage to the hatching (A); in different adult tissues (B); and in gonad tissues at different developmental stages (C). Data are shown as mean± standard error of mean (n=3). Different superscripts indicate significant differences P<0.05.
3.3 Localization of Pofigla in Gonad Tissues by ISH
In the ovary,mRNA signals could be specially found in the cytoplasm of oocytes but not in oogonia. In testis, the signals were mainly concentrated in the sperma- togonia and primary spermatocytes. No signals could be detected in the somatic cells around the germ cells both in ovary and testis, which indicatemay be a germ cell specific transcription factor in(as shown in Fig.5).
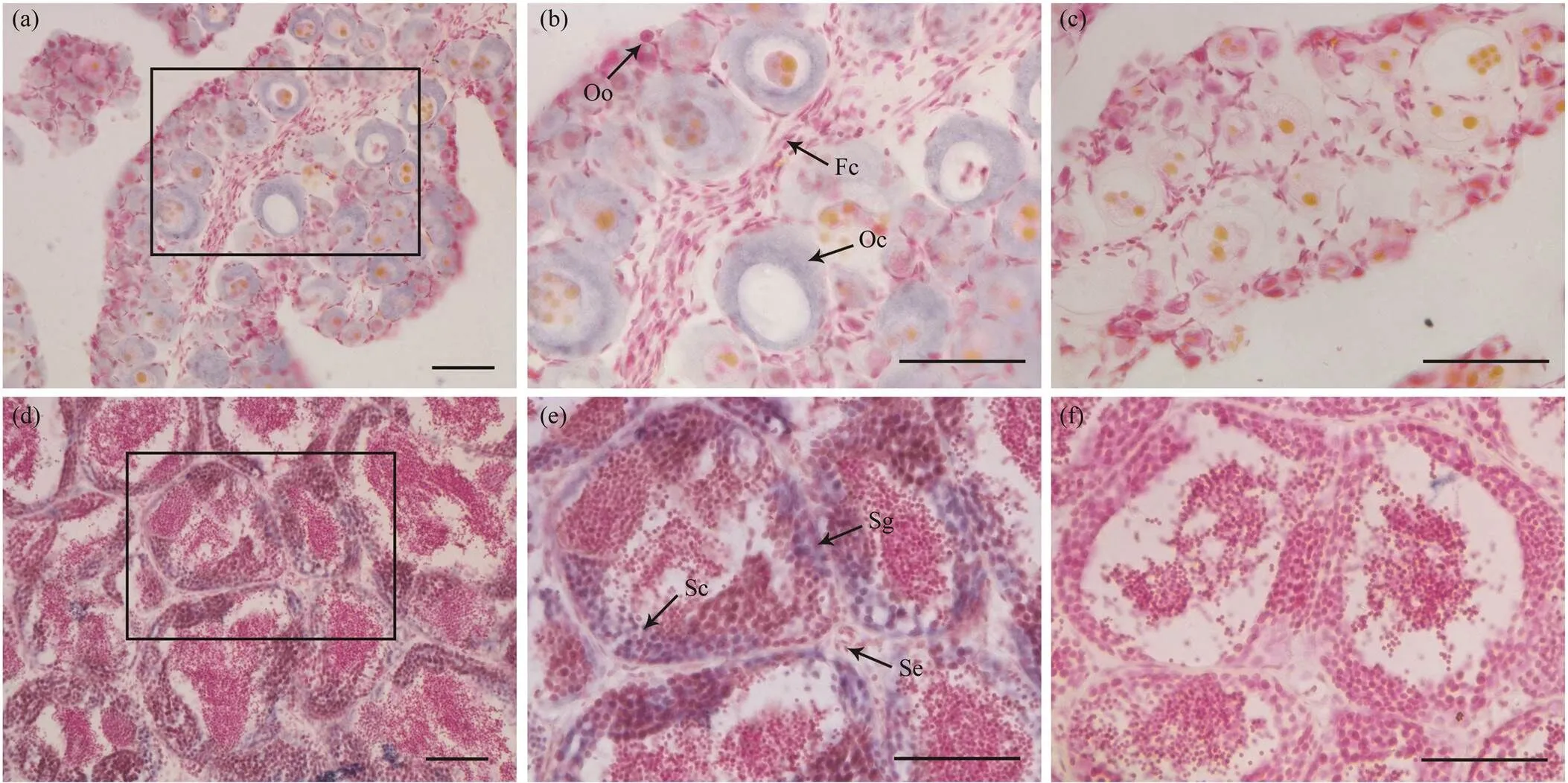
Fig.5 The expression of Pofigla in gonad tissues determined by ISH. The positive signals were stained as purple or blue (a,b,d,e), while the negative control with sense probe hybridization was unstained (c,f). Panels (b) and (e) are magnifica- tions of panels (a) and (d), respectively. The lower left cornerpart is magnifications of black box in panel (e). Oo, oogonia; Oc, oocytes; Fc, Follicular cell; Sg, spermatogonia; Sc, spermatocyte; Se, Sertoli cells. Scale bars=50μm.
3.4 Functional Promoter Regions Characterization of Pofigla
In order to find the cis-elements responsible for promo- ter activity of, we constructed a series of plasmids con- taining GFP driven by the sequentially deleted promoters of. A 2923bp fragment (?2966 to ?44) of the5’flanking region was cloned and sequenced. These deletionconstructs were transiently transfected into HEK 293T cells,and the green fluorescence intensity was observed after 48h (Fig.6A–I). The results showed that thepromo- ter fragments ?251/?44 (Fig.6B), ?444/?44 (Fig.6C), ?1137/?44 (Fig.6E), ?1729/?44 (Fig.6F) and ?2126/?44 (Fig.6G) could hardly induce GFP expression in HEK 293T cells, while the region ?772/?44 (Fig.6D) and ?2966/?44 (Fig.6H) could evidently drive GFP expression. This indicated that there may be some strongly positively regulated cis-ele- ments binding in the fragments ?772/?444 and 2966/?2126. The fluorescence intensity of allpromoter fragments were weaker than those of CMV promoters (Fig.6I).
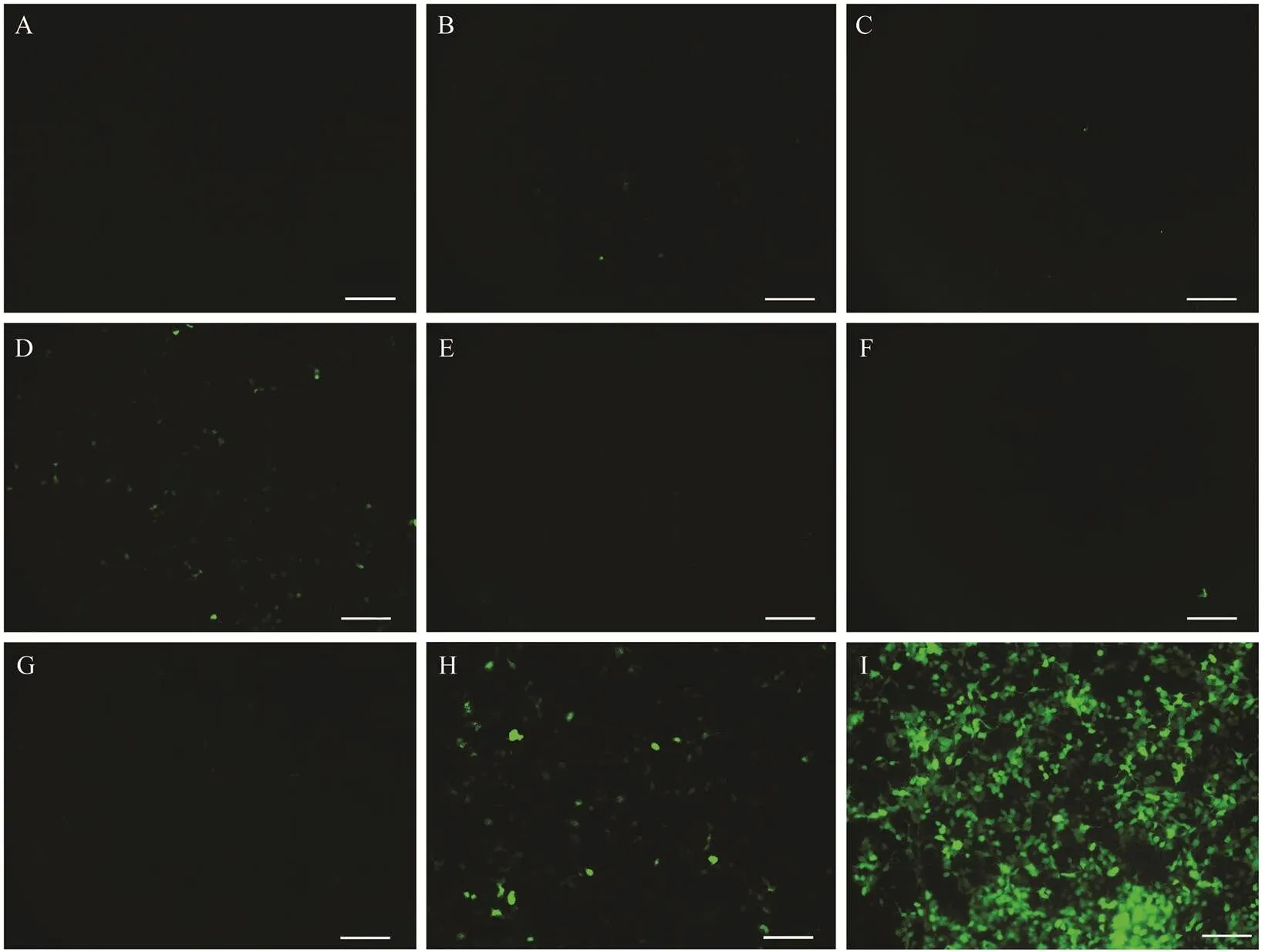
Fig.6 Functional analysis of Pofigla promoter. (A) HEK 293T cells transfected with pEGFP-1. (B–H) HEK 293T cells trans- fected with plasmids containing EGFP driven by the sequentially deleted promoters pEGFP-251/?44 (B), pEGFP?444/?44 (C), pEGFP?772/?44 (D), pEGFP?1137/?44 (E), pEGFP?1729/?44 (F), pEGFP?2126/?44 (G), and pEGFP?2966/?44 (H), respectively. (I) HEK 293T cells transfected with pEGFP-C1. Scale bars=100μm.
3.5 Identification of Cis-Acting Elements Regulating Pofigla Promoter Activity
In the present study, ?772/?444 and ?2966/?2126 are two highly active promoter regions of. To identify re- gulatory cis-acting elements in these promoter regions, se- quence analysis was performed by the MatInspector pro- gram with a cutoff value over 95%. Some putative bind- ing sites of the transcription factor motifs, including GATA1,FOXP1, LEF1, SMAD1 in ?2966/?2126 and SOX5, GATA1, YY2 in ?772/?444, respectively, were identified(Fig.7A). Furthermore, by removing the motifs that lack experimen- tal basis provided in MatInspector, we set the research tar- gets at FOXP1 motif located at ?2428 to ?2412, LEF1 mo-tif located at ?2218 to ?2202, SOX5 motif located at ?738 to ?716, and GATA1 motif located at ?654 to ?642 (Figs.7A, B). To define the functional status of the four poten- tial four regulatory elements within the promoter, we per- formed four site-directed plasmid mutations, namely, ?2966/?44-ΔFOXP1, ?2966/?44-ΔLEF1, ?772/?44-ΔSOX5 and ?772/?44-ΔGATA1, respectively (Fig.7A). Then, we eva- luated the function of these cis-acting elements through dual-luciferase reporter assay in HEK 293T cells. The re- lative luciferase activities of ?2966/?44-ΔFOXP1, ?2966/?44-ΔLEF1 and ?772/?44-ΔSOX5 exhibited statistically lower level compared with that of the wild-type construct pGL3?2966/?44 or pGL3?772/?44 (shown in Fig.7C). In contrast, ?772/?44-ΔGATA1 showed the significantly high-er relative luciferase level compared with construct pGL3-?772/?44 (Fig.7C). These results indicated that FOXP1, LEF1 and SOX5 were effective to regulatepromoter transcriptional activity, while GATA1 might play a nega- tive regulatory role fortranscription in ?772/?44 re- gion.
3.6 Regulation of Pofigla Transcription by the Wnt Signaling Pathway
The above results illustrated thatplays non-ignor- able role in activating the transcription of. Given the positive transcription regulation effect ofon, we explore whether there is a co-localization between them. When HEK 293T cells were transfected with LEF1-pEGFP-N1 and Figla-pmCherry-N1, the green and red fluo- rescence were coincided mostly in the nuclear with each other. While in other groups (Fig.8), there was no apparent merge phenomenon of nuclear localization. These results indicated the close relationship betweenand. A previous study has showncould regulate themRNA level ofin half-smooth tongue sole (Zhu., 2017). Given thatandare pivotal members of the canonical Wnt signaling pathway (Wong., 2002; Deroo., 2004; Pilon., 2006), it is reasonable to speculate that the transcription ofmay be regulated by this pathway. To test this hypothesis, we examined the effects ofandon the transcription activity of thepromoter separately or by co-transfecting the over- expression vector into the HEK 293T cells. The dual-luci- ferase reporter assays showed that both B-catenin and LEF1could enhance transcriptional activity ofin a dose-dependent manner (Fig.9A). Mutant LEF1 binding site in- hibited theoroverexpression-induced up- regulation of thepromoter activity (Fig.9B). Then we explored whethermRNA could change in ovar- ian cells with the activator and inhibitor of Wnt signaling pathway. qRT-PCR showed that BIO (activator) could ap- parently improve the mRNA expression of,and(Figs.9C–E), while XAV-939 (inhibitor) could sig- nificantly inhibit the mRNA expression of,and showed an insignificant inhibitory effect onex- pression (Figs.9C–E)These results suggested that the pu- tative binding sites for LEF1 are required for the transcrip- tionactivation ofand implied that the regulation of the canonical Wnt signaling pathway is significant (Fig.9F).
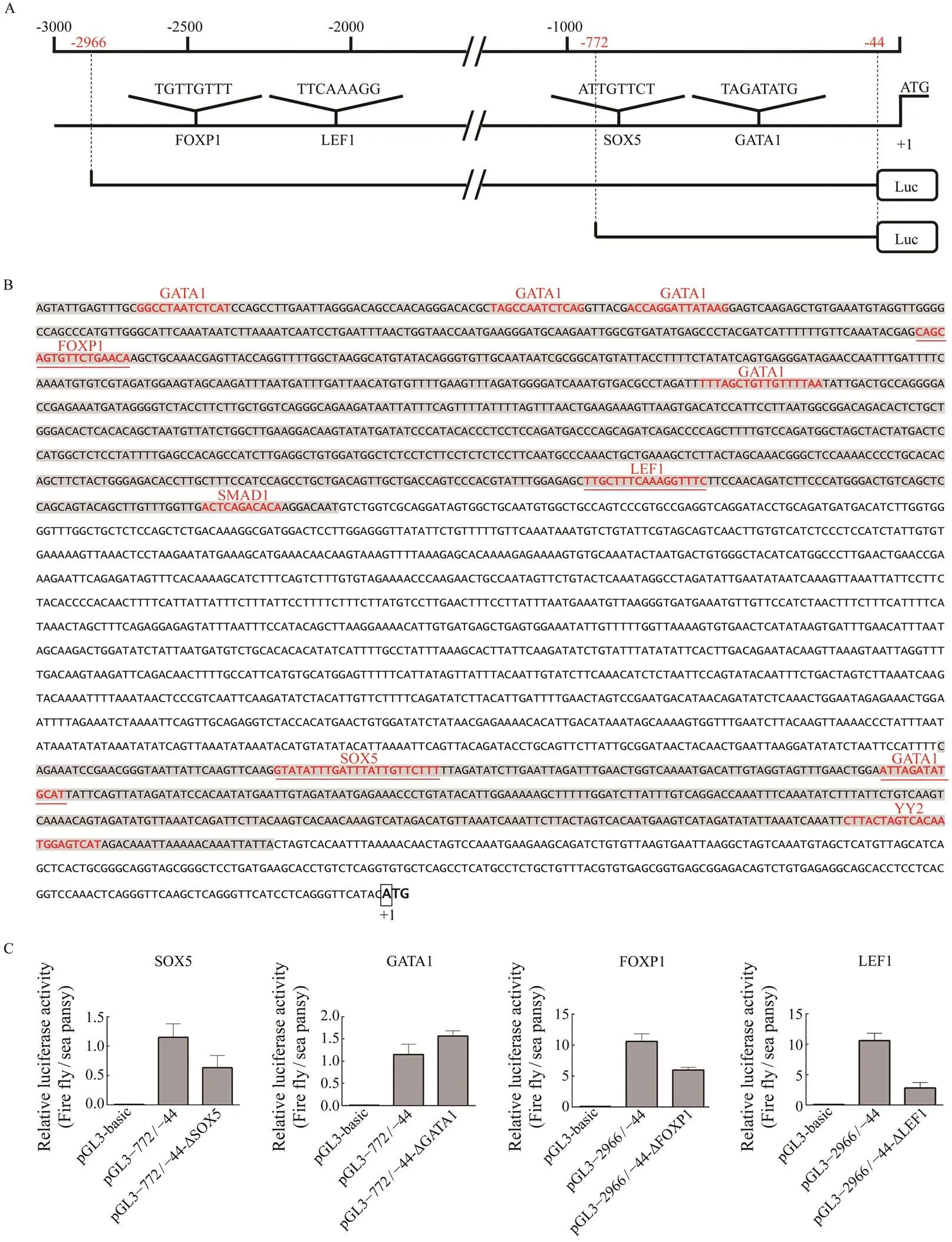
Fig.7 Putative binding motif of SOX5, GATA1, FOXP1 and LEF1 and site-directed mutations assays in the figla promoter. (A) A schematic diagram of putative SOX5, GATA1, FOXP1 and LEF1 binding elements in the figla promoter. The ini- tiation codon (ATG) is designated as +1. The scale is given above and the names of the potential TF binding sites are pro- vided at the bottom. The core motif sequence of each transcription factor is shown just above their names. Extremities of the two control plasmid locations are marked with red numbers in the scale. (B) Nucleotide sequences and putative regu- latory elements of the Pofigla promoter regions. The ?2966/?2126 and ?772/?444 regions are represented with a gray background. The putative regulatory element motif sequences are in red font. The candidate four motifs are underlined in red and ATG is defined as +1 position. (C) The effect of mutagenesis of transcription factor binding sites of SOX5, GATA1, FOXP1 and LEF1 on the figla promoter. Fragments indicated in red demonstrate the success of mutation assays. Data are shown as mean±SEM (n=3 for each group). Different superscripts indicate that P<0.05.

Fig.8 Co-localization of LEF1 and Figla. HEK 293T cells were transfected with different combination of recombinant pEGFP and pmCherry plasmids. The left-hand panels depict GFP staining, the second panels from the left depict mCherry staining, the third panels from the left depict DAPI staining, and the right-hand panels depict merged GFP/mCherry/DAPI staining. The upper panels depict localization of the GFP and mCherry negative control (A), and the 2–4 panels depict lo- calization of the pEGFP-N1 and PoFigla-pmCherry-N1 (B), PoLEF1-pEGFP-N1 and pmCherry-N1 (C), PoLEF1-pEGFP- N1 and PoFigla-pmCherry-N1 proteins (D), respectively. The nuclei were stained as blue by DAPI. Scale bars=20μm.
4 Discussion
Gonadal differentiation and development determine re- productive potential and concomitantly affect growth insome fishes. As an important gene associated with ovarian development and female fertility,has been clonedand characterized in many species, including mouse (Liang., 1997), bovine (Tripurani., 2013), zebrafish (Qin., 2018), half-smooth tongue (Li., 2016) and Nile Tilapia (Qiu., 2015). Althoughhas been identi- fied in some teleosts, the clues about its transcription re- gulation is still unclear. To date, it was found thatcan directly targetto regulate the meiotic process of oocytes in mouse (Grive., 2016). It is meaningful to explore the transcriptional mechanism ofin teleosts, which helps us understand the internal molecular mecha- nisms of gonadal development from the perspective of gene regulation.
In this study, the core sequence ofgene was ob- tained from ovary cDNA library in Japanese flounder. Thededuced amino acid sequence ofshowed typical cha- racteristic features of the bHLH family with conserved DNA-binding region (Liu and Zhao, 2010) and followed by a helix-loop-helix domain responsible for interacting with other bHLH proteins (Wang., 2015). Yeast two-hy- brid assay indicated Figla probably regulates downstream genes as a heterodimers with other transcription factor in Japanese flounder. As an important gene for fertility,has been well studied in human (Bayne., 2004). Once there was one or several mutations in the exon ofgene, most women will suffer the severe premature ovar- ian failure (Tosh., 2015). With the rapid development of high-throughput sequencing technology,has been considered as a female-biased gene in gonad of several te- leosts, such as spotted scat (He., 2019), black porgy (Zhang., 2019), and Japanese eel (Jeng., 2018). In this study,was expressed extremely high in the ovary and it was specifically distributed in the cytoplasm of oocytes in Japanese flounder (Fig.5), which was con- sistent with the study by Liang. (2020). In bovine,might be a maternal gene with high mRNA expres- sion from pronuclear to eight-cell stage but is barely de- tectable at morula and blastocyst stages (Tripurani., 2013). Here,was highly expressed from 1 cell stage to low blastula stage (Fig.4), which also showed a mater- nal expression pattern in Japanese flounder.
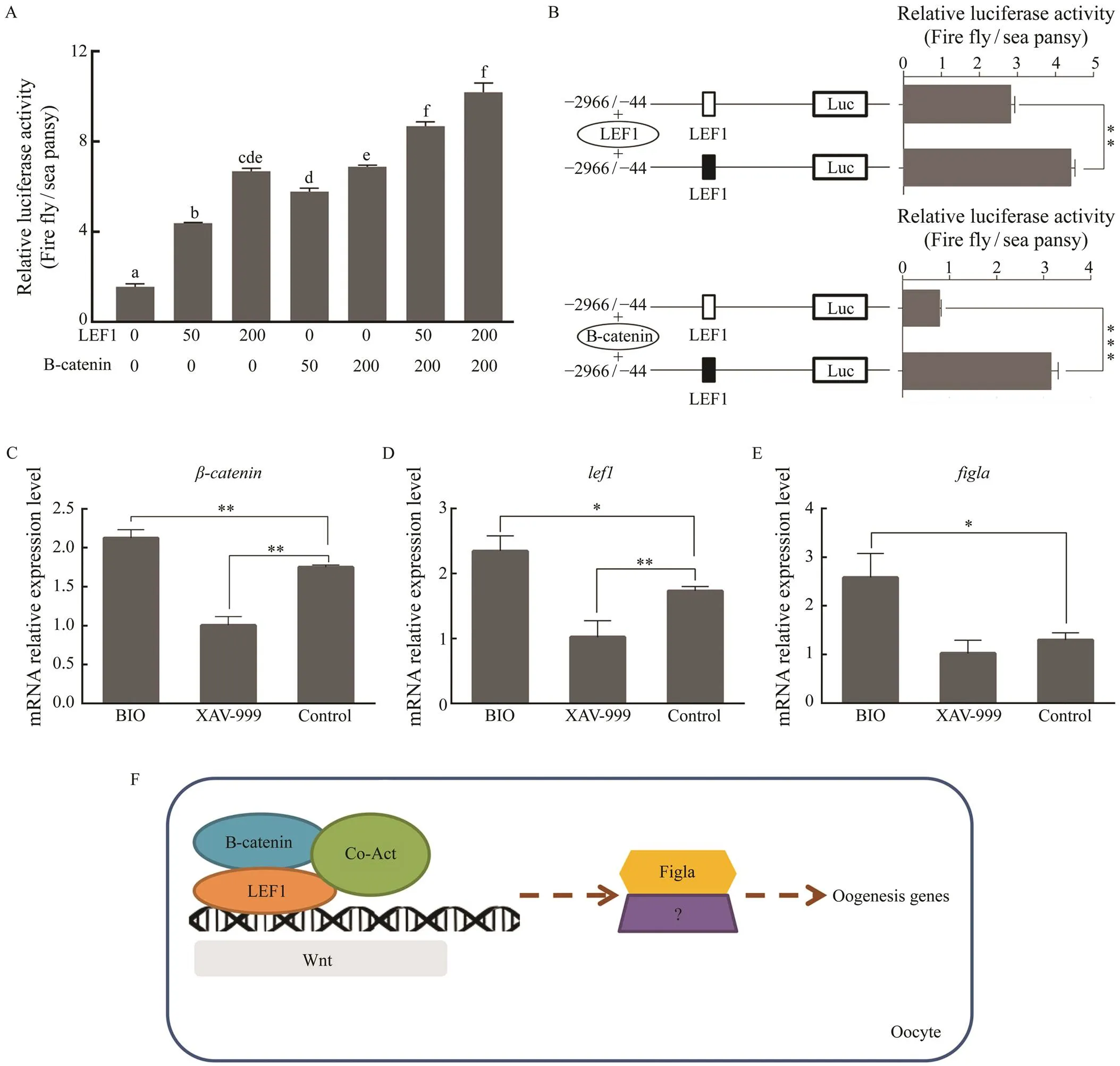
Fig.9 The effect of Wnt signaling pathway on the transcription of Pofigla. (A) The luciferase activity of Pofigla promoter ?2966/?44 region after overexpressing lef1 and β-catenin with different amount (ng). (B) Effects of lef1 or β-catenin over- expression on the activity of the Pofigla promoter ?2966/?44 region with a mutation in the binding site for lef1. Black and white box indicated the normal and mutational binding motif of LEF1, respectively. (C–E) qRT-PCR analyses for ex- pressions of β-catenin (C), lef1 (D), and figla (E) in Japanese flounder ovarian cells stimulated by BIO or XAV-939. (F) Hypothetical model of figla regulated by Wnt signaling in Japanese flounder oocytes. Data are shown as mean±SEM (n=3 for each group). * indicates significant difference (P<0.05), while ** indicates highly significant difference (P<0.01).
One interesting phenomenon was thatwas also highly expressed in testis compared with other tissues ex- cept for ovaries, which is different from the results by Liang. ( 2020). In their results, the expression ofwas nearly not detectable in the testis throughout the develop- mental cycle. While in our study, the higher expression ofin testis was detected at 8-month-old stage, comparedwith 1-year-old and 1.5-year-old stage. And the smallest ex- pression difference, less than a 2-fold gap, between testis and ovary was found at 8-month-old stage. It is worth re- minding that in half-smooth tongue sole,was uni-quely expressed in the female ovary, whilewas mainly expressed in the testis of pseudo male (Li., 2016). Therefore, different developmental backgrounds of the male fish (genetic background, presence of adulterated pseudo males, different sampling season) could contribute to these different results. We suspected thatgene not only plays a role in the ovary, but also participates in the development of the testis at early developmental stage.
Here, we identified and characterized the highly active promoter regions ofand predicted the putative im- portant transcription factor according to their binding mo-tif sequences. In addition to the identified two promoter re-gions (?772/?444 and ?2966/?2126) with positive regula- tory cis-elements, the results also showed that there may be strong negative regulating elements in the fragments ?1137/?772 and ?444/?251. Lack of transcription factors that can activate theexpression in the HEK 293T cells may also contribute to this result. By dual luciferase assay, we concluded that SOX5, LEF1 and FOXP1 may play im- portant roles in the transcriptional activation of, while GATA1 may play a negative role in the regulation ofexpression.was reported to regulateex- pression to promote gonad development in protogynous hermaphrodite red-spotted grouper and(Huang., 2009; Chen., 2014). In medaka,was involved in germ-cell regulation and sex determina- tion (Schartl., 2018). We speculated thatis also involved in the development of the gonads, through regu- latingin Japanese flounder.
Another transcription factor that has a significant posi- tive regulatory effect onis. As a target gene of Wnt,is generally considered to be an important indi- cator of whether Wnt is activated (Clevers, 2006). In mouse, there is a female-specific regulation ofsplicing, and this regulation may enhance Wnt signaling activity in fe- male gonads (Zhao., 2018). In addition, it has been reported recently that the canonical Wnt signaling path-way regulated gonad differentiation and development inzebrafish (Sreenivasan., 2014; Safian., 2018).Once Wnt was inhibited during gonad differentiation in zeb- rafish, it will lead to more male individuals and decrease the expression of gonadal aromatase gene () (Sreeni-vasan., 2014). In our study, we proved thatandhave positive regulatory effects on the promoter activity of. It is reasonable to think that there are some genes or other pathways that compensate for the expres- sion ofwhen Wnt was inhibited in Japanese flounder.andindeed play an important role in thetranscriptional activation of. Hence Wnt signaling may regulate the expression ofgene in the ovary of Japa- nese flounder, which was similar to the ‘pro-female’ path- way of Wnt/signaling in zebrafish (Sreenivasan., 2014). We have found many vital members of Wnt signaling pathway such as,,,,,,,and, were ovary-biased genes in our previous RNA-seq data (Zhang., 2016). Taken together, all the results implied that transcription level ofmight be activated by canonical Wnt signaling path- way based on potential binding site of LEF1 on the pro- moter ofin Japanese flounder (Fig.9F).
5 Conclusions
In summary, the cDNA sequence ofwas identified in. Amino acid alignment and chromosome synteny analysis showed thatwas conserved in evo- lution and held conserved structure between fish and te- trapods. qRT-PCR assay demonstrated thatwas sig- nificantly expressed in gonad, with a higher expression in ovary than in testis. ISH results revealed thatwas spe- cifically distributed in the spermatogonia and primary sper- matocyte of male testis and oocytes of female ovary. Screen-ing and analyzing promoter regions ofshowed itstranscription could be regulated by SOX5, GATA1, FOXP1, LEF1 and Wnt signaling pathway. All the results in this study provided the first evidence thatmay not onlyserves as a considerable role in the ovary development, but also plays some important functions in testis development and/or male germ cell differentiation. Moreover, Wnt sig- nal inducesexpression in oocytes in Japanese floun- der.
Abbreviations
,;, the factor in the gerline alpha;,; cDNA, complementary deoxyribonucleic acid; UTR, un- translated region; ORF, open reading frame; qRT-PCR, quantitative real-time polymerase chain reaction; ISH,hybridization;, serpin family B member 6c;, Rho GTPase activating protein 20;, PDZ domain containing 1; NALP, NACHT, LRR and pyrin do- mains containing protease; E12, helix-loop-helix protein E12; CMV, human cytomegalovirus; HEK, human em- bryonic kidney; GATA1, transcription factor GATA-1; FOXP1, Forkhead box P1; SOX5, SRY-box transcription factor 5; LEF1, lymphoid enhancer binding factor 1; SMAD1, mothers against decapentaplegic homolog 1; YY2,YY2 transcription factor; Frizzled-1, frizzled class re- ceptor 1; Dsh, dishevelled; Wnt1, Wnt family member 1; Lrp5, LDL receptor related protein 5; Lrp6, LDL receptor related protein 6; RTK, receptor protein tyrosine kinase; ROR2, receptor tyrosine kinase like orphan receptor 2; BCL9, BCL9 transcription coactivator.
Acknowledgement
This study was supported by the National Key R&D Pro- gram of China (No. 2018YFD0901205).
Bayne, R. A., Martins da Silva, S. J., and Anderson, R. A., 2004. Increased expression of the FIGLA transcription factor is as- sociated with primordial follicle formation in the human fetal ovary.,10(6): 373-381, DOI:10.1093/molehr/gah056.
Blumel, R., Zink, M., Klopocki, E., and Liedtke, D., 2019. On the traces of: Investigation of the gene expression pattern du-ring development and cranial suture patterning in zebrafish ().,14(6): e0218286, DOI:10.1371/journal.pone.0218286.
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A.,, 2005. MatInspector and beyond: Pro- moter analysis based on transcription factor binding sites.,21(13): 2933-2942, DOI:10.1093/bioinformatics/bti473.
Chen, X. W., Jiang, S., Gu, Y. F., and Shi, Z. Y., 2014. Molecu- lar characterization and expression ofgene in.,85(2): 516-522, DOI:10.1111/jfb.12418.
Chiba, Y., He, B., Yoshizaki, K., Rhodes, C., Ishijima, M., Bleck, C. K. E.,., 2019. The transcription factor AmeloD stimu- lates epithelial cell motility essential for tooth morphology.,294(10): 3406-3418, DOI:10.1074/jbc.RA118.005298.
Clevers, H., 2006. Wnt/beta-catenin signaling in development and disease.,127(3): 469-480, DOI:10.1016/j.cell.2006.10.018.
Deroo, T., Denayer, T., Van Roy, F., and Vleminckx, K., 2004. Global inhibition of Lef1/Tcf-dependent Wnt signaling at its nuclear end point abrogates development in transgenicembryos.,279(49): 50670-50675, DOI:10.1074/jbc.M408969200.
Gao, J., Wang, J., Jiang, J., Fan, L., Wang, W., Liu, J.,., 2013. Identification and characterization of ahomolog in Ja- panese flounder ().,531(2): 411-421, DOI:10.1016/j.gene.2013.08.030.
Grive, K. J., Gustafson, E. A., Seymour, K. A., Baddoo, M., Schorl, C., Golnoski, K.,., 2016. TAF4b regulates oocyte-speci- fic genes essential for meiosis.,12 (6): e1006128, DOI: 10.1371/journal.pgen.1006128.
He, F. X., Jiang, D. N., Huang, Y. Q., Mustapha, U. F., Yang, W., Cui, X. F.,., 2019. Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat ()., 45(6): 1963-1980, DOI:10.1007/s10695-019-00693-8.
Hu, W., Gauthier, L., Baibakov, B., Jimenez-Movilla, M., and Dean, J., 2010. FIGLA, a basic helix-loop-helix transcription factor, balances sexually dimorphic gene expression in post- natal oocytes.,30(14): 3661-3671, DOI:10.1128/mcb.00201-10.
Huang, W., Zhou, L., Li, Z., and Gui, J. F., 2009. Expression pat- tern, cellular localization and promoter activity analysis of ova-rian aromatase (Cyp19a1a) in protogynous hermaphrodite red-spotted grouper.,307(1-2): 224-236, DOI:10.1016/j.mce.2009.04.003.
Jeng, S. R., Wu, G. C., Yueh, W. S., Kuo, S. F., Dufour, S., and Chang, C. F., 2018. Gonadal development and expression of sex-specific genes during sex differentiation in the Japanese eel., 257: 74-85, DOI:10.1016/j.ygcen.2017.07.031.
Joshi, S., Davies, H., Sims, L. P., Levy, S. E., and Dean, J., 2007. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor.,7: 67, DOI:10.1186/1471-213x-7-67.
Li, H., Xu, W., Zhang, N., Shao, C., Zhu, Y., Dong, Z.,., 2016. Twohomologues have disparate functions during sex dif-ferentiation in half-smooth tongue sole ().,6: 28219, DOI:10.1038/srep28219.
Liang, L., Soyal, S. M., and Dean, J., 1997. FIGalpha, a germ cell specific transcription factor involved in the coordinate expres- sion of the zona pellucida genes.,124(24): 4939-4947.
Liang, S., Wang, W., Wang, L., Wu, Z., Zou, Y., Tan, X.,, 2020.gene roles in the proliferation of oocytes in the olive flounder.,528: 735493, DOI:10.1016/j.aquaculture.2020.735493.
Liu, W. Y., and Zhao, C. J., 2010. Genome-wide identification andanalysis of the chicken basic helix-loop-helix factors., 2010: 682095, DOI:10.1155/2010/682095.
Liu, X. T., Wang, Y., Wang, X. H., Tao, X. F., Yao, Q., and Chen, K. P., 2014. A genome-wide identification and classification of basic helix-loop-helix genes in the jewel wasp,(Hymenoptera: Pteromalidae).,57(10): 525-536, DOI:10.1139/gen-2014-0171.
Niu, J., Guan, J., Li, R., Li, X., Zhai, J., Qi, J.,., 2018.regulates embryo development byinhibiting the Wnt/beta-catenin signaling pathway.,19(7): 1915, DOI:10.3390/ijms19071915.
Pilon, N., Oh, K., Sylvestre, J. R., Bouchard, N., Savory, J., and Lohnes, D., 2006.is a direct target of the canonical Wnt pathway.,289(1): 55-63, DOI:10.1016/j.ydbio.2005.10.005.
Qin, M., Zhang, Z., Song, W., Wong, Q. W., Chen, W., Shirgaon-kar, N.,., 2018. Roles of Figla/in juvenile ovary de- velopment and follicle formation during zebrafish gonadoge- nesis.,159(11): 3699-3722, DOI:10.1210/en.2018-00648.
Qiu, Y., Sun, S., Charkraborty, T., Wu, L., Sun, L., Wei, J.,.,2015. Figla favors ovarian differentiation by antagonizing sper- matogenesis in a teleosts, Nile tilapia ().,10(4): e0123900, DOI:10.1371/journal.pone.0123900.
Safian, D., Bogerd, J., and Schulz, R. W., 2018. Igf3 activates beta-catenin signaling to stimulate spermatogonial differenti- ation in zebrafish.,238(3): 245-257, DOI:10.1530/joe-18-0124.
Schartl, M., Schories, S., Wakamatsu, Y., Nagao, Y., Hashimoto, H., Bertin, C.,, 2018.is involved in germ-cell re- gulation and sex determination in medaka following co-option of nested transposable elements.,16(1): 16, DOI:10.1186/s12915-018-0485-8.
Soyal, S. M., Amleh, A., and Dean, J.,2000. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation.,127(21): 4645-4654.
Sreenivasan, R., Jiang, J., Wang, X., Bartfai, R., Kwan, H. Y., Christoffels, A.,., 2014. Gonad differentiation in zebra- fish is regulated by the canonical Wnt signaling pathway., 90(2): 45, DOI:10.1095/biolreprod.113.110874.
Tan, X., Xu, P., Zhang, Y., and Zhang, P. J., 2019. Olive flounder () myogenic regulatory factor 4 and its muscle-specific promoter activity..,236: 110310, DOI:10.1016/j.cbpb.2019.110310.
Tarczewska, A., and Greb-Markiewicz, B., 2019. The signifi- cance of the intrinsically disordered regions for the functions of the bHLH transcription factors.,20(21): 5306, DOI:10.3390/ijms20215306.
Tosh, D., Rani, H. S., Murty, U. S., Deenadayal, A., and Grover, P., 2015. Mutational analysis of thegene in women with idiopathic premature ovarian failure.,22(5): 520-526, DOI:10.1097/gme.0000000000000340.
Tripurani, S. K., Wee, G., Lee, K. B., Smith, G. W., Wang, L., andYao, J. B., 2013. MicroRNA-212 post-transcriptionally re-gulates oocyte-specific basic-helix-loop-helix transcription fac-tor, factor in the germline alpha (FIGLA), during bovine early embryogenesis.,8(9): e76114, DOI:10.1371/journal.pone.0076114.
Wang, L., Zhu, Y., Xu, W., Shao, C., Dong, Z., Li, H.,., 2017. Molecular characterization ofduring sex develop-ment in Chinese tongue sole ().,494(3-4): 714-718, DOI:10.1016/j.bbrc.2017.10.126.
Wang, X. H., Wang, Y., Liu, A. K., Liu, X. T., Zhou, Y., Yao, Q.,., 2015. Genome-wide identification and analysis of basic helix-loop-helix domains in dog,familiaris.,290(2): 633-648, DOI:10.1007/s00438-014-0950-1.
Wong, M. H., Huelsken, J., Birchmeier, W., and Gordon, J. I., 2002. Selection of multipotent stem cells during morphoge- nesis of small intestinal crypts of Lieberkuhn is perturbed by stimulation of Lef-1/beta-catenin signaling.,277(18): 15843-15850, DOI:10.1074/jbc.M200184200.
Yamamoto, E., 1999. Studies on sex-manipulation and produc- tion of cloned populations in hirame,(Temminck et Schlegel).,173(1): 235-246, DOI:10.1016/S0044-8486(98)00448-7.
Zhang, D., Li, G., and Wang, Y., 2017. A genome-wide identifi- cation and analysis of basic helix-loop-helix transcription fac- tors in cattle.,626: 241-250, DOI:10.1016/j.gene.2017.05.036.
Zhang, J., Hu, Y. H., Sun, B. G., Xiao, Z. Z., and Sun, L., 2013. Selection of normalization factors for quantitative real time RT-PCR studies in Japanese flounder () and turbot () under conditions of viral infection.,152(3-4): 303-316, DOI:10.1016/j.vetimm.2012.12.018.
Zhang, K., Xu, J., Zhang, Z., Huang, Y., Ruan, Z., Chen, S.,.,2019. A comparative transcriptomic study on developmental gonads provides novel insights into sex change in the pro- tandrous black porgy ().,111(3): 277-283, DOI:10.1016/j.ygeno.2018.11.006.
Zhang, W., Liu, Y., Yu, H., Du, X., Zhang, Q., Wang, X.,., 2016. Transcriptome analysis of the gonads of olive flounder ().,42(6): 1581-1594, DOI:10.1007/s10695-016-0242-2.
Zhao, L., Wang, C., Lehman, M. L., He, M., An, J., Svingen, T.,., 2018. Transcriptomic analysis of mRNA expression and alternative splicing during mouse sex determination.,478: 84-96, DOI:10.1016/j.mce.2018.07.010.
Zhu, Y., Hu, Q., Xu, W., Li, H., Guo, H., Meng, L.,, 2017. Identification and analysis of thegene in half-smooth tongue sole ().,12(5): e0176122, DOI:10.1371/journal.pone.0176122.
January 2, 2021;
March 4, 2021;
August17, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
* Correspondence author. E-mail: qzhang@ouc.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- Complete Mitochondrial Genome of Myra affinis (Decapoda:Brachyura: Leucosiidae) and Its Phylogenetic Implications for Brachyura
- Multisource Target Classification Based on Underwater Channel Cepstral Features
- Joint Model of Wind Speed and Corresponding Direction Based on Wind Rose for Wind Energy Exploitation
- Elastic-Wave Reverse Time Migration Random Boundary-Noise Suppression Based on CycleGAN
- Identification, Phylogeny and Expressional Profiles of Peptidoglycan Recognition Protein (PGRP) Gene Family in Sinonovacula constricta
- Expressions of Toll Like Receptor (TLR) Genes in Paralichthys olivaceus After Induced by Different Extracts of Edwardsiella tarda
