Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants
Muhammad Tahir ul Qamar,Safar M.Alqahtani,Mubarak A.Alamri,Ling-Ling Chen,*
aCollege of Life Science and Technology,Guangxi University,Nanning,530004,PR China
bHubei Key Laboratory of Agricultural Bioinformatics,College of Informatics,Huazhong Agricultural University,Wuhan,430070,PR China
cDepartment of Pharmaceutical Chemistry,College of Pharmacy,Prince Sattam Bin Abdulaziz University,11323,Alkarj,Saudi Arabia
Keywords:
Coronavirus
SARS-CoV-2
COVID-19
Natural products
Protein homology modelling
Molecular docking
Molecular dynamics simulation
ABSTRACT
The recent pandemic of coronavirus disease 2019(COVID-19)caused by SARS-CoV-2 has raised global health concerns.The viral 3-chymotrypsin-like cysteine protease(3CLpro)enzyme controls coronavirus replication and is essential for its life cycle.3CLprois a proven drug discovery target in the case of severe acute respiratory syndrome coronavirus(SARS-CoV)and Middle East respiratory syndrome coronavirus(MERS-CoV).Recent studies revealed that the genome sequence of SARS-CoV-2 is very similar to that of SARS-CoV.Therefore,herein,we analysed the 3CLprosequence,constructed its 3D homology model,and screened it against a medicinal plant library containing 32,297 potential anti-viral phytochemicals/traditional Chinese medicinal compounds.Our analyses revealed that the top nine hits might serve as potential anti-SARS-CoV-2 lead molecules for further optimisation and drug development process to combat COVID-19.
1.Introduction
A novel coronavirus strain linked with fatal respiratory illness was reported in late 2019[1].Swift actions were taken by the Centre for Disease Control and Prevention(CDC),Chinese health authorities,and researchers.The World Health Organization(WHO)temporarily named this pathogen 2019 novel coronavirus(2019-nCoV)[2].On January 10,2020,the first whole-genome sequence of 2019-nCoV was released,which helped researchers to quickly identify the virus in patients using reverse-transcription polymerase chain reaction(RT-PCR)methods[3].On January 21,the first article relatedto2019-nCoV waspublished,which revealed that 2019-nCoV belongs to the beta-coronavirus group,sharing ancestry with batcoronavirus HKU9-1,similar to SARS-coronaviruses,and that despite sequence diversity its spike protein interacts strongly with the human ACE2 receptor[1].On January 30,the WHO announced a Public Health Emergency of International Concern(PHEIC)for the 2019-nCoV outbreak.Later,the human-to-human transmission was confirmed.As of January 31,51 whole-genome sequences of 2019-nCoV from different laboratories and regions had been submitted to GISAID database[4].On February 12,the WHO permanently named the 2019-nCoV pathogen as SARS-CoV-2 and the causing disease as coronavirus disease 2019(COVID-2019).The Chinese government's swift actions helped them to control COVID-19 in China.However,SARSCoV-2 affected several countries world-wide.On March 11,the WHO formally recognized the COVID-19 as a pandemic.By March 19,2020,the global death toll reached 9,913,with 2,42,650 laboratory-confirmed cases.The case fatality rate among infected people varies in different countries.However,global case fatality rate is presently around 3.92% (calculated as deaths/[deaths+laboratory confirmed cases]).

Fig.1.(A)Phylogenetic tree inferred from closest homologs of SARS-CoV-2 3CLpro.The maximum likelihood method was used to construct this tree.(B)Multiple sequence alignment of closest homologs of SARS-CoV-2 3CLprosharing≥70% sequence identity.(C)Cartoon representation of the SARS-CoV-2 3CLprohomodimer.Chain-A(protomer-A)is in multicolour and Chain-B(protomer-B)is in dark blue.The N-finger that plays an important role in dimerization maintaining the active conformation is shown in hot pink,domain I is coloured cyan,domain II is shown in green,and domain III is coloured yellow.The N-and C-termini are labelled.Residues of the catalytic dyad(Cys-145 and His-41)are highlighted in yellow and labelled.(D)Cartoon representation of the 3CLpromonomer model(chain/protomer-A)of SARS-CoV-2 superimposed with the SARS-CoV 3CLprostructure.The SARS-CoV 3CLprotemplate is coloured cyan,the SARS-CoV-2 3CLprostructure is coloured grey,and all identified mutations are highlighted in red.(E)Docking of 5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone inside the receptor-binding site of SARS-CoV-2 3CLpro,showing hydrogen bonds with the catalytic dyad(Cys-145 and His-41).The 3CLprostructure is coloured dark blue,the 5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone is orange,and hydrogen coloured maroon.
Coronaviruses are single-stranded positive-sense RNA viruses that possess large viral RNA genomes[5].Recent studies showed that SARS-CoV-2 has a similar genomicorganization tothatof other beta-coronaviruses,consisting of a 5′-untranslated region(UTR),a replicasecomplex (orf1ab)encoding non-structuralproteins(nsps),a spike protein(S)gene,envelope protein(E)gene,a membrane protein(M)gene,a nucleocapsid protein(N)gene,3′-UTR,and several unidentified non-structural open reading frames[3].Although SARS-CoV-2 is classified into the beta-coronaviruses group,it is different from MERS-CoV and SARS-CoV.Recent studies highlighted that SARS-CoV-2 genes share<80% nucleotide identity and 89.10% nucleotide similarity with SARS-CoV genes[6,7].Usually,beta-coronaviruses produce a~800 kDa polypeptide upon transcription of the genome.This polypeptide is proteolytically cleaved to generate various proteins.The proteolytic processing is mediated by papain-like protease(PLpro)and 3-chymotrypsin-like protease(3CLpro).The 3CLprocleaves the polyprotein at 11 distinct sites to generate various non-structural proteins that are important for viral replication[8].3CLproplays a critical role in the replication of virus particles and unlike structural/accessory protein-encoding genes,it is located at the 3′end which exhibits excessive variability.Therefore,it is a potential target for anti-coronaviruses inhibitors screening[9].Structurebased activity analyses and high-throughput studies have identified potential inhibitors for SARS-CoV and MERS-CoV 3CLpro[10—12].Medicinal plants,especially those employed in traditional Chinese medicine,have attracted significant attention because they include bioactive compounds that could be used to develop formal drugs against several diseases with no or minimal side effects[13].Therefore,the present study was conducted to gain structural insights into the SARS-CoV-2 3CLproand to discover potent anti-COVID-19 natural compounds.
2.Materials and methods
2.1.Data collection
Whole-genome sequences of all SARS-CoV-2 isolates available till January 31,2020,were downloaded from GISAID database(accession numbers and details are given in Table S1)[4].The genome sequence of BetaCoV/Kanagawa/1/2020(GISAID:EPI_-ISL_402126)was incomplete,and the genome sequence of Beta-CoV/bat/Yunnan/RaTG13/2013 (EPI_ISL_402131)was an old sequence(2013);therefore,these sequences were not included in our analyses.Gene sequences of 3CLprowere extracted from the whole-genome sequences and translated into protein sequences using the translate tool of the ExPASy server[14].The first SARSCoV-2 sequence(Wuhan-Hu-1;GSAID:EPI_ISL_402125)was used as a reference in our analysis.

Table 1 Physicochemical parameters of SARS-CoV-2 3CLpro.

Table 2 Summary of top ranked phytochemicals screened against SARS-CoV-2 3CLproreceptor binding site with their respective structures,docking score,binding affinity and interacting residues.

Table 2(continued)
2.2.Sequence analyses
In order to identify similar sequences and key/conserved residues,and to infer phylogeny,multiple sequence alignment of SARSCoV-2 3CLprofollowed byphylogenetictreeanalyseswere performed using T-Coffee[15]and the alignment figure was generated using ESPript3[16].Physicochemical parameters of SARS-CoV-2 3CLproincluding isoelectric point,instability index,grand average of hydropathicity(GRAVY),and amino acid and atomic composition were investigated using the ProtParam tool of ExPASy[14].
2.3.Structural analyses
To probe the molecular architecture of SARS-CoV-2 3CLpro,comparative homology modelling was performed using Modeller v9.11[17].To select closely-related templates for modelling,PSIBLAST was performed against all known structures in the protein databank(PDB)[18].Chimera v1.8.1[19]and PyMOL educational version[20]were used for initial quality estimation,energy minimisation,mutation analyses,and image processing.
2.4.Ligand database preparation and molecular docking
A comprehensive medicinal plant library containing 32,297 potential anti-viral phytochemicals and traditional Chinese medicinal compounds was generated from our previously collected data and studies[13,21—23],and screened against the predicted SARS-CoV-2 3CLprostructure.Molecular operating environment(MOE)[24]was used for molecular docking,ligand-protein interaction and drug likeness analyses.All analyses were performed using the same protocols that are already described in our previous studies[13,25,26].The qualitative assessment of absorption,deposition,metabolism,excretion and toxicity(ADMET)profile of selected hits were predicted computationally by using ADMETsar server[27].
2.5.Molecular dynamics simulations
Explicit solvent molecular dynamics(MD)simulations were performed to verify docking results and to analyse the binding behaviour and stability of potential compounds using the predicted SARS-CoV-2 3CLprohomology model.GROMACS v5.1.4,GROMOS96 and the PRODRG server were employed to run 50 ns MD simulations[28,29]following the same protocol as described in our previous studies[13,30].
3.Results and discussion
3.1.Sequence and structural analyses
Multiple sequence alignment results revealed that 3CLprowas conserved,with 100% identity among all SARS-CoV-2 genomes.Next,the SARS-CoV-2 3CLproprotein sequence was compared with its closest homologs(Bat-CoV,SARS-CoV,MERS-CoV,Human-CoV and Bovine-CoV).The results revealed that SARS-CoV-2 3CLproclusters with bat SARS-like coronaviruses and shares 99.02% sequence identity(Fig.1A).Furthermore,it shares 96.08% ,87.00% ,90.00% and 90.00% sequence identity with SARS-CoV,MERS-CoV,Human-CoV and Bovine-CoV homologs,respectively(Fig.1B).These findings were consistent with those of initial studies reporting that SARS-CoV-2 is more similar to SARS-CoV than to MERS-CoV,and shares a common ancestor with bat coronaviruses[1,3,31].Analysis of physicochemical parameters revealed that the SARS-CoV-2 3CLpropolypeptide is 306 amino acids long with a molecular weight of 33,796.64 Da and a GRAVY score of-0.019,categorising the protein as a stable,hydrophilic molecule capable of establishing hydrogen bonds(Table 1).
Next,for comparative modelling,BLAST[32]search identified SARS-CoV 3CLpro(PDB ID:3M3V)as the best possible match in the PDB,with 100% query coverage,an E-value of 0.00,and 96.08% sequence identity.There were 12-point mutations(Val35Thr,Ser46Ala,Asn65Ser,Val86Leu,Lys88Arg,Ala94Ser,Phe134His,Asn180Lys,Val202Leu,Ser267Ala,Ser284Ala and Leu286Ala)between SARS-CoV and SARS-CoV-2 3CLproenzymes(Fig.S1).Except for replacement of Leu with Ala at position 286,all other replacements conserve polarity and hydrophobicity.However,these mutations may affect 3CLprostructure and function.Therefore,the 3D structure of SARS-CoV-2 3CLprowas predicted.Firstly,a single chain monomericmodelcomprisingalldomains(Domain I=residues 8—100;Domain II=residues 101—183;Domain III=residues 200—303)was built(Fig.S2).N-terminal amino acids 1 to 7 form the N-finger that plays a significant role in dimerization and formation of the active site of 3CLpro.Domains I and II,collectively referred to as the N-terminal domain,include an antiparallelβ-sheet structurewith 13β-strands.The binding site for the substrate is situated in a cleft between domains I and II.A loop from residues 184 to 199 joins the N-terminal domain and domain III,which is also called the C-terminal domain and comprises an antiparallel cluster of fiveα-helices.The overall molecular architecture of SARS-CoV-2 3CLprowas in consistent with the crystal structure of SARS-CoV(PDB ID:3M3V);the root mean square deviation(RMSD)between the homology model and the template was 0.629 ?.Structural and Ramachandran plot analyses revealed that 99% of residues are in favourable regions.
After quality assessment,individual chains were combined to form a homodimeric 3D structure,as shown in Fig.1C.To facilitate other researchers,the predicted 3D model has been submitted to the Protein Model Database(PMDB)[33],and anyone can download/use the SARS-CoV-2 3CLprofinal structure using PMDB ID:PM0082635.Furthermore,mutational analyses depicted none of the mutations affected the overall structure of SARS-CoV-2,which fully superimposed on the SARS-CoV 3CLprostructure(Fig.1D).The results also revealed that SARS-CoV-2 has a Cys-His catalytic dyad(Cys-145 and His-41),consistent with that of SARS 3CLpro(Cys-145 and His-41),TGEV 3CLpro(Cys-144 and His-41)and HCoV 3CLpro(Cys-144 and His-41)[34].These results revealed that the SARSCoV-2 3CLproreceptor-binding pocket conformation resembles that of the SARS-CoV 3CLprobinding pocket and raises the possibility that inhibitors intended for SARS-CoV 3CLpromay also inhibit the activity of SARS-CoV-2 3CLpro.
3.2.Molecular docking
To test this hypothesis,we docked(R)-N-(4-(tert-butyl)phenyl)-N-(2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl)furan-2-carboxamide),a potential noncovalent inhibitorof SARS-CoV 3CLpronamed ML188[35],with the SARS-CoV-2 3CLprohomology model.We also docked ML188 with the SARS-CoV 3CLprostructure(PDB ID:3M3V)as a reference,and ML188 bound strongly tothe receptor binding site of SARS-CoV 3CLpro.The inhibitor targets the Cys-His catalytic dyad(Cys-145 and His-41)along with the other residues,and the docking score(S=-12.27)was relatively high.However,surprisingly,ML188 did not show significant binding to the catalytic dyad(Cys-145 and His-41)of SARS-CoV-2,and the docking score(S=-8.31)was considerably lower(Fig.S3).These results indicated that the 12-point mutations identified at previous step may disrupt important hydrogen bonds and alter the receptor binding site,thereby affecting its ability to bind with the SARS-CoV inhibitors.
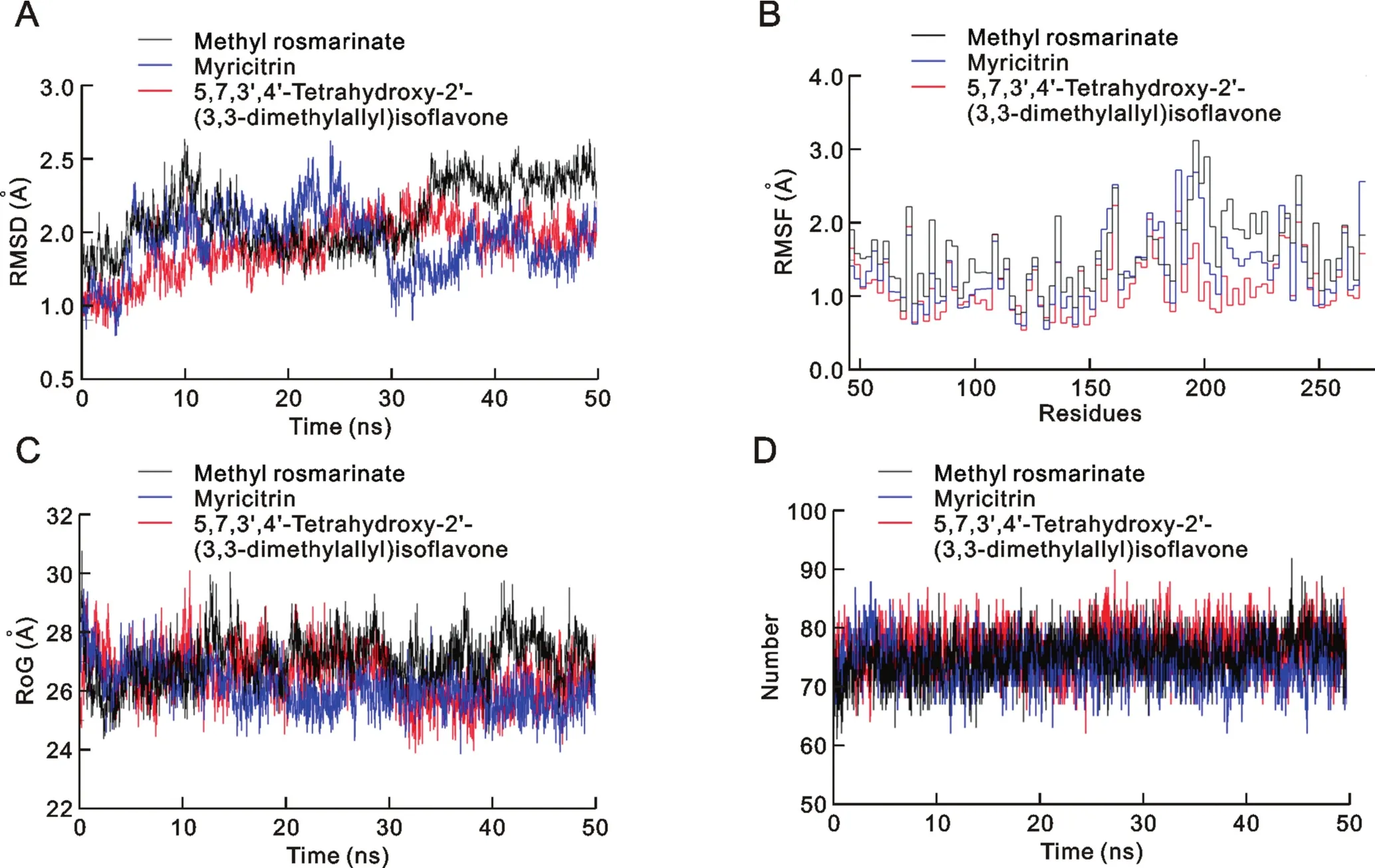
Fig.2.(A)Root mean square deviation(RMSD),(B)root mean squarefluctuation(RMSF),(C)potential energy and(D)Hydrogen Bond interactions for all three complexes over the 50 ns simulation.
Therefore,it is essential to discover novel compounds that may inhibit SARS-CoV-2 3CLproand serve as potential anti-COVID-19 drug compounds.We developed a library from our previously published studies that contains numerous natural compounds possessing potential anti-viral activities and screened it against the SARS-CoV-2 3CLprohomology model.Recent drug repurposing studies proposed few drugs that target SARS-CoV-2 3CLproand suggested that they could be used to treat COVID-19.Herein,we selected the best of these(Nel finavir,Prulifloxacin and Colistin)from three different drug repurposing studies[36,37]and docked them as controls in the present study(Fig.S4).Our analyses identified nine novel non-toxic,druggable natural compounds that are predicted to bind with the receptor binding site and catalytic dyad(Cys-145 and His-41)of SARS-CoV-2 3CLpro(Table 2;Fig.S5).ADMET profiling of the selected hits is given in Table S2.Among these screened phytochemicals,5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone is an iso flavone extracted from Psorothamnus arborescens[38]that exhibited the highest binding affinity(-29.57 kcal/mol)and docking score(S=-16.35),and formed strong hydrogen bonds with the catalytic dyad residues(Cys-145 and His-41)as well as significant interactions with the receptor-binding residues Thr24,Thr25,Thr26,Cys44,Thr45,Ser46,Met49,Asn142,Gly143,His164,Glu166 and Gln189(Fig.1E).A literature review revealed that 5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone has been successfully used as an antileishmanial agent[38],and it is also found in traditional Chinese medicine records[39].Our screened phytochemicals displayed higher docking scores,stronger binding energies,and closer interactions with the conserved catalytic dyad residues(Cys-145 and His-41)than Nel finavir,Prulifloxacin and Colistin.These results suggested that natural products identified in our study may prove more useful candidates for COVID-19 drug therapy.
3.3.MD simulations
To further investigate the molecular docking results,the top three phytochemical complexes,namely 5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone,myricitrin,and methyl rosmarinate,were subjected to 50 ns MD simulation.The root mean square deviation(RMSD),root mean squarefluctuation(RMSF),radius of gyration(RoG)and hydrogen bond parameters were calculated.RMSD is an indicator of the stability of ligand-protein complexes.None of the complexes showed any obvious fluctuations,and all three were stable,with average RMSD values of 1.6 ± 0.02 ?,1.5 ± 0.02 ? and 1.7 ± 0.02 ? for 5,7,3′,4′-tetrahydroxy-2'-(3,3-dimethylallyl)iso flavone,myricitrin,and methyl rosmarinate,respectively(Fig.2A).RMSF is an indicator of residual flexibility.Minimal fluctuations were observed for myricitrin and methyl rosmarinate,and the overallcomplexes remained stable throughout the simulations.The functionally important catalytic dyad residues(Cys-145 and His-41)displayed stable behaviour,and fluctuations were observed toward the C-terminal end of the SARSCoV-2 3CLpromolecule(Fig.2B).RoG is an indicator of protein compactness,stability,and folding,and the results suggested normal behaviour for all three complexes;all remained compact and stable throughout the 50 ns simulations(Fig.2C).In addition,hydrogen bonds,which are the main stabilising interactions factors in proteins,suggested thattheSARS-CoV-2 3CLprointernal hydrogen bonds remain stable throughout the simulation,with no obvious fluctuations(Fig.2D).These results confirmed our findings and further indicated that these compounds may serve as potential anti-COVID-19 drug sources.
4.Conclusion
In conclusion,our study revealed that 3CLprois conserved in SARS-CoV-2.It is highly similar to bat SARS-like coronavirus 3CLpro,with some differences from other beta-coronaviruses.We predicted the 3D structure of the SARS-CoV-2 3CLproenzyme,and the findings may help researchers working on COVID-19 drug discovery.Despite significant overall similarity with the SARS-CoV 3CLprostructure,the SARS-CoV-2 3CLprosubstrate binding site had some key differences,which highlighted the need for rapid drug discovery to address the alarming COVID-19 pandemic.Medicinal plant compounds have already been used to successfully treat numerous viral diseases.Herein,we screened a medicinal plant database containing 32,297 potential anti-viral phytochemicals and selected the top nine hits that may inhibit SARS-CoV-2 3CLproactivity and hence virus replication.Further in-vitro and in-vivo analyses are required to transform these potential inhibitors into clinical drugs.We anticipate that the insights gained in the present study may prove valuable for exploring and developing novel natural anti-COVID-19 therapeutic agents in the future.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China(2020YFC0845600),the Hubei Provincial Natural Science Foundation of China(2019CFA014),the Starting Research Grant for High-level Talents from Guangxi University,Nanning,China and Postdoctoral Research Platform Grant of Guangxi University,Nanning,China.We also acknowledge all the authors and laboratories mentioned in Table S1 for their sampling,analysis,and genome sequencing efforts.In addition,we acknowledge GISAID(https://www.gisaid.org/)for facilitating open data sharing.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.03.009.
 Journal of Pharmaceutical Analysis2020年4期
Journal of Pharmaceutical Analysis2020年4期
- Journal of Pharmaceutical Analysis的其它文章
- Comparative pharmacokinetics of six major compounds in normal and insomnia rats after oral administration of Ziziphi Spinosae Semen aqueous extract
- Electrochemical and in silico approaches for liver metabolic oxidation of antitumor-active triazoloacridinone C-1305☆
- Magnetic metal organic framework for pre-concentration of ampicillin from cow milk samples
- An integrated spectroscopic strategy to trace the geographical origins of emblic medicines:Application for the quality assessment of natural medicines
- TGA/Chemometrics addressing innovative preparation strategies for functionalized carbon nanotubes
- Identification of impurities in nafamostat mesylate using HPLC-ITTOF/MS:A series of double-charged ions
