Rapid mapping of candidate genes for cold tolerance in Oryza rufipogon Griff. by QTL-seq of seedlings
LUO Xiang-dong, LIU Jian, ZHAO Jun, DAI Liang-fang, CHEN Ya-ling, ZHANG Ling, ZHANG Fantao, HU Biao-lin, XIE Jian-kun
1 College of Life Science, Jiangxi Normal University, Nanchang 330022, P.R.China
2 Rice Research Institure, Jiangxi Academy of Agricultural Sciences, Nanchang 330200, P.R.China
1. Introduction
Rice (Oryza sativaL.) is one of the world’s most important crop plants because it is a staple food for more than 50% of the global population (Tanet al. 2007; Zhanget al. 2009).Cold stress is one of the major limiting factors affecting global rice production because it can lead to poor germination,slow growth, discoloration, withering, and anther injury in rice plants (Glaszmannet al. 1990). Around 15 million hectares of rice in 24 countries are under threat from cold stress, especially in Japan, Korea, and China (Zeng and Pu 2006; Louet al. 2007). Rice is very susceptible to cold stress at the seedling stage, which can seriously reduce rice yields. In temperate areas, rice cultivation usually involves transplanting seedlings from nurseries to paddy fields to protect seedlings from the effects of low temperature.However, this cultivation approach is expensive, timeconsuming, and labor-intensive. Consequently broadening our understanding of the genetic variation related to cold tolerance in rice as well as understanding the genetic basis and molecular mechanisms behind cold tolerance would facilitate development of unique rice varieties with high cold tolerance at seedling stage (CTSS).
CTSS in rice is a complex quantitative trait that is controlled by multiple genes (Andaya and Tai 2007). One of the traditional methods for genetic studies of quantitative traits is quantitative trait locus (QTL) mapping. QTL mapping provides the basis for map-based cloning of related genes and for marker-assisted selection (MAS) in crop breeding.At present, more than 90 QTLs for CTSS in rice have been identified by QTL mapping (Andaya and Mackill 2003;Kosekiet al. 2010; Kimet al. 2014; Xiaoet al. 2014; Zhanget al. 2014). Using a set of recombinant inbred lines (RILs)derived from a cross between M202 and IR50 (indicavarieties that are highly sensitive to cold stress), Andaya and Mackill (2003) mapped a major QTL (qCTS12a) on chromosome 12 that accounted for 41% of the phenotypic variation in seedling growth after cold stress. Zhanget al.(2005) detected 3 QTLs for cold tolerance at the early seedling stage on chromosomes 3, 7, and 11 using RILs derived from a cross between a tropicaljaponicacultivar and anindicacultivar. Louet al. (2007) identified a major QTL,qCTS-2, on chromosome 2 in doubled haploid (DH)lines that were derived from a cross between a cold tolerantjaponicavariety and a cold sensitiveindicacultivar. Using genome-wide association studies (GWAS), a total of 132 loci were identified at the seedling stage (Lüet al. 2016). Most of these QTLs analyses, however, were conducted inindicarice subspecies orjaponicasubspecies populations (Zhanget al. 2014), and most of the QTLs were located in the large physical intervals, making subsequent fine mapping difficult(Lüet al. 2016). Moreover, the major QTL of cold tolerance in cultivated rice is limited, and some genes for cold tolerance in cultivated rice have been lost because of longterm directional selection and domestication. Therefore, it is important to broaden the genetic variation in cultivated rice using related wild species. Accurate mapping and analysis of the genes for cold tolerance in wild rice might be helpful for cloning and using these genes to confer cold tolerance to cultivated rice.
Traditional QTL mapping is usually conducted by genotyping a large number of individuals in segregating populations derived from bi-parental crosses, which is labor-intensive, time-consuming, and sometimes costly(Lüet al. 2016). Bulked segregant analysis (BSA) can effectively and rapidly detect linked markers for target genes or QTLs from a couple of pooled DNA samples that are composed of two sets of individuals with distinct or opposite extreme phenotypes (Michelmoreet al. 1991).With the rapid development of next-generation sequencing(NGS) technologies, new strategies have been proposed to take advantage of the power of BSA and NGS-aided high-throughput genotyping, which have been used to identify major QTLs in yeast (Ehrenreichet al. 2010; Wengeret al. 2010; Swinnenet al. 2012),Arabidopsis thaliana(Schneebergeret al. 2009), rice (Oryza sativaL.) (Abeet al. 2012; Yanget al. 2013), and sunflower (Helianthus annuusL.) (Livajaet al. 2013). Recently, Takagiet al. (2013)successfully identified QTLs for partial resistance to fungal rice blast disease and seedling vigor using QTL-seq.
Dongxiang wild rice (Oryza rufipogonGriff., hereafter referred to as DWR) exists in the northernmost habitats in the world for common wild rice (O.rufipogon) populations,and belongs to one of the second classes of national protected plants (Chenet al. 2008; Xieet al. 2010). DWR possesses many genes that positively affect rice growth,including genes for strong cold tolerance (Liuet al. 2003;Luoet al. 2012), drought tolerance (Zhanget al. 2005; Xieet al. 2010), and fertility restoring gene for male sterility(Chenet al. 2008). Among these agronomic characteristics,cold tolerance is the most significant factor for improving rice production (Luoet al. 2012). DWR can overwinter successfully in Wuhan City, Hubei Province, China, where the minimum temperature reaches –12°C in winter (Liuet al. 2003), and because of this, DWR has become important for studying cold tolerance in rice. Kosekiet al.(2010) mapped a major QTL,qCtss11, to chromosome 12 for CTSS in a genetic mapping population derived from a cross between a cold-tolerant wild rice and a cold-sensitiveindicacultivar. Maoet al. (2015) identified at least 13 QTLs on chromosomes 2, 3, 7, 8, 9, 11, and 12 for seedling cold tolerance in DWR through a combination of interval mapping and single locus analyses in two genetic populations. So far, about 25 QTLs conferring cold-tolerance have been identified in DWR (Liuet al. 2003; Xiaet al. 2010; Maoet al. 2015). However, to date, the genetic and molecular mechanisms for cold tolerance in DWR are still unclear,resulting in limited applications of DWR in rice breeding programs.
In order to identify major or novel alleles conferring cold tolerance in wild rice, and ultimately to transfer these genes into cultivated rice (O.sativa), we developed a set of BC1F10backcross inbred lines (BILs) of 229 lines using cultivated rice as the recipient and DWR as the donor. In this study, the 229 previously constructed BILs were used to establish extremely cold resistant bulks (R-bulks) and cold sensitive bulks (S-bulks). Whole genome resequencing of R- and S-bulks was conducted to detect cold tolerance candidate genes in DWR at the early seedling stage using sequencing-based BSA with the QTL-seq method. The different expression patterns of candidate genes in DWR were further verified by quantitative real-time PCR (qRTPCR) analysis. Candidate genes were further verified using linked molecular markers. These results would be helpful for cloning and transferring genes for cold tolerance from common wild rice into cultivated rice.
2. Materials and methods
2.1. Plant materials
The mapping population consisted of 229 BILs derived from an inter-specific cross of Xieqingzao B//Xieqingzao B/DWR(Luoet al. 2012).O. sativassp.indicavar. Xieqingzao B(hereafter referred to as XB) is the maintainer line of the dwarf-abortive cytoplasmic male sterile line Xieqingzao A,and DWR is an accession ofO.rufipogonfrom Dongxiang County, Jiangxi Province, China. Cultivated rice XB (the recurrent parent) and common wild rice DWR (the donor parent) were used to produce F1hybrids. Hybrid F1plants were backcrossed to XB to create the BC1F1population.We consecutively selfed the BC1F1population to obtain the BC1F10RIL population using the single-seed descent method of plant breeding.
2.2. ldentification of extreme phenotypes for cold tolerance in BILs
About 50 seeds were selected from each of the BC1F10BILs.Seeds were stored at 50°C for 3 d to break dormancy. After 3 d, 0.6% sodium hypochlorite solution was used to surfacesterilize seeds for 15 min, seeds were rinsed 3 times with sterile distilled water, and were then placed equidistantly in 90 mm×10 mm glass Petri dishes with 20 mL distilled water in a chamber at 28°C for 3 d. Afterwards, germinated seeds with coleoptiles 8–12 mm in length were supplemented with MS solution (Murashige and Skoog 1962) and the solution was renewed every 7 d. At the 3 and a half leaves stage,weak and dead plants were removed and healthy, growing uniform seedlings were retained for cold treatment at 8°C for 7 d. After cold treatment, the temperature was adjusted back to 25°C to start the recovery process for 7 d. The seed survival percentage (SP) was used as indicator of cold tolerance. The survival percentage (%)=Number of surviving seedlings/Number of total seedlings×100. The experiment was conducted using a randomized complete block design with 3 replications. When the SP was more than 80%, the BIL individual was identified as extremely resistant to cold stress (hereafter referred to R). When the SP was lower than 20%, the BIL individual was identified as extremely sensitive to cold stress (hereafter referred to S). R and S individuals were selected to establish R- and S-bulks, respectively.
2.3. DNA extraction and lllumina sequencing
DNA from young leaf tissues of R-bulks (20 R individuals),S-bulks (20 S individuals), and their parents (DWR and XB)were extracted using the cetyltrimethyl ammonium bromide(CTAB) method (Porebskiet al. 1997). DNA pools were purified using RNA enzymes. The quality and concentration of DNA were examined by electrophoresis on 1% agarose gels with λDNA ladders. Tested and qualified DNA samples were sent to Novogene (Beijing, China) to build libraries with the Illumina TruSeq DNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA) from different DNA pools.Libraries were sequenced using an Illumina HiSeq 2500 sequencing platform. Raw data from the electropherogram acquired after sequencing were transformed by base calling from raw reads and data were stored in FASTQ format. Raw reads were subjected to a quality check using SeQC v2.1(Genotypic Proprietary Tool) (Mudalkaret al. 2014). Clean reads were obtained after error rate distribution checks, GC content distribution checks, and sequencing data filtering.
2.4. Analysis of next generation sequencing (NGS)data
Analysis of filtered clean reads from NGS data included the following four steps: (1) SNP-calling: Short, clean reads from R- and S-bulks were aligned to the DWR genome and to the reference genome of Nipponbare (japonica) in the Burrows-Wheeler Aligner (BWA ver. 0.7.10) (Li and Durbin 2009). SNP-calling was performed using SAM tools (Li and Durbin 2009). Low-quality SNPs with a base quality value<20 and a read depth<7 or those with >32× coverage were excluded because these SNPs may be false positives due to repetitive sequence, or due to errors in sequencing or alignment (Takagiet al. 2013). (2) Calculation of SNP-index and Δ(SNP-index): SNP-index and Δ(SNP-index) were calculated to identify candidate regions for cold tolerance QTLs (Abeet al. 2012; Takagiet al. 2013). The SNP-index is the proportion of reads harboring the SNP that are different from the reference sequence. If the SNP in all short reads from R- or S-bulks was the same as DWR, then the SNP-index was 0; if the SNP in all short reads in R- or S-bulks was the same as XB, then the SNP-index was 1. The Δ(SNP-index) was obtained by subtracting the SNP-index of the S-bulks from that of the R-bulks. (3) Constructing a graph of the SNP-index: An average SNP-index was computed over a 1-Mb interval using a 10-kb sliding window (Takagiet al. 2013). In order to visually reflect the distribution of offspring SNP-index on chromosomes, the distribution of SNP-index in chromosomes was graphed; theX-axis was the physical location of chromosomes and theY-axis was the average SNP-index. (4) Constructing a graph of the ΔSNP-index: The ΔSNP-index graph was created according to the method used for the SNP-index graph.
Statistical confidence intervals of the Δ(SNP-index) were calculated for all SNP positions with given read depths under the null hypothesis of no QTL, and plotted along with the Δ(SNP-index). For each read depth, 99 and 95% confidence intervals of the Δ(SNP-index) were obtained according to Takagiet al. (2013). When the Δ(SNP-index)=1, the locus of bulked DNA was entirely from the DWR genome; the Δ(SNP-index)=–1 suggested that the locus of bulked DNA was entirely from the XB genome; the Δ(SNP-index)=0 suggested that the parents had the same SNP-index at this genomic region; therefore, if the Δ(SNP-index) were closer to 1, it would be a prime candidate region for cold tolerance genes.
2.5. ldentification of candidate genes by real-time PCR
Candidate cold tolerance genes were verified by qRT-PCR according to our previous method (Daiet al. 2015). Total RNA of each sample (XB and DWR at 0 d and after 3 d of cold stress at 8°C) was isolated using TRIzol (Invitrogen, USA),and purified with DNase I (TaKaRa, Dalian, China). RNA was then extracted using phenol-chloroform, precipitated with 70% ethanol, dissolved in diethy pyrocarbonate(DEPC)-treated ddH2O, and then reverse transcribed to cDNA using the First-Strand Synthesis of cDNA Kit(Promega, Peking, China) according to the manufacturer’s instructions. Specific primers used in subsequent qRT-PCR were designed according to the coding sequence (CDS)sequences of candidate genes, and are listed in Table 1. RTPCR amplification was performed in 20 μL reaction volumes,which contained SYBR?PremixEx TaqII (2×), 10 μmol L–1forward primers, 10 μmol L–1reverse primers, and ROX reference dye (50×). The housekeeping geneActinwas used as an internal control to normalize the concentration of mRNA present in each sample. Each sample was measured 3 times and each time have 3 duplications. qRT-PCR was performed using a StepOnePlusTMReal-Time PCR System as follows: initial denaturation at 95°C for 30 s, 40 cycles at 95°C for 5 s, 60°C for 30 s, 60°C for 1 min, and melt curve stage at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s.
2.6. Verification of candidate regions
In order to verify the candidate regions identified by QTL-seq,insertion-deletion (InDel) makers were designed based on the sequence of Nipponbare (japonica) andindiciarice 93-11 insertions or deletions using Premier 5.0. The Nipponbare related sequences were download from the Gramene website, then to findindicarice 93-11 sequence on the http://www.ncbi.nlm.nih.gov/website, through the analysis to find the sequence insertions or deletions (generally vary more than 4 bp) between Nipponbare and 93-11. Polymorphic InDel markers between DWR and XB were then used for PCR amplification and electrophoretic analysis in BILs (including R and S individuals). Amplification was performed with a thermocycler (PTC-200; MJ Research Inc., Chatham, New Jersey, USA) using the following procedure: samples were denatured at 94°C for 4 min, then 35 cycles at 94°C for 60 s,55°C for 30 s, 72°C for 60 s, and the last cycle had an extra 8 min extension period at 72°C. PCR products were separated by electrophoresis using 4% agarose gels. In the case of null alleles or novel bands in BILs, PCR amplification was repeated and failed PCR reactions were excluded.
3. Results
3.1. ldentification of extreme phenotypes for cold tolerance in BILs
Cold resistance in 229 BILs and their parents was evaluated based on SP after cold stress and recovery. The cold experiment was conducted 3 times to ensure the correct phenotype of each BIL individual. The donor parent DWR had higher cold tolerance than the recurrent parent XB.For the donor parent DWR, the value of SP is 100% and for the recurrent parent XB, the value of SP is 8.78%. The SP values in BIL individuals had clear differences, rangingfrom 100 to 0%, with an average value of 29.21% (Fig. 1).Statistical analysis showed that the frequency of the SP value had a skew normal distribution, which agrees with our previous results (Luoet al. 2016).

Table 1 Sequences of quantitative real-time PCR (qRT-PCR) primers
According to the SP, 20 BIL individuals consistently showed high resistance to cold in 3 cold stress experiments,with SP values>80%. The 20 cold resistant BILs were used as R-bulks. Another set of 20 BILs consistently showed high susceptibility to cold, with SP<20%; these individuals were used as S-bulks (Fig. 2). Genomic DNA of R individuals was bulked in an equal ratio to generate R-bulks DNA, and that of S individuals was bulked similarly to generate S-bulks DNA.
3.2. Quality evaluation using Illumina sequencing
The results of Illumina sequencing and related data analyses are shown in Table 2. Using high-throughput sequencing for R- and S-bulks, a total of 21.805 Gb clean data were obtained, 10 933.824 Mb from R-bulks (Q20≥94.3%;GC content, 45.06%) and 11 153.471 Mb from S-bulks(Q20≥94.47%; GC content, 42.77%) (Table 2). For the donor parent DWR, the total amount of clean data were 25 414.545 Mb (Q20≥96.36%; GC content, 43.05%). For the recurrent parent XB, the total amount of clean data were 24 561.463 Mb (Q20≥97.12%, GC content, 42.76%)(Table 2). After trimming and filtering, over 95% of the clean reads were selected and aligned to the reference genome.The contrast ratio of all genome samples were between 95.74 and 96.82%; the average depth of sequencing for the four samples (excluding the N area) was between 27.2 to 59.97×. 1× coverage, the percent of at least one base covered loci of reference genome in the whole genome,was more than 96.95%; and 4× coverage, the percent of at least four bases covered loci of reference genome in the whole genome, was more than 90.27% (Table 2). The Illumina sequencing data for each sample were enough, the sequencing quality was qualified, and the GC distribution was normal. These results suggested that Illumina sequencing was successful and met the criteria for mutation detection and analysis.

Fig. 1 The frequency distribution of survival percent (SP) in backcross inbred lines (BILs) population after cold stress. XB,Xieqingzao B; DWR, Dongxiang wild rice.

Fig. 2 Seedlings vigor of cold resistant bulks (R, left) and cold sensitive bulks (S, right) after cold stress and recovering.

Table 2 The quality and depth of sequencing data1)
3.3. Variance analysis of progeny SNP frequency
Clean reads of R- and S-bulks were aligned to the DWR and reference genomes using BWA (Li and Durbin 2009), and the SNP-index of the two bulks were calculated and analyzed. A total of 1 290 297 SNPs were obtained by filtering raw SNPs.SNP-index and Δ(SNP-index) were calculated according to Abeet al. (2012) and Takagiet al. (2013), respectively.Graphs of the SNP-index and the Δ(SNP-index) that reflect their distributions on chromosomes were created (Fig. 3).
We selected 99 and 95% confidence levels as screening thresholds to analyze the significant difference area of the SNP-index for the two bulks. After 1 000 permutations,a SNP-index locus of approximately 1 for S-bulks and a SNP-index locus of approximately 0 for R-bulks (i.e., the Δ(SNP-index) was nearly to 1) were selected, these areas according with the characteristics may be associated with cold tolerance QTL loci.
A total of 2 333 SNPs loci were obtained at the 99%confidence level in intervals of 3.58–7.01 Mb and 14.00–19.88 Mb on chromosome 12, (Fig. 3-D). ANNOVAR annotation results found 2 333 candidate polymorphic markers on chromosome 12. Among them, there are 24 non-synonymous mutations involved in 16 differential genes,one of them contain terminator mutation. In order to detect more potential differential genes, the 95% confidence level was used as a screening threshold. On chromosomes 1, 3,4, 6, 7, 8, 9, 10, and 12, an additional 276 SNPs were found between R- and S-bulks, which were involved in other 42 differential genes. Based on the above results, there were 58 potential candidate genes in the DWR genome, including 2 genes on chromosome 1; 2 genes on chromosome 3; 2 genes on chromosome 4; 3 genes on chromosome 6; 2 genes on chromosome 7; 4 genes on chromosome 8; 2 genes on chromosome 9; 1 gene on chromosome 10; and 40 genes on chromosome 12. The expression patterns of these potential candidate genes for cold tolerance were further verified by RT-PCR.
3.4. Expression analysis of candidate genes
The results of qRT-PCR analysis showed that 5 out of 58 potential candidate genes were significantly different between DWR and XB after cold stress (Fig. 4). There was no significant difference in the relative expression of the 5 candidate genes between DWR and XB at a normal temperature condition. Of the 5 genes, the expression levels of 4 genes (DX-CTS-08-01,DX-CTS-12-14,DX-CTS-12-22,andDX-CTS-12-38) were significantly increased in DWR after cold stress (Fig. 4). The expression levels of the 4 genes were not changed in XB after cold stress. These results indicated that the 4 genes may be inherent cold tolerance-related genes, because the induced expression of these genes was more noticeable in DWR. For the 5th candidate gene (DX-CTS-09-01), the opposite expression pattern was observed: The expression level was significantly increased in XB after cold stress (Fig. 4), which suggested that the gene might play a negative role in regulating cold tolerance by indirectly inhibiting the expression of some cold-tolerance genes.

Fig. 3 QTL-seq analysis on R (resistant)- and S (susceptible)-bulks for identification of quantitative trait loci (QTLs) conferring cold tolerance in Dongxiang wild rice (DWR). A, single nucleotide polymorphism (SNP)-index plots of R-bulks. B, single nucleotide polymorphism SNP-index plots of S-bulks (top). C, ?(SNP-index) graph of all chromosomes. D, ?(SNP-index) graph of chromosome 12.R1 and R2 are the regions of candidate genes for cold tolerance in DWR. Statistical confidence intervals under the null hypothesis of no QTL (blue, P<0.05; pink, P<0.01).

Fig. 4 Real-time PCR analysis of 5 candidate genes in Oryza sativacv. Xieqing zao B (XB) and Oryza rufipogon Griff. (Dongxiang wild rice, DWR). Bars mean SE.
3.5. Functional annotation and verification of candidate regions
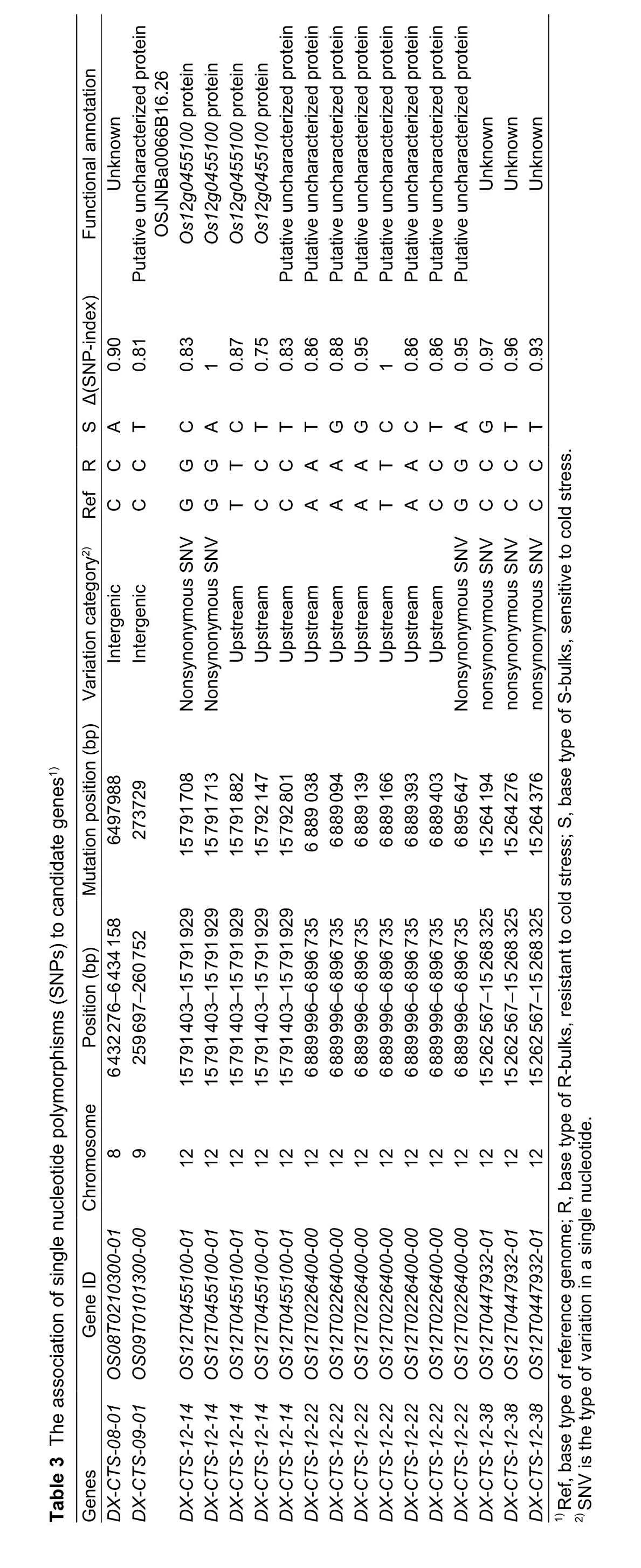
Structural variation and functional annotation of the 5 verified candidate genes based on QTL-seq analysis are shown in Table 3. Three candidate genes were on chromosome 12, and the other 2 genes were on chromosomes 8 and 9. The types of variation among these candidate genes were intergenic, nonsynonymous, and upstream mutations.Several mutations were detected in the candidate genes on chromosome 12. All 5 candidate genes were previously unknown or uncharacterized.
Further analysis showed that the physical position ofDXCTS-08-01was between base pairs 6 432 276 and 6 434 158 on chromosome 8, andDX-CTS-09-01was located between base pairs 259 697 and 260 752 on chromosome 9. The other 3 candidate genes (DX-CTS-12-14,DX-CTS-12-22andDX-CTS-12-38) were located between base pairs 6 889 996 and 15 791 929 on chromosome 12. Four out offive of these candidate genes (except forDX-CTS-09-01on chromosome 9) were located in the cold tolerance QTL interval found in our previous study (Luoet al. 2016).
In order to verify the candidate regions of cold tolerance in DWR, 30 pairs of InDel markers were developed in the target QTL intervals on chromosomes 8 and 12. Out of the 30 InDel markers, 3 showed clear polymorphisms between the parents (DWR and XB). The 3 polymorphic markers were used to genotype the 229 BILs, including the R and S individuals (Fig. 5). The result showed that the band pattern of most R individuals (16 out of 20 R individuals) amplified by InDel 12-7 and 12-16 was similar to that of DWR. InDel markers 12-7 and 12-16 were located in R1 and R2 intervals,respectively (Fig. 3-D), which suggested that InDel markers 12-7 and 12-16 linked with genes for cold tolerance on chromosome 12 in DWR. These data further confirmed that the candidate genes identified in the present study were valid. Based on these results, the candidate region of cold tolerance genes should be between 5.5 and 7.5 Mb(R1 interval) and between 15 and 17.5 Mb (R2 interval) on chromosome 12 (Fig. 3-D).
4. Discussion
CTSS is a very important agronomic trait in rice.Understanding the genetic mechanism of CTSS and identifying genes related to cold tolerance are key steps for improving cold tolerance of rice. Although numbers of QTLs conferring cold tolerance in rice were detected by traditional QTL mapping, the QTLs located in the large physical intervals made subsequent fine mapping and illuminating the regulation network of CTSS difficult (Xiaoet al. 2014;Lüet al. 2016). In this study, the QTL-seq method was employed (Takagiet al. 2013) to identify cold tolerancerelated genes in DWR using a BC1F10population. This method took advantage of high-throughput whole genome resequencing and BSA. In addition, use of the SNP-index allowed accurate quantitative evaluation of the frequencies of parental alleles as well as the genomic contribution from the two parents to offspring. These features of QTL-seq mean that it is a rapid and efficient way to identify genomic regions harboring the major QTL of a target gene. QTL-seq only needs two DNA bulks of progeny with extreme phenotypic variation, meaning that it is cheaper, and takes less time and energy than traditional QTL mapping. A total of 58 candidate genes were selected in a preliminary sequencing analysis;these genes were distributed on chromosomes 1, 3, 4,6, 7, 8, 9, 10, and 12. Our results have greatly narrowed the scope for exploring cold tolerance related genes in DWR, and have provided useful information for further identification and cloning of cold tolerance related genes in a wild rice species.
Previous studies indicated that cold tolerance in rice is a quantitative trait controlled by complex genetic factors. Previously, researchers identified more than 90 QTLs for cold tolerance on all 12 chromosomes using different bi-parental mapping populations and evaluation indexes; these QTLs explained 2.7–49.30%of the phenotypic variation in response to cold (Hanet al. 2007; Kosekiet al. 2010; Xiaoet al. 2014, Zhanget al. 2014; Yanget al. 2015). Out of these QTLs, onlyqCTS12(Andaya and Tai 2006),Ctss11(Kosekiet al.2010),qSCT11(Kimet al. 2014),qLOP2, andqPSR2-1(Xiaoet al. 2015) were finely mapped. Recently, a regulator of G-protein signaling (COLD1) conferring cold tolerance in rice was cloned. The quantitative trait locusCOLD1of chromosome 4 has been proven to be important for plant adaptation (Maet al. 2015).In this study, QTL-seq analysis and molecular marker verification confirmed that the cold tolerance candidate genes of DWR were mainly located in the 3.58–7.01 Mb interval and the 14.00–19.88 Mb interval of chromosome 12, in the 3.18–8.43 Mb interval of chromosome 8,and in the 21.16–22.32 Mb interval of chromosome 9(Fig. 3-D). A total of 5 cold tolerance genes were identified from 58 potential candidate genes through qRT-PCR analysis. In addition, the structural variation and functional annotation of the 5 candidate genes have been analyzed. These reliable and accurate information is very important for further investigation.
?
Compared with the physical positions of related QTLs for seedling cold tolerance in rice, 4 out 5 candidate genes (except forDX-CTS-09-01on chromosome 9)detected in this study were located in a known QTL interval.DX-CTS-09-01(located between base pairs 259 697 and 260 752 on chromosome 9) was very far away from previously identified QTLs on chromosome 9 and might be a new regulatory factor from XB. In the 3.0 to 8.0 Mb interval of chromosome 8, 4 QTLs of seedling cold tolerance were detected in previous studies,including a QTL for germination cold tolerance and a QTL for reproductive stage cold tolerance (Yanget al. 2015). In our previous study, one QTL was also detected in the RM407 to RM152 interval on chromosome 8 (Luoet al. 2016).These results suggested that the position ofDX-CTS-08-01found in this study was close to previously identified cold tolerance QTLs, which might mean it is a specific and reliable tolerance-related gene. On chromosome 12, there were several cold tolerance QTLs detected in a previous study,such as a QTL for budding cold tolerance (14.00–18.00 Mb) and 3 QTLs for seedling cold tolerance (3.00–7.00 Mb)(Yanget al. 2015), includingqCTS12.2(Maoet al. 2015) andqCTS12(Andaya and Tai 2006). Recently, we also detected 4 QTLs on chromosome 12 by traditional QTL mapping(Luoet al. 2016). Comparative analysis suggested that the physical position of the candidate regions (DX-CTS-12-14,DX-CTS-12-22, andDX-CTS-12-38) agreed with our pervious results. In or near the candidate regions, some QTLs for cold tolerance genes in rice have been detected, such asqCTS12(Andaya and Tai 2006),qCTS12.2(Maoet al. 2015),qLTG-12(Liet al. 2013), andqCT-12-1(Zhuet al. 2015).In the present study, 2 molecular markers that were tightly linked with cold tolerance candidate genes in DWR were also identified. Therefore, the candidate regions (R1 and R2 of chromosome 12, Fig. 3-D) are worthy of attention. Our results are potentially very helpful for isolating and identifying major genes for cold tolerance in DWR, illuminating the genetic basis and molecular mechanisms of DWR in low-temperature environments. In addition, our research provides data that are useful for developing elite introgression lines or unique rice varieties containing positive QTLsviaMAS based on the information of QTL analysis for cold-tolerance.
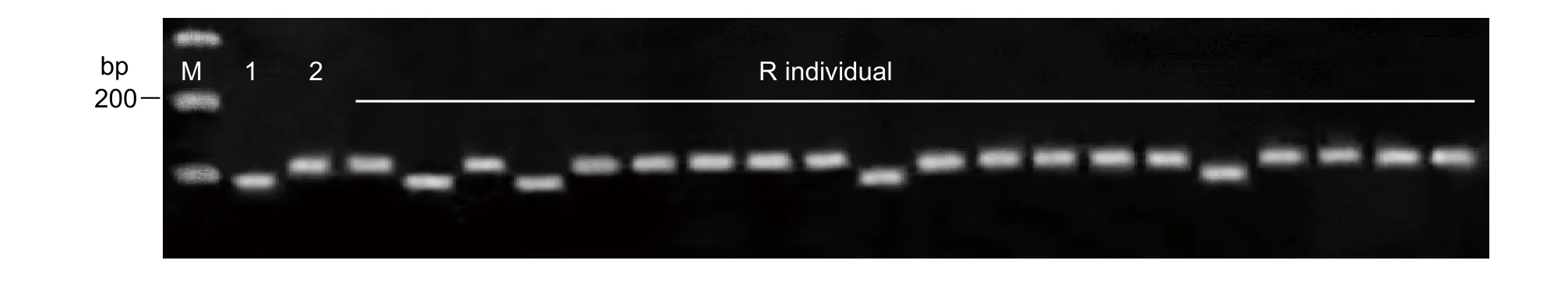
Fig. 5 PCR products of some BC1F10 lines and their parents amplified by primer insertion-deletion (InDel) marker 12-7. M,marker; 1, Oryza sativa L. (Xieqingzao B), 2, Oryza rufipogon (Dongxiang wild rice, DWR). R individual, cold resistant individual of backcross inbred line (BIL).
5. Conclusion
Using high-throughput sequencing-based bulked segregant analysis of QTL-seq method for the extremely resistant (R)and susceptible (S) bulks of BILs and their parents, a total of 2 609 candidate SNPs were obtained, including 58 potential candidate genes for cold tolerance. The variation types of these candidate genes were intergenic, nonsynonymous,and upstream mutations. Quantitative RT-PCR analysis revealed that 5 out of 58 potential candidate genes had significantly different expression betweenO.sativaandO.rufipogon. All 5 candidate genes were unknown or uncharacterized genes. Two InDel markers (12-7 and 12-16) were tightly linked with the candidate genes on chromosome 12 in DWR. These results are helpful for cloning and using cold tolerance genes from common wild rice in cultivated rice.
Acknowledgements
This research was partially supported by the National Natural Science Foundation of China (31260255, 31360147 and 31660384), the Natural Science Foundation of Jiangxi Province, China (20151BAB204008), the Scientific Planning Project of Jiangxi Provincial Education Department, China(GJJ12184 and KJLD12059), and the Major Projects in Jiangxi Province, China (20161ACF60022).
Abe A S, Kosugi K, Yoshida S, Natsume H, Takagi H, Kanzaki H, Matsumura K, Yoshida C, Mitsuoka M, Tamiru H, Innan L, Cano S, Kamoun R, Terauchi. 2012. Genome sequencing reveals agronomically-important loci in rice from mutant populations.Nature Biotechnology, 30, 174–178.
Andaya V C, Mackill D J. 2003. Mapping of QTLs associated with cold tolerance during the vegetative stage in rice.Journal of Experimental Botany, 54, 2579–2585.
Andaya V C, Tai T H. 2006. Fine mapping of theqCTS12locus,a major QTL for seedling cold tolerance in rice.Theoretical and Applied Genetics, 113, 467–475.
Andaya V C, Tai T H. 2007. Fine mapping of theqCTS4locus associated with seedling cold tolerance in rice (Oryza sativaL.).MolecularBreeding, 20, 349–358.
Chen X R, Yang K S, Fu J R, Zhu C L, Peng X S, He X P, He H H. 2008. Identification and genetic analysis of fertility restoration ability in Dongxiang wild rice (Oryza rufipogon).Rice Science, 15, 21–28.
Dai L F, Chen Y L, Luo X D, Wen X F, Cui F L, Zhang F T, Zhou Y, Xie J K. 2015. Level and pattern of DNA methylation changes in rice cold tolerance introgression lines derived fromOryza rufipogonGriff.Euphytica, 205, 73–83.
Ehrenreich M I, Torabi N, Jia Y, Kent J, Martis S, Shapiro J A, Gresham D, Caudy A A, Kruglyak L. 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants.Nature, 446, 1039–1042.
Glaszmann J C, Kaw R N, Khush G S. 1990. Genetic divergence among cold tolerant rices (Oryza sativa).Euphytica, 45,95–104.
Han L Z, Qiao Y L, Zhang S Y, Gao G L, Kim J H, Lee K, Koh H. 2007. Identification of quantitative trait loci for cold response of seedling vigor traits in rice.Journal of Genetics and Genomics, 34, 239–246.
Kim S M, Suh J P, Lee C K, Lee J H, Kim Y G, Jena K K. 2014.QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativaL.).Molecular Genetics and Genomics,289, 333–343.
Koseki M, Kitazawa N, Yonebayashi S, Maehara Y, Wang Z X,Minobe Y. 2010. Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage.Molecular Genetics and Genomics, 284, 45–54.
Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform.Bioinformatics, 25,1754–1760.
Li L F, Liu X, Xie K, Wang Y H, Liu F, Lin Q Y, Wang W Y, Yang C Y, Lu B Y, Liu S J, Chen L M, Jiang L, Wan J M. 2013.qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativaL.).Theoretical and Applied Genetics, 126, 2313–2322.
Liu F X, Sun C Q, Tan L B, Fu Y C, Li D J, Wang X K. 2003.Identification of QTL for cold tolerance at booting andflowering stage in Jiangxi Dongxiang wild rice.Chinese Science Bulletin, 48, 1864–1867. (in Chinese)
Livaja M, Wang Y, Wieckhorst S, Haseneyer G, Seidel M, Hahn V, Knapp S J, Taudien S, Sch?n C C, Bauer E. 2013. BSTA:A targeted approach combines bulked segregant analysis with nextgeneration sequencing andde novotranscriptome assembly for SNP discovery in sunflower.BMC Genomics,14, 628.
Lou Q J, Chen L, Sun Z X. 2007. A major QTL associated with cold tolerance at seedling stage in rice (Oryza sativaL.).Euphytica, 158, 87–94.
Lü Y, Guo Z L, Li X K, Ye H Y, Li X H, Xiong L Z. 2016. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis.Plant Cell and Environment, 39, 556–570.
Luo X D, Dai L F, Cao J F, Jian S R, Chen Y L, Hu B L, Xie J K. 2012. Identification and molecular cytology analysis of cold tolerance introgression lines derived fromOryza sativaL. mating withO.rufipogonGriff.Euphytica, 187, 461–469.
Luo X D, Zhao J, Dai L F, Zhang F T, Zhou Y, Wan Y, Xie J K.2016. Linkage map construction and QTL mapping for cold tolerance inOryza rufipogonGriff. at early seedling stage.Journal of Integrative Agriculture, 15, 2703–2711.
Ma Y, Dai X Y, Xu Y Y, Luo W, Zheng X M, Zeng D L, Pan Y J, Lin X L, Liu H H, Zhang D J, Xiao J, Guo X Y, Xu S J,Niu Y D, Jin J B, Zhang H, Xu X, Li L G, Wang W, Qian Q,et al. 2015.COLD1confers chilling tolerance in rice.Cell,160, 1209–1221.
Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, Zhang D, Chen C. 2015. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat.Theoretical and Applied Genetics, 128, 1359–1371.
Michelmore R W, Paran I, Kesseli R V. 1991 Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations.Proceedings of the National Academy of Sciences of the United States of America, 88, 9828–9832.
Mudalkar S, Golla R, Ghatty S, Reddy A R. 2014.De novotranscriptome analysis of an imminent biofuel crop,Camelina sativaL. using Illumina GAIIX sequencing platform and identification of SSR markers.Plant Molecular Biology, 84, 159–171.
Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures.Physiologia Plantarum, 15, 473–497.
Porebski S, Bailey L G, Bernard R. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components.Plant Molecular Biology Reporter, 15, 8–15.
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen A H, Nielsen K L, Jorgensen J E, Weigel D, Andersen S U.2009. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing.Nature Methods, 6,550–551.
Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G,Yang Y D, Demeke M, Foulquié-Moreno M R, Goovaerts A, Souvereyns K, Clement L, Dumortier F, Thevelein J M.2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis.Genome Research, 22, 975–984.
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H L,Cano M, Kamoun S, Terauchi R. 2013. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations.The Plant Journal, 74, 174–183.
Tan L B, Liu F X, W Xue, Wang G J, Ye S, Zhu Z F, Fu Y C,Wang X K, Sun C Q. 2007. Development ofOryza rufipogonandO.sativaintrogression lines and assessment for yieldrelated quantitative trait loci.Journal of Integrative Plant Biology, 49, 871–884.
Wenger W J, Schwartz K, Sherlock G. 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene fromSaccharomyces cerevisiae.PLos Genetics, 6, e1000942.
Xiao N, Huang W N, Li A H, Gao Y, Li Y H, Pan C H, Ji H J,Zhang X X. 2015. Fine mapping of theqLOP2andqPSR2-1loci associated with chilling stress tolerance of wild rice seedlings.Theoretical and Applied Genetics, 128, 173–185.
Xiao N, Huang W N, Zhang X X, Gao Y, Li A L, Dai Y, Yu L,Liu G Q, Pan C H, Li Y H, Dai Z Y, Chen J M. 2014. Fine mapping ofqRC10-2, a quantitative trait locus for cold tolerance of rice roots at seedling and mature stages.PLoS ONE, 9, e96046.
Xie J K, Hu B L, Wan Y, Zhang T, Li X, Liu R N, Huang Y H, Dai L F, Luo X D. 2010. Comparison of the drought resistance characters at seedling stage between Dongxiang Common Wild rice (Oryza rufipogonGriff.) and cultivars (Oryza sativaL.).Acta Eclolgica Sinica, 30, 1665–1674. (in Chinese)
Yang T F, Zhang S H, Zhao J L, Huang Z H, Zhang G Q, Liu B.2015. Meta-analysis of QTLs underlying cold tolerance in Rice (Oryza sativaL.).Molecular Plant Breeding, 13, 1–15.
Yang Z M, Huang D Q, Tang W Q, Zheng Y, Liang K J,Cutler A J, Wu W R. 2013. Mapping of quantitative trait loci underlying cold tolerance in rice seedlingsviahighthroughput sequencing of pooled extremes.PLoS ONE,8, e68433.
Zeng Y W, Pu X Y. 2006. Genetic analysis for cold tolerance at booting stage forjaponicarice (Oryza sativaL.).Indlan Journal of Genetics and Plant Breeding, 66, 100–102.
Zhang Q, Chen Q H, Wang S L, Hong Y H, Wang Z L. 2014. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci.Rice, 7, 1–12.
Zhang Y S, Luo L J, Liu T M, Xu C G,Xing Y Z. 2009. Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds.Theoretical and Applied Genetics,118, 1035–1044.
Zhang Z H, Su L, Li W, Chen W, Zhu Y G. 2005. A major QTL conferring cold tolerance at the early seedling stage using recombinant inbred lines of rice (Oryza sativaL.).Plant Science, 168, 527–534.
Zhu Y, Chen K, Mi X, Chen T, Ali J, Ye G, Xu J L, Li Z K.2015. Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice.PLoS ONE,10, e0145704.
 Journal of Integrative Agriculture2018年2期
Journal of Integrative Agriculture2018年2期
- Journal of Integrative Agriculture的其它文章
- Experimental infectivity of Theilerialuwenshuni and Theileria uilenbergi in Chinese Kunming mice
- Development of a sensitive and reliable droplet digital PCR assay for the detection of ‘Candidatus Liberibacter asiaticus’
- One size fits all? Contract farming among broiler producers in China
- Designing price-contingent vegetable rotation schedules using agent-based simulation
- The effects of aeration and irrigation regimes on soil CO2 and N2O emissions in a greenhouse tomato production system
- The efficiency of long-term straw return to sequester organic carbon in Northeast China’s cropland
