基于機(jī)器學(xué)習(xí)方法的丙型肝炎病毒非結(jié)構(gòu)蛋白5B聚合酶抑制劑活性預(yù)測(cè)
呂 巍 薛 英
(1山東農(nóng)業(yè)大學(xué)生命科學(xué)學(xué)院,作物生物學(xué)國(guó)家重點(diǎn)實(shí)驗(yàn)室,山東泰安271018;2四川大學(xué)化學(xué)學(xué)院,教育部綠色化學(xué)與技術(shù)重點(diǎn)實(shí)驗(yàn)室,成都610064;3四川大學(xué)生物治療國(guó)家重點(diǎn)實(shí)驗(yàn)室,成都610041)
基于機(jī)器學(xué)習(xí)方法的丙型肝炎病毒非結(jié)構(gòu)蛋白5B聚合酶抑制劑活性預(yù)測(cè)
呂 巍1薛 英2,3,*
(1山東農(nóng)業(yè)大學(xué)生命科學(xué)學(xué)院,作物生物學(xué)國(guó)家重點(diǎn)實(shí)驗(yàn)室,山東泰安271018;2四川大學(xué)化學(xué)學(xué)院,教育部綠色化學(xué)與技術(shù)重點(diǎn)實(shí)驗(yàn)室,成都610064;3四川大學(xué)生物治療國(guó)家重點(diǎn)實(shí)驗(yàn)室,成都610041)
在丙型肝炎病毒(HCV)的基因復(fù)制和蛋白質(zhì)成熟的過程中,非結(jié)構(gòu)蛋白5B(NS5B)作為RNA依賴的RNA聚合酶起到了重要的作用.抑制NS5B聚合酶可以阻止丙型肝炎病毒的RNA復(fù)制,因此成為一種治療丙型肝炎的有效方法.通過計(jì)算機(jī)方法進(jìn)行虛擬篩選和預(yù)測(cè)NS5B聚合酶抑制劑已經(jīng)變得越來越重要.本文主要采用機(jī)器學(xué)習(xí)方法(支持向量機(jī)(SVM)、k-最近相鄰法(k-NN)和C4.5決策樹(C4.5 DT))對(duì)已知的丙型肝炎病毒NS5B蛋白酶抑制劑與非抑制劑建立分類預(yù)測(cè)模型.1248個(gè)結(jié)構(gòu)多樣性化合物(552個(gè)NS5B抑制劑與696個(gè)非NS5B抑制劑)被用于測(cè)試分類預(yù)測(cè)系統(tǒng),并用遞歸變量消除法選擇與NS5B抑制劑相關(guān)的性質(zhì)描述符以提高預(yù)測(cè)精度.獨(dú)立驗(yàn)證集的總預(yù)測(cè)精度為84.1%-85.0%,NS5B抑制劑的預(yù)測(cè)精度為81.4%-91.7%,非NS5B抑制劑的預(yù)測(cè)精度為78.2%-87.2%.其中支持向量機(jī)給出最好的NS5B抑制劑預(yù)測(cè)精度(91.7%);C4.5決策樹給出最好的非NS5B抑制劑預(yù)測(cè)精度(87.2%);k-最近相鄰法給出最好的總預(yù)測(cè)精度(85.0%).研究表明機(jī)器學(xué)習(xí)方法可以有效預(yù)測(cè)未知數(shù)據(jù)集中潛在的NS5B抑制劑,并有助于發(fā)現(xiàn)與其相關(guān)的分子描述符.
機(jī)器學(xué)習(xí)方法;分子描述符;遞歸變量消除法; 支持向量機(jī);丙型肝炎病毒
1 Introduction
Hepatitis C virus(HCV)is a positive strand RNA virus of the Flaviviridae family.1The capsid and envelope are composed of four structural proteins and there are five non-structural proteins which play important roles in protein maturation and gene replication.HCV is responsible for a variety of clinical conditions ranging from acute viral hepatitis to chronic liver disease and cirrhosis.2It is the major cause of liver cancer and about two thirds of all liver transplants are a result of HCV infection.Since the HCV cDNA was cloned successfully by Choo et al.in 1989,3there have been many researches about the genome,function of protein,and biological function of HCV.In 2000,according to the World Health Organization survey,there are estimated 170 million people worldwide chronically infected by the HCV and an estimated 3-4 million new infections annually.4Currently,there is no effective vaccine to prevent hepatitis.Therefore,drug development for the treatment of HCV has become a hot spot for many scientists.The current standard of care for HCV,which is based on a combination of Interferon(IFN)and Ribavarin,can cure HCV infections,but is often inadequate.5In addition,there will be serious side effects and a low success rate in the main viral genotype. The need for improved therapies is pressing because although the incidence of new HCV infections is declining,mortality is expected to increase into the middle of the next decade.6
The lack of a highly effective and safe treatment option for the HCV highlights the necessity of developing more efficient means of combating and curing this viral disease ultimately.A primary focus is currently on finding new inhibitors of the NS5B polymerase.As we all know,NS5B polymerase has been identified as an RNA dependent RNA polymerase,which can be essential for replication since it both affects the synthesis of a(-)-stranded HCV RNA template and regenerates the (+)-stranded genomic RNA.7Inhibiting NS5B polymerase will prevent the RNA replication,so it has a significant effect on the treatment of HCV.In recent years,some X-ray crystallographic structures of NS5B have been determined and their resolutions are more accurate too,such as the structures(PDB ID: 3mwv,3mww,3gyn,etc.).8,9The modulation of NS5B inhibitors has become more explicit10and more databases of commercially available compounds were generated.In this situation, the area of NS5B inhibitor predicted by the non-structurebased computational methods has developed too.
The machine learning(ML)methods are very important computational methods and efficacious tools in the virtual screening and the computer-aided drug design.They have been applied in drug pharmacodynamics,pharmacokinetics,and toxicology,11-13with much achievement.Recursive feature elimination(RFE)method,14,15which has been extensively used in the feature selecting,is employed in this research for selecting the most relevant molecular descriptors to NS5B inhibitory liability.To assess the prediction accuracy of the models in this research,two different evaluation methods have been employed which are valid for estimating drug prediction models.One is five-fold cross validation16and the other is evaluation by an independent validation set.
In this paper,we use ML methods,such as support vector machines(SVM),k-nearest neighbor(k-NN),and C4.5 decision tree(C4.5 DT),to study NS5BIs and non-NS5BIs for developing a fast and cost-efficient tool for facilitating NS5BIs prediction and design,and we employ the RFE method to select the descriptors which are most relative to the discrimination of NS5BIs and non-NS5BIs for improving the accuracy of the prediction models.
2 Methodology
2.1 Selection of NS5BIs and non-NS5BIs
Atotal of 1313 compounds about NS5B with known IC50values were selected from a number of published papers9,17-70(See Supporting Information:Table S1).Based on the tested experimental data in prevenient researches,35when the IC50value is lower than 300 nmol·L-1,the molecule has good activity.And in the other papers,24,56it was indicated that when the IC50value is between 400 and 600 nmol·L-1,the molecule has inhibitory potential too,but not strong.For example,the compounds(07-1-20 and 07-1-21)have the activity of inhibiting NS5B and their IC50values are 200 and 300 nmol·L-1,respectively.35The compound(08-9-4c)with IC50value of 440 nmol·L-1can inhibit NS5B activity,however,methylation of the R3 sulfonamide moiety present in 08-9-4b(27 nmol·L-1)resulted in significant loss of NS5B inhibition properties.56The phenyl ethyl amide compounds(05-3-43 and 05-3-44)with substituents at R3 with Cl and F are little potential at inhibiting NS5B and the IC50values are 500 and 600 nmol·L-1,respectively.24The compound (09-15-11f)with the IC50value of 685 nmol·L-1is a weaker NS5B inhibitor.61For compounds(09-12-4n and 09-12-4s) with the IC50values of 780 and 730 nmol·L-1,respectively,the inhibiting activity for NS5B is very weak because of an αbranched in the R1 moiety.58In this work,based on experimental data mentioned above,one method was applied to assign compounds as NS5BIs and non-NS5BIs.71In all molecules collected,we can divide them into three sets based on the IC50values of these molecules.One set includes 552 inhibitors(IC50≤400 nmol·L-1),the second set includes 696 non-inhibitors (IC50≥600 nmol·L-1).The last set includes 65 molecules(400 nmol·L-1<IC50<600 nmol·L-1)which are ambiguous between inhibitors and non-inhibitors.In these three sets,we only choose the first two sets to test.
The two-dimensional(2D)structure of each of the compounds was generated by using ChemDraw72and was subsequently converted into three-dimensional(3D)structure by using Corina73for calculating the quantum chemical properties. The 3D structure of each compound was manually inspected to ensure that the chirality of each chiral agent is properly generated.All the generated geometrics have been fully optimized without symmetry restrictions.
Firstly,all compounds were divided into training set,testing set,and independent validation set according to their distributions in the chemical space defined by their structural and chemical features.74Compounds of similar structural and chemical features were evenly assigned into separate sets.For those compounds without enough structurally and chemically similar counterparts,they were assigned,in order of priority,to the training set and then the testing set,respectively.The ID of compound in every subset is supplied in Table S2 in Supporting Information.The training set was used by SVM to develop a statistical model.The testing set was used by SVM to optimize the parameters of SVM classification algorithm,and the independent validation set was used for assessing the classification accuracy of the model.Then,all compounds in training set and testing set were randomly divided into five subsets of approximately equal size.After training the SVM with a collection of four subsets,the performance of the SVM was tested against the fifth subset.This process was repeated five times, so that every subset was once used as the test data.
2.2 Molecular descriptors
Molecular descriptors were used to routinely represent quantitatively structural and physicochemical properties of molecules,which have been extensively applied in the structureactivity relationship(SAR),14quantitative structure-activity relationship(QSAR),75and other computational researches of pharmaceutical agents.76,77In this work,a total of 198 molecular descriptors listed in Table S3 in Supporting Information were used,which were selected from more than 1000 descriptors described in the literature by eliminating those descriptors that are obviously redundant.78The resulting 198 molecular descriptors include 18 descriptors in the class of simple molecular properties,27 descriptors in the class of molecular connectivity and shape,97 descriptors in the class of electro-topological state,31 descriptors in the class of quantum chemical properties,and 25 descriptors in the class of geometrical properties. They were computed from the 3D structure of each compound by using the molecular descriptor computing program.79The irrelevant and redundant descriptors to NS5BIs and non-NS5BIs were further eliminated by using feature selection method.11,80
2.3 Feature selection method
In a dataset with a fixed number of samples,excessive descriptors may cause a predicting model to be over-fitted to affect its precision.Therefore,feature selection methods have become increasingly prevalent.It is good at enhancing the performance of ML methods by eliminating the molecular descriptors which are redundant and irrelevant to the discrimination of different datasets.12Recursive feature elimination(RFE),one of the feature selection methods,has been widely acknowledged because of its high efficacy manifested in discovering informative feature molecular descriptors most relevant to the cancer classification,13prediction of P-glycoprotein substrates,81prediction of tetrahymena pyriformis toxicity chemicals,12and the drug activity analysis.11RFE with SVM is used to reduce the other redundant and unrelated descriptors.For a fixed parameter σ,in the first step,the SVM builds a model with the complete set of descriptors.In the second step,the contribution of the descriptors is ranked in the datasets based on a criterion score which is calculated by a scoring function.In the third step,the m ranked lowest descriptors are washed out.Finally, the SVM classifier is retrained by using the remaining descriptors,and the corresponding prediction accuracy is computed by means of five-fold cross validation.All the four steps are then repeated for other σ until all descriptors have been removed. After the completion of these procedures,the set of descriptors and parameter σ which give the best prediction accuracy are selected.
The choice of the parameter m affects the performance of SVM as well as the speed of feature selection.To control the size of the selected descriptors,we only consider the number of descriptors smaller than one-fifth of the whole descriptors.78Our earlier studies suggested that the performance of a SVM system with m=5 is only reduced by a few percentages smaller than that with m=1,which is consistent with the findings from other studies.15In this work,m=5 is used for the sake of computational efficiency.
2.4 Machine learning methods
There are a number of downloadable ML methods software packages.For example,PHAKISO(http://www.phakiso.com/ index.htm)and WEKA(http://www.cs.waikato.ae.nz/~ml/weka)for a collection of ML methods software,82NeuNet(http:// www.cormactech.com/neunet/index.html)for neural network, SVM-Light(http://svmlight.joachims.org)for SVM software were used in many researches.We used our in-house program to build SVM model83for predicting the compounds from NS5BIs and non-NS5BIs.And we also use the other ML methods to predict them,for example,k-NN84and C4.5.85Then the results calculated by these ML methods are compared.
2.5 Performance evaluation
As in the case of all discriminative methods,86the performance of ML methods can be measured by the quantity of true positives(TP),true negatives(TN),false positives(FP),and false negatives(FN),which are the number of NS5BIs predicted as NS5BIs,non-NS5BIs predicted as non-NS5BIs,non-NS5BIs predicted as NS5BIs,NS5BIs predicted as non-NS5BIs,respectively.There are several accuracy functions for measuring prediction performance,which include sensitivity SE=(TP/(TP+FN))×100%(prediction accuracy for NS5BIs), specificity SP=(TN/(TP+FN))×100%(prediction accuracy for non-NS5BIs),the overall prediction accuracy(Q),and Matthews correlation coefficient(C)are given by Eq.(1)and Eq.(2),respectively.
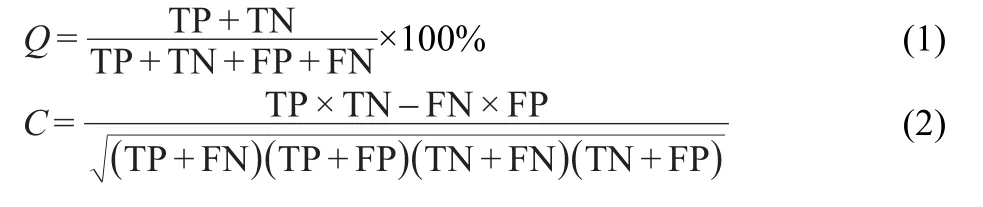
3 Results and discussion
3.1 Overall prediction accuracies and merit of the machine learning methods
SVM prediction of NS5BIs is evaluated by the method of 5-fold cross validation.Through comparing the accuracies of SVM,which used 5-fold cross validation with and without theuse of RFE of feature selection method,we find that the feature selection method plays an important role in the performance of SVM for the prediction of NS5BIs and non-NS5BIs. The results are listed in Table 1.Through this method,we find 24 descriptors which are critical for SVM model.The 24 descriptors are listed in Table 2.The accuracies of SVM with RFE are 81.8%for NS5BIs and 81.8%for non-NS5BIs;and the accuracies of SVM without RFE are 90.2%for NS5BIs and 53.1%for non-NS5BIs.The average accuracies with and without RFE are 82.0%and 69.8%,respectively.It obviously indicates that the method with RFE is substantially better than that derived from SVM without RFE,especially for NS5BIs.This suggests that RFE is useful in selecting the proper set of molecular descriptors for the prediction of NS5BIs.The results show that the selection of appropriate molecular descriptors is important for the improvement of average prediction accuracy,but more important for implying which pharmacological features are more propitious to distinguish NS5BIs and non-NS5BIs.
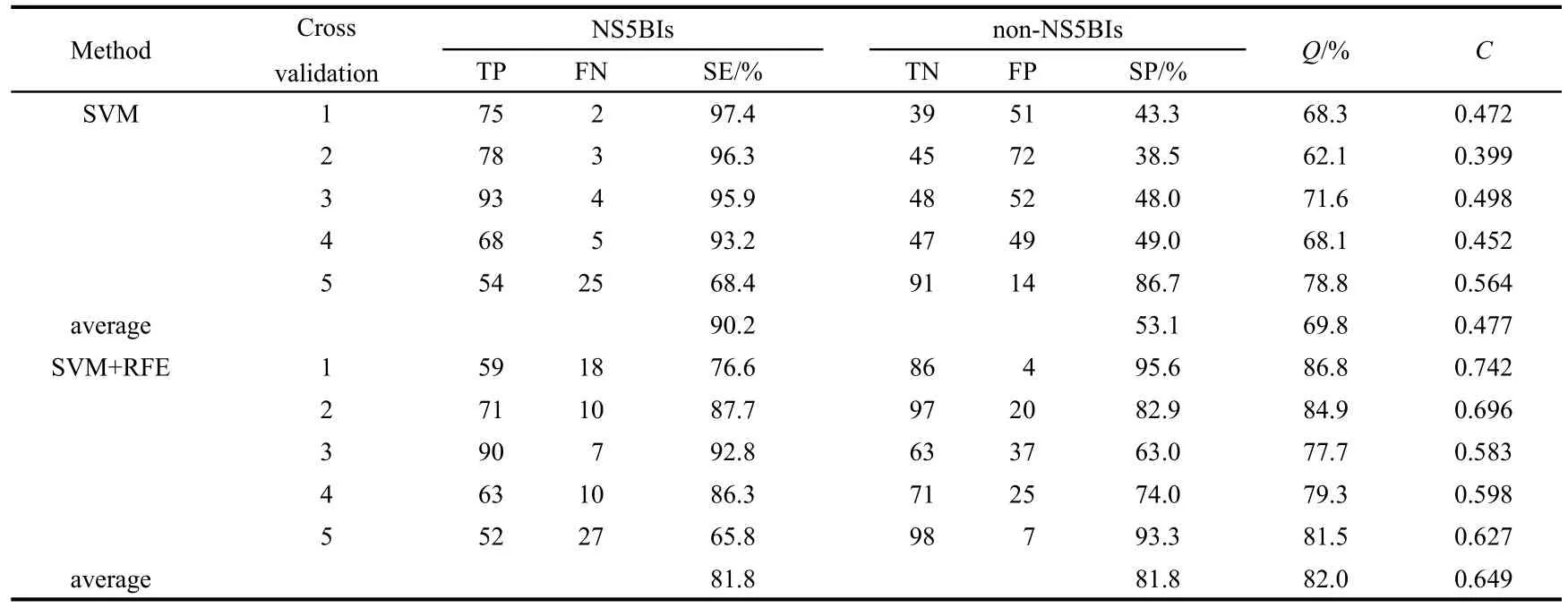
Table 1 Accuracies of NS5BIs and non-NS5BIs derived from SVM without and with the use of the RFE method (SVM+RFE)by using five-fold cross validation

Table 2 The 24 molecular descriptors selected from the RFE method for the classification of NS5BIs and non-NS5BIs
Table 3 gives the prediction accuracies of NS5BIs and non-NS5BIs derived from other two machine learning methods(k-NN and C4.5 DT)by using the RFE selected descriptors and five-fold cross validation method.For comparison,those results from SVM are also labeled in Table 3.By comparing the prediction accuracies from the three methods,we have obtained several results.For NS5BIs,the accuracies of these methods are in the range of 81.4%-91.7%with SVM givingthe best accuracy at 91.7%.For non-NS5BIs,the accuracies are in the range of 78.2%-87.2%with C4.5 DT giving the best accuracy at 87.2%.Lastly,for both NS5BIs and non-NS5BIs, the average accuracies are in the range of 84.1%-85.0%with k-NN giving the best accuracy at 85.0%,C4.5 DT giving the second best accuracy at 84.7%and SVM giving the worst accuracy at 84.1%.
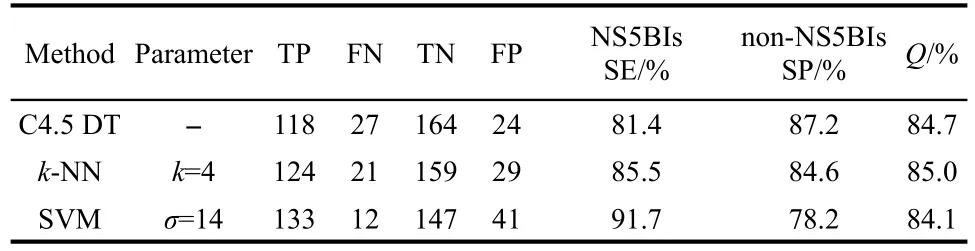
Table 3 Comparison of the prediction accuracies of NS5BIs and non-NS5BIs derived from different machine learning methods by using independent validation sets
A frequently used method for checking whether a prediction system is over-fitting is to compare the prediction accuracies determined by using cross validation methods and independent validation sets.Since descriptor selection is performed by using the cross validation method as the modeling testing sets,an over-fitted classification system is expected to have much higher prediction accuracy for the cross validation sets than that for the independent validation sets.As shown in Table 1 and Table 3,the predication accuracies of the SVM systems based on the five-fold cross validation method and those based on independent validation sets are similar.This shows that the SVM classi-fication systems in this work are unlikely over-fitted.
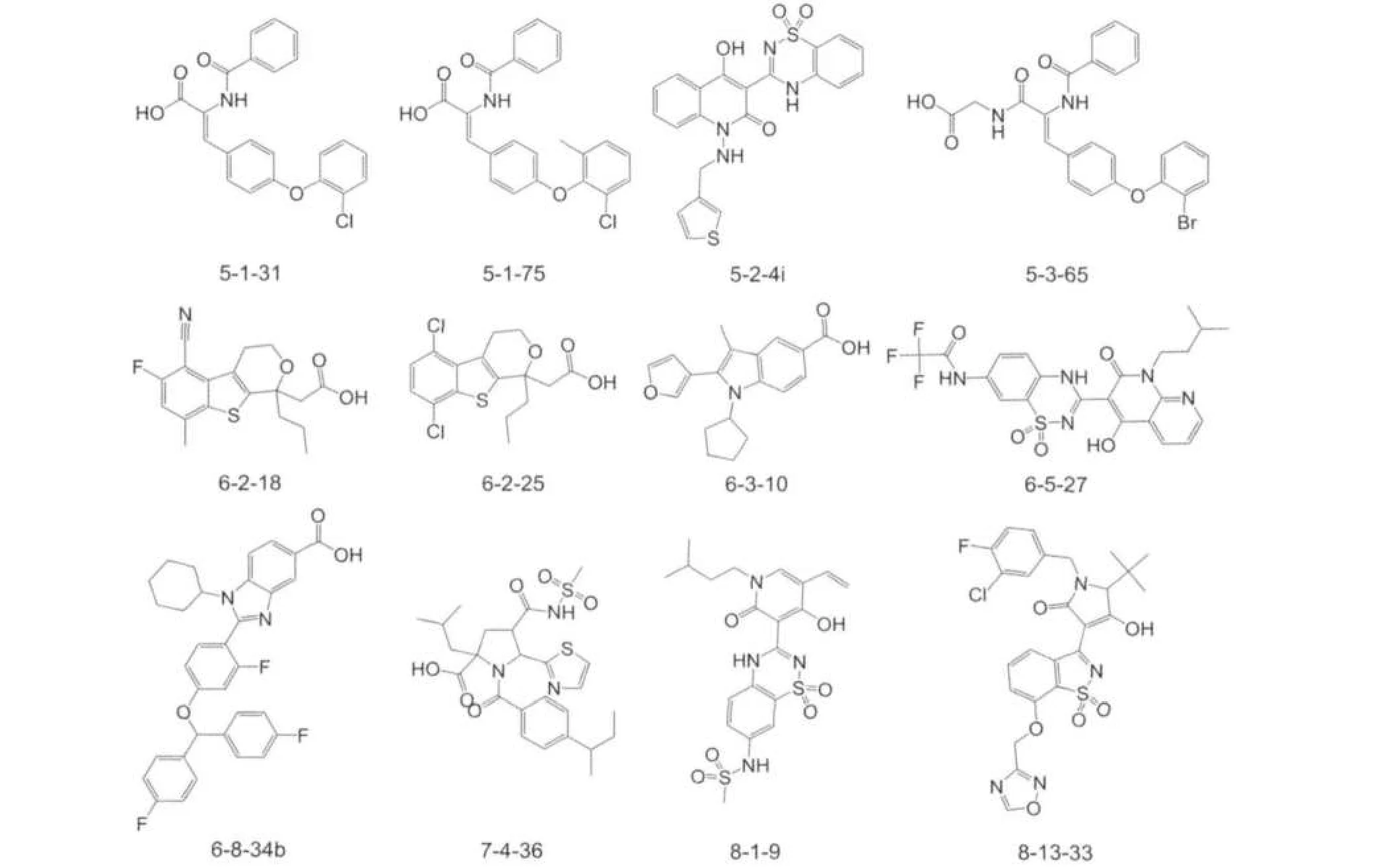
Fig.1 Structures of the part of misclassified NS5BIs
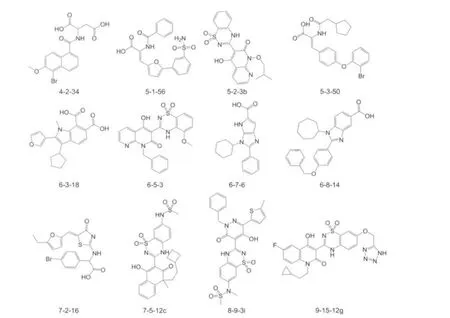
Fig.2 Structures of the part of misclassified non-NS5BIs
Overall,our study suggests that ML methods are useful for facilitating the prediction of novel NS5BIs from compounds with diverse structures.Another advantage of the SVM studied in this work is that they do not require the knowledge about the molecular mechanism or structure-activity relationship of a particular drug property.
3.2 Molecular descriptors associated with the diversity between NS5BIs and non-NS5BIs
Selecting molecular descriptors which are most relevant to the prediction of NS5BIs is important for optimizing the prediction models and for elucidating the molecular factors contributing to NS5BIs.Commonly,QSAR models particularly design a group of specific descriptors to represent the studied NS5BIs which have similar structural groups or structural alerts.34In this research,a total of 24 molecular descriptors are selected by RFE.These descriptors,given in Table 2,represent the structural and physicochemical properties associated with the diversity between NS5BIs and non-NS5BIs.All of them are found to match or partially match those descriptors used in the published NS5BIs QSAR models.34The physicochemical properties,such as steric,electrostatic,hydrophobic,hydrogen bond acceptor,and hydrogen bond donor,are incorporated in the comparative molecular similarity indices analysis(CoMSIA)and comparative molecular field analysis(CoMFA)methods for the studies of NS5B polymerase inhibitors.34In our work,the descriptors selected by RFE method are the same as the results in other researches.34For example,topological state descriptors including S(1),S(5),S(8),S(13),S(25),S(28), S(76)and Tbmdd are selected which are related with steric property;χen(electronegativity index),QH,Max(most positive charge on H atoms),QC,Max(most positive charge on C atoms), QN,SS(sum of squares of charges on N atoms),QO,SS(sum of squares of charges on O atoms),Rpc(relative positive charge), Rnc(relative negative charge),Svpc(sum of van der Waals surface areas of positively charged atoms)and Svpcw(sum of charge weighted van der Waals surface areas of positively charged atoms)are related to electrostatic;and the descriptors of Shpb(hydrophobic region)and Hiwpb(hydrophobic integy moment)are selected to descript the hydrophobic property in our work.In addition,Srivastava et al.87constructed the QSAR model of NS5BIs with several molecular descriptors including electronegativity(χeq)which is selected as χen(electronegativity index)in our work.
3.3 Misclassified compounds in the independent validation set
There are 53 molecules incorrectly classified by our SVM system with the independent validation set method.The predic-tion accuracy is 91.7%for NS5BIs,78.2%for non-NS5BIs, and 84.1%for all of them.And for NS5BIs set,which is comprised of 145 molecules,there are 12 molecules which are predicted to non-NS5BIs,on the other hand,for non-NS5BIs set, which is comprised of 188 molecules,there are 41 molecules which are predicted to NS5BIs.All of these misclassified molecules are shown in Fig.1,Fig.2 and Fig.S1 in Supporting Information.From these figures,we can see that the misclassified agents are mainly the compounds with multiple and dense rings.It suggests that using current molecular descriptors may not be sufficient to properly show the molecular features.So it implies that further improvement and refinement of our molecular descriptors may be needed.
4 Conclusions
This study shows that machine learning methods,especially SVM,are useful for facilitating the prediction of NS5BIs without the knowledge of mechanisms but only with the choice of specific molecular descriptors.However,the current ML methods are limited in their ability to facilitate the study of the mechanism of predicted properties.Nevertheless,we believe in the near future,this weakness may be partially overcome by the development of regression-based ML methods.In addition, our study indicates that prediction accuracy of this model is affected by the molecular descriptors selected by RFE which can further help to optimally select molecular descriptors.To conclude,the availability of more extensive information about various NS5BIs and associated mechanisms will facilitate the development of machine learning methods into practical tools for the prediction of different types of NS5BIs in the early stage of drug development.
Supporting Information Available:The information of the investigated dataset is provided in Tables S1,S2,S3 and Fig.S1. This information is available free of charge via the internet at http://www.whxb.pku.edu.cn.
(1) Bréchot,C.Digest.Dis.Sci.1996,41,6S.
(2) Hoofnagle,J.H.Hepatology 1997,26,15S.
(3) Choo,Q.L.;Kuo,G.;Weiner,A.J.;Overby,L.R.;Bradley,D. W.;Houghton,M.Science 1989,244,359.
(4)World Health Organization(WHO)Hepatitis C Fact Sheet No. 164,Rev,October,2000.
(5)Cornberg,M.;Wedemeyer,H.;Manns,M.P.Curr. Gastroenterol.Rep.2002,4,23.
(6) Garber,K.;Arbor,A.Nat.Biotechnol.2007,25,1379.
(7) Appel,N.;Schaller,T.;Penin,F.;Bartenschlager,R.J.Biol. Chem.2006,281,9833.
(8) Laplante,S.R.;Gillard,J.R.;Jakalian,A.;Aubry,N.; Coulombe,R.;Brochu,C.;Tsantrizos,Y.S.;Poirier,M.; Kukolj,G.;Beaulieu,P.L.J.Am.Chem.Soc.2010,132,15204.
(9) Ellis,D.A.;Blazel,J.K.;Tran,C.V.;Ruebsam,F.;Murphy,D. E.;Li,L.S.;Zhao,J.;Zhou,Y.;McGuire,H.M.;Xiang,A.X.; Webber,S.E.;Zhao,Q.;Han,Q.;Kissinger,C.R.;Lardy,M.; Gobbi,A.;Showalter,R.E.;Shah,A.M.;Tsan,M.;Patel,R.A.; LeBrun,L.A.;Kamran,R.;Bartkowski,D.M.;Nolan,T.G.; Norris,D.A.;Sergeeva,M.V.;Kirkovsky,L.Bioorg.Med. Chem.Lett.2009,19,6047.
(10)Biswal,B.K.;Wang,M.;Cherney,M.M.;Chan,L.; Yannopoulos,C.G.;Bilimoria,D.;Bedard,J.;James,M.N.G. J.Mol.Biol.2006,361,33.
(11) Xue,Y.;Li,Z.R.;Yap,C.W.;Sun,L.Z.;Chen,X.;Chen,Y.Z. J.Chem.Inf.Comput.Sci.2004,44,1630.
(12)Xue,Y.;Li,H.;Ung,C.Y.;Yap,C.W.;Chen,Y.Z.Chem.Res. Toxicol.2006,19,1030.
(13)Lin,H.H.;Han,L.Y.;Yap,C.W.;Xue,Y.;Liu,X.H.;Zhu,F.; Chen,Y.Z.J.Mol.Graph.Model.2007,26,505.
(14)Yu,H.;Yang,J.;Wang,W.;Han,J.Proc.IEEE Comput.Soc. Bioinformatics Conf.2003,220.
(15) Furlanello,C.;Serafini,M.;Merler,S.;Jurman,G.Neural Networks 2003,16,641.
(16) Trotter,M.W.B.;Holden,S.QSAR Comb.Sci.2003,22,533.
(17) Pace,P.;Nizi,E.;Pacini,B.;Pesci,S.;Matassa,V.;De-Francesco R.;Altamura S.;Summa V.Bioorg.Med.Chem.Lett.2004,14, 3257.
(18) Gopalsamy,A.;Lim,K.;Ellingboe,J.W.;Krishnamurthy,G.; Orlowski,M.;Feld,B.;van Zeijlb,M.;Howe,A.Y.M.Bioorg. Med.Chem.Lett.2004,14,4221.
(19) Stansfield,I.;Avolio,S.;Colarusso,S.;Gennari,N.;Narjes,F.; Pacini,B.;Ponzi,S.;Harper,S.Bioorg.Med.Chem.Lett.2004, 14,5085.
(20) Chan,L.;Pereira,O.;Reddy,T.G.;Das,S.K.;Poisson,C.; Courchesne,M.;Proulx,M.L.;Siddiqui,A.;Yannopoulos,C. G.;Nguyen-Ba,N.;Roy,C.;Nasturica,D.;Moinet,C.;Bethell, R.;Hamel,M.;Heureux,L.L.;David,M.;Nicolas,O.; Courtemanche-Asselin,P.;Brunette,S.;Bilimoria,D.;Bédard, J.Bioorg.Med.Chem.Lett.2004,14,797.
(21) Chan,L.;Das,S.K.;Reddy,T.G.;Poisson,C.;Proulx,M.L.; Pereira,O.;Courchesne,M.;Roy,C.;Wang,W.Y.;Siddiqui,A.; Yannopoulos,C.G.;Nguyen-Ba,N.;Labrecque,D.;Bethell,R.; Hamel,M.;Courtemanche-Asselin,P.;Heureux,L.L.;David, M.;Nicolas,O.;Brunette,S.;Bilimoria,D.;Bédard,J.Bioorg. Med.Chem.Lett.2004,14,793.
(22) Pfefferkorn,J.A.;Greene,M.L.;Nugent,R.A.;Gross,R.G.; Mitchell,M.A.;Finzel,B.C.;Harris,M.S.;Wells,P.A.; Shelly,G.A.;Anstadt,R.A.;Kilkuskie,B.E.;Koptab,L.A.; Schwendea,F.J.Bioorg.Med.Chem.Lett.2005,15,2481.
(23) Pratt,J.K.;Donner,P.;McDaniel,K.F.;Maring,C.J.;Kati,W. M.;Mo,H.M.;Middleton,T.;Liu,Y.Y.;Ng,T.;Xie,Q.H.; Zhang,R.;Montgomery,D.;Molla,A.;Kempf,D.J.; Kohlbrenner,W.Bioorg.Med.Chem.Lett.2005,15,1577.
(24) Pfefferkorn,J.A.;Nugent,R.;Gross,R.J.;Greene,M.; Mitchell,M.A.;Reding,M.T.;Funk,L.A.;Anderson,R.; Wells,P.A.;Shelly,J.A.;Anstadt,R.;Finzel,B.C.;Harris,M. S.;Kilkuskie,R.E.;Koptab,L.A.;Schwendea,F.J.Bioorg. Med.Chem.Lett.2005,15,2812.
(25)Shipps,G.W.;Deng,Y.Q.;Wang,T.;Popovici-Muller,J.; Curran,P.J.;Rosner,K.E.;Cooper,A.B.;Girijavallabhan,V.; Butkiewiczb,N.;Cableb,M.Bioorg.Med.Chem.Lett.2005, 15,115.
(26) LaPorte,M.G.;Lessen,T.A.;Leister,L.;Cebzanov,D.; Amparo,E.;Faust,C.;Ortlip,D.;Bailey,T.R.;Nitz,T.J.; Chunduru,S.K.;Young,D.C.;Burns,J.C.Bioorg.Med. Chem.Lett.2006,16,100.
(27) Gopalsamy,A.;Aplasca,A.;Ciszewski,G.;Park,K.;Ellingboe, J.W.;Orlowski,M.;Feldb,B.;Howeb,A.Y.M.Bioorg.Med. Chem.Lett.2006,16,457.
(28) Beaulieu,P.L.;Gillard,J.;Bykowski,D.;Brochu,C.; Dansereau,N.;Duceppe,J.S.;Haché,B.;Jakalian,A.;Lagacé, L.;LaPlante,S.;McKercher,G.;Moreau,E.;Perreault,S.P.; Stammers,T.;Thauvette,L.;Warrington,J.;Kukolj,G.Bioorg. Med.Chem.Lett.2006,16,4987.
(29)Krueger,A.C.;Madigan,D.L.;Jiang,W.W.;Kati,W.M.;Liu, D.C.;Liu,Y.Y.;Maring,C.J.;Masse,S.;McDaniel,K.F.; Middleton,T.;Mo,H.M.;Molla,A.;Montgomery,D.;Pratt,J. K.;Rockway,T.W.;Zhang,R.;Kempf,D.J.Bioorg.Med. Chem.Lett.2006,16,3367.
(30) Rockway,T.W.;Zhang,R.;Liu,D.C.;Betebenner,D.A.; McDaniel,K.F.;Pratt,J.K.;Beno,D.;Montgomery,D.;Jiang, W.W.;Masse,S.;Kati,W.M.;Middleton,T.;Molla,A.; Maring,C.J.;Kempf,D.J.Bioorg.Med.Chem.Lett.2006,16, 3833.
(31) Gopalsamy,A.;Shi,M.X.;Ciszewski,G.;Park,K.;Ellingboe, J.W.;Orlowski,M.;Feldb,B.;Howeb,A.Y.M.Bioorg.Med. Chem.Lett.2006,16,2532.
(32) Ontoria,J.M.;Hernando,J.I.M.;Malancona,S.;Attenni,B.; Stansfield,I.;Conte,I.;Ercolani,C.;Habermann,J.;Ponzi,S.; Filippo,M.D.;Koch,U.;Rowley,M.;Narjes,F.Bioorg.Med. Chem.Lett.2006,16,4026.
(33) Ishida,T.;Suzuki,T.;Hirashima,S.;Mizutani,K.;Yoshida,A.; Ando,J.;Ikeda,S.;Adachic,T.;Hashimotoa,H.Bioorg.Med. Chem.Lett.2006,16,1859.
(34) Li,H.;Tatlock,J.;Linton,A.;Gonzalez,J.;Borchardt,A.; Dragovich,P.;Jewell,T.;Prins,T.;Zhou,R.;Blazel,J.;Parge, H.;Love,R.;Hickey,M.;Doan,C.;Shi,S.;Duggal,R.;Lewisc, C.;Fuhrmana,S.Bioorg.Med.Chem.Lett.2006,16,4834.
(35)Yan,S.Q.;Appleby,T.;Gunic,E.;Shim,J.H.;Tasu,T.;Kim, H.;Rong,F.;Chen,N.H.;Hamatake,R.;Wu,J.Z.;Hong,Z.; Yao,N.H.Bioorg.Med.Chem.Lett.2007,17,28.
(36)Yan,S.Q.;Larson,G.;Wu,J.Z.;Appleby,T.;Ding,Y.L.; Hamatake,R.;Hong,Z.;Yao,N.H.Bioorg.Med.Chem.Lett. 2007,17,63.
(37)Yan,S.Q.;Appleby,T.;Larson,G.;Wu,J.Z.;Hamatake,R.K.; Hong,Z.;Yao,N.H.Bioorg.Med.Chem.Lett.2007,17,1991.
(38) Burton,G.;Ku,T.W.;Carr,T.G.;Kiesow,T.;Sarisky,R.T.; Lin-Goerke,J.L.;Hofmann,G.A.;Slater,M.G.;Haigh,D.; Dhanak,D.;Johnson,V.K.;Parryb,N.R.;Thommesb,P. Bioorg.Med.Chem.Lett.2007,17,1930.
(39) Krueger,A.C.;Madigan,D.L.;Green,B.E.;Hutchinson,D. K.;Jiang,W.W.;Kati,W.M.;Liu,Y.Y.;Maring,C.J.;Masse, S.V.;McDaniel,K.F.;Middleton,T.R.;Mo,H.M.;Molla,A.; Montgomery,D.A.;Ng,T.I.;Kempf,D.J.Bioorg.Med.Chem. Lett.2007,17,2289.
(40)Rong,F.;Chow,S.;Yan,S.Q.;Larson,G.;Hong,Z.;Wu,J. Bioorg.Med.Chem.Lett.2007,17,1663.
(41) Ding,Y.L.;Smith,K.L.;Varaprasad,C.V.N.S.;Chang,E.; Alexander,J.;Yao,N.H.Bioorg.Med.Chem.Lett.2007,17, 841.
(42) Dragovich,P.S.;Blazel,J.K.;Ellis,D.A.;Han,Q.;Kamran, R.;Kissinger,C.R.;LeBrun,L.A.;Li,L.S.;Murphy,D.E.; Noble,M.;Patel,R.A.;Ruebsam,F.;Sergeeva,M.V.;Shah,A. M.;Showalter,R.E.;Tran,C.V.;Tsan,M.;Webber,S.E.; Kirkovsky,L.;Zhou,Y.F.Bioorg.Med.Chem.Lett.2008,18, 5635.
(43) Hutchinson,D.K.;Rosenberg,T.;Klein,L.L.;Bosse,T.D.; Larson,D.P.;He,W.P.;Jiang,W.W.;Kati,W.M.; Kohlbrenner,W.E.;Liu,Y.Y.;Masse,S.V.;Middleton,T.; Molla,A.;Montgomery,D.A.;Beno,D.W.A.;Stewart,K.D.; Stoll,V.S.;Kempf,D.J.Bioorg.Med.Chem.Lett.2008,18, 3887.
(44) Rawal,R.K.;Katti,S.B.;Kaushik-Basu,N.;Arora,P.;Pan,Z. H.Bioorg.Med.Chem.Lett.2008,18,6110.
(45)Kim,S.H.;Tran,M.T.;Ruebsam,F.;Xiang,A.X.;Ayida,B.; McGuire,H.;Ellis,D.;Blazel,J.;Tran,C.V.;Murphy,D.E.; Webber,S.E.;Zhou,Y.F.;Shah,A.M.;Tsan,M.;Showalter,R. E.;Patel,R.;Gobbi,A.;LeBrun,L.A.;Bartkowski,D.M.; Nolan,T.G.;Norris,D.A.;Sergeeva,M.V.;Kirkovsky,L.; Zhao,Q.;Han,Q.;Kissinger,C.R.Bioorg.Med.Chem.Lett. 2008,18,4181.
(46) Evans,K.A.;Chai,D.P.;Graybill,T.L.;Burton,G.;Sarisky,R. T.;Lin-Goerke,J.;Johnstonb,V.K.;Riveroa,R.A.Bioorg. Med.Chem.Lett.2006,16,2205.
(47) Bosse,T.D.;Larson,D.P.;Wagner,R.;Hutchinson,D.K.; Rockway,T.W.;Kati,W.M.;Liu,Y.Y.;Masse,S.;Middleton, T.;Mo,H.;Montgomery,D.;Jiang,W.;Koev,G.;Kempf,D.J.; Molla,A.Bioorg.Med.Chem.Lett.2008,18,568.
(48) Donner,P.L.;Xie,Q.H.;Pratt,J.K.;Maring,C.J.;Kati,W.; Jiang,W.;Liu,Y.Y.;Koev,G.;Masse,S.;Montgomery,D.; Molla,A.;Kempf,D.J.Bioorg.Med.Chem.Lett.2008,18, 2735.
(49) Liu,Y.Y.;Donner,P.L.;Pratt,J.K.;Jiang,W.W.;Ng,T.; Gracias,V.;Baumeister,S.;Wiedeman,P.E.;Traphagen,L.; Warrior,U.;Maring,C.;Kati,W.M.;Djuric,S.W.;Molla,A. Bioorg.Med.Chem.Lett.2008,18,3173.
(50) Li,L.S.;Zhou,Y.F.;Murphy,D.E.;Stankovic,N.;Zhao,J.J.; Dragovich,P.S.;Bertolini,T.;Sun,Z.X.;Ayida,B.;Tran,C. V.;Ruebsam,F.;Webber,S.E.;Shah,A.M.;Tsan,M.; Showalter,R.E.;Patel,R.;LeBrun,L.A.;Bartkowski,D.M.; Nolan,T.G.;Norris,D.A.;Kamran,R.;Brooks,J.;Sergeeva, M.V.;Kirkovsky,L.;Zhao,Q.;Kissinger,C.R.Bioorg.Med. Chem.Lett.2008,18,3446.
(51) Zhou,Y.F.;Webber,S.E.;Murphy,D.E.;Li,L.S.;Dragovich, P.S.;Tran,C.V.;Sun,Z.X.;Ruebsam,F.;Shah,A.M.;Tsan, M.;Showalter,R.E.;Patel,R.;Li,B.;Zhao,Q.;Han,Q.; Hermann,T.;Kissinger,C.R.;LeBrun,L.;Sergeeva,M.V.; Kirkovsky,L.Bioorg.Med.Chem.Lett.2008,18,1413.
(52) Ruebsam,F.;Webber,S.E.;Tran,M.T.;Tran,C.V.;Murphy, D.E.;Zhao,J.J.;Dragovich,P.S.;Kim,S.H.;Li,L.S.;Zhou, Y.F.;Han,Q.;Kissinger,C.R.;Showalter,R.E.;Lardy,M.; Shah,A.M.;Tsan,M.;Patel,R.;LeBrun,L.A.;Kamran,R.; Sergeeva,M.V.;Bartkowski,D.M.;Nolan,T.G.;Norris,D. A.;Kirkovsky,L.Bioorg.Med.Chem.Lett.2008,18,3616.
(53) Sergeeva,M.V.;Zhou,Y.F.;Bartkowski,D.M.;Nolan,T.G.; Norris,D.A.;Okamoto,E.;Kirkovsky,L.;Kamran,R.; LeBrun,L.A.;Tsan,M.;Patel,R.;Shah,A.M.;Lardy,M.; Gobbi,A.;Li,L.S.;Zhao,J.J.;Bertolini,T.;Stankovic,N.; Sun,Z.X.;Murphy,D.E.;Webber,S.E.;Dragovich,P.S. Bioorg.Med.Chem.Lett.2008,18,3421.
(54) Zhou,Y.F.;Li,L.S.;Dragovich,P.S.;Murphy,D.E.;Tran,C. V.;Ruebsam,F.;Webber,S.E.;Shah,A.M.;Tsan,M.;Averill, A.;Showalter,R.E.;Patel,R.;Han,Q.;Zhao,Q.;Hermann,T.; Kissinger,C.R.;LeBrun,L.;Sergeeva,M.V.Bioorg.Med. Chem.Lett.2008,18,1419.
(55) Ruebsam,F.;Sun,Z.X.;Ayida,B.K.;Webber,S.E.;Zhou,Y. F.;Zhao,Q.;Kissinger,C.R.;Showalter,R.E.;Shah,A.M.; Tsan,M.;Patel,R.;LeBrun,L.A.;Kamran,R.;Sergeeva,M. V.;Bartkowski,D.M.;Nolan,T.G.;Norris,D.A.;Kirkovsky, L.Bioorg.Med.Chem.Lett.2008,18,5002.
(56) Ellis,D.A.;Blazel,J.K.;Webber,S.E.;Tran,C.V.;Dragovich, P.S.;Sun,Z.X.;Ruebsam,F.;McGuire,H.M.;Xiang,A.X.; Zhao,J.J.;Li,L.S.;Zhou,Y.F.;Han,Q.;Kissinger,C.R.; Showalter,R.E.;Lardy,M.;Shah,A.M.;Tsan,M.;Patel,R.; LeBrun,L.A.;Kamran,R.;Bartkowski,D.M.;Nolan,T.G.; Norris,D.A.;Sergeeva,M.V.;Kirkovsky,L.Bioorg.Med. Chem.Lett.2008,18,4628.
(57) Hendricks,R.T.;Spencer,S.R.;Blake,J.F.;Fell,J.B.;Fischer, J.P.;Stengel,P.J.;Leveque,V.J.P.;LePogam,S.;Rajyaguru, S.;Najera,I.;Josey,J.A.;Swallow,S.Bioorg.Med.Chem.Lett. 2009,19,410.
(58) Ruebsam,F.;Tran,C.V.;Li,L.S.;Kim,S.H.;Xiang,A.X.; Zhou,Y.F.;Blazel,J.K.;Sun,Z.X.;Dragovich,P.S.;Zhao,J. J.;McGuire,H.M.;Murphy,D.E.;Tran,M.T.;Stankovic,N.; Ellis,D.A.;Gobbi,A.;Showalter,R.E.;Webber,S.E.;Shah, A.M.;Tsan,M.;Patel,R.A.;LeBrun,L.A.;Hou,H.Y.J.; Kamran,R.;Sergeeva,M.V.;Bartkowski,D.M.;Nolan,T.G.; Norris,D.A.;Kirkovsky,L.Bioorg.Med.Chem.Lett.2009,19, 451.
(59) deVicente,J.;Hendricks,R.T.;Smith,D.B.;Fell,J.B.;Fischer, J.;Spencer,S.R.;Stengel,P.J.;Mohr,P.;Robinson,J.E.; Blake,J.F.;Hilgenkamp,R.K.;Yee,C.;Zhao,J.P.;Elworthy, T.R.;Tracy,J.;Chin,E.;Li,J.;Lui,A.;Wang,B.;Oshiro,C.; Harris,S.F.;Ghate,M.;Leveque,V.J.P.;Najera,I.;Pogam,S. L.;Rajyaguru,S.;Ao-Ieong,G.;Alexandrova,L.;Fitch,B.; Brandl,M.;Masjedizadeh,M.;Wua,S.Y.;de Keczer,S.; Voronin,T.Bioorg.Med.Chem.Lett.2009,19,5648.
(60) Pacini,B.;Avolio,S.;Ercolani,C.;Koch,U.;Migliaccio,G.; Narjes,F.;Pacini,L.;Tomei,L.;Harper,S.Bioorg.Med.Chem. Lett.2009,19,6245.
(61) Shaw,A.N.;Tedesco,R.;Bambal,R.;Chai,D.P.;Concha,N. O.;Darcy,M.G.;Dhanak,D.;Duffy,K.J.;Fitch,D.M.;Gates, A.;Johnston,V.K.;Keenan,R.M.;Lin-Goerke,J.;Liu,N.; Sarisky,R.T.;Wiggall,K.J.;Zimmerman,M.N.Bioorg.Med. Chem.Lett.2009,19,4350.
(62) Habermann,J.;Capitò,E.;Ferreira,M.R.R.;Koch,U.;Narjes, F.Bioorg.Med.Chem.Lett.2009,19,633.
(63) Muller,J.P.;Shipps,G.W.,Jr.;Rosner,K.E.;Deng,Y.Q.; Wang,T.;Curran,P.J.;Brown,M.A.;Siddiqui,M.A.;Cooper, A.B.;Duca,J.;Cable,M.;Girijavallabhan,V.Bioorg.Med. Chem.Lett.2009,19,6331.
(64) deVicente,J.;Hendricks,R.T.;Smith,D.B.;Fell,J.B.;Fischer, J.;Spencer,S.R.;Stengel,P.J.;Mohr,P.;Robinson,J.E.; Blake,J.F.;Hilgenkamp,R.K.;Yee,C.;Adjabeng,G.; Elworthy,T.R.;Li,J.;Wanga,B.;Bamberg,J.T.;Harris,S.F.; Wonga,A.;Leveque,V.J.P.;Najera,I.;Pogam,S.L.;Rajyaguru, S.;Ao-Ieong,G.;Alexandrova,L.;Larrabee,S.;Brandl,M.; Briggs,A.;Sukhtankar,S.;Farrell,R.Bioorg.Med.Chem.Lett. 2009,19,5652.
(65) deVicente,J.;Hendricks,R.T.;Smith,D.B.;Fell,J.B.;Fischer, J.;Spencer,S.R.;Stengel,P.J.;Mohr,P.;Robinson,J.E.; Blake,J.F.;Hilgenkamp,R.K.;Yee,C.;Adjabeng,G.; Elworthy,T.R.;Tracy,J.;Chin,E.;Li,J.;Wanga,B.;Bamberg, J.T.;Stephenson,R.;Oshiro,C.;Harris,S.F.;Ghate,M.; Leveque,V.;Najera,I.;Pogam,S.L.;Rajyaguru,S.;Ao-Ieong, G.;Alexandrova,L.;Larrabee,S.;Brandl,M.;Briggs,A.; Sukhtankar,S.;Farrell,R.;Xu,B.Bioorg.Med.Chem.Lett. 2009,19,3642.
(66)Wang,G.Y.;Lei,H.X.;Wang,X.F.;Das,D.;Hong,J.; Mackinnon,C.H.;Coulter,T.S.;Montalbetti,C.A.G.N.; Mears,R.;Gai,X.J.;Bailey,S.E.;Ruhrmund,D.;Hooi,L.; Misialek,S.;Rajagopalan,P.T.R.;Cheng,R.K.Y.;Barker,J. J.;Felicetti,B.;Sch?nfeld,D.L.;Stoycheva,A.;Buckman,B. O.;Kossen,K.;Seiwert,S.D.;Beigelman,L.Bioorg.Med. Chem.Lett.2009,19,4480.
(67)Wanga,G.Y.;Zhang,L.G.;Wu,X.M.;Das,D.;Ruhrmund,D.; Hooi,L.;Misialek,S.;Rajagopalan,P.T.R.;Buckman,B.O.; Kossen,K.;Seiwert,S.D.;Beigelman,L.Bioorg.Med.Chem. Lett.2009,19,4484.
(68)Wanga,G.Y.;He,Y.Z.;Sun,J.;Das,D.;Hu,M.G.;Huang,J. H.;Ruhrmund,D.;Hooi,L.;Misialek,S.;Rajagopalan,P.T.R.; Stoycheva,A.;Buckman,B.O.;Kossen,K.;Seiwert,S.D.; Beigelman,L.Bioorg.Med.Chem.Lett.2009,19,4476.
(69)McGowan,D.;Nyanguile,O.;Cummings,M.D.;Vendeville, S.;Vandyck,K.;den Broeck,W.V.;Boutton,C.W.;Bondt,H. D.;Quirynen,L.;Amssoms,K.;Bonfanti,J.F.;Last,S.; Rombauts,K.;Tahri,A.;Hu,L.L.;Delouvroy,F.;Vermeiren, K.;Vandercruyssen,G.;Van der Helm,L.;Cleiren,E.; Mostmans,W.;Lory,P.;Pille,G.;Van Emelen,K.;Fanning,G.; Pauwels,F.;Lin,T.I.;Simmen,K.;Raboisson,P.Bioorg.Med. Chem.Lett.2009,19,2492.
(70) Hendricks,R.T.;Fell,J.B.;Blake,J.F.;Fischer,J.P.; Robinson,J.E.;Spencer,S.R.;Stengel,P.J.;Bernacki,A.L.; Leveque,V.J.P.;Pogam,S.L.;Rajyaguru,S.;Najera,I.;Josey, J.A.;Harris,J.R.;Swallow,S.Bioorg.Med.Chem.Lett.2009, 19,3637.
(71)Lv,W.;Xue,Y.Eur.J.Med.Chem.2010,45,1167.
(72) ChemDraw,version 9.0;Cambridge Soft Corporation: Cambridge,USA,2004.
(73)Corina,Version 3.4;Molecular Networks GmbH Computerchemie:Erlangen,Germany,2006.
(74) Todeschini,R.;Consonni,V.Handbook of Molecular Descriptors;Wiley-VCH:New York,2000.
(75) Hasegawa,K.J.Chem.Inf.Comput.Sci.1999,39,112.
(76) Byvatov,E.;Fechner,U.;Sadowski,J.;Schneider,G.J.Chem. Inf.Comput.Sci.2003,43,1882.
(77) He,L.;Jurs,P.C.;Custer,L.L.;Durham,S.K.;Pearl,G.M. Chem.Res.Toxicol.2003,16,1567.
(78) Lü,W.;Xue,Y.Acta Phys.-Chim.Sin.2010,26,471. [呂 巍,薛 英.物理化學(xué)學(xué)報(bào),2010,26,471.]
(79) Degroeve,S.;de Baets,B.;van de Peer,Y.;Rouze,P. Bioinformatics 2002,18,S75.
(80)Xue,Y.;Yap,C.W.;Sun,L.Z.;Cao,Z.W.;Wang,J.F.;Chen, Y.Z.J.Chem.Inf.Comput.Sci.2004,44,1497.
(81) Leach,A.R.;Gillet,V.J.An Introduction to Chemoinformatics; Springer:New York,2007.
(82) Garner,S.R.Weka,version 3.4.12;University of Waikato:New Zealand,2005.
(83) Vapnik,V.N.The Nature of Statistical Learning Theory; Springer-Verlag:New York,1995.
(84) Johnson,R.A.;Wichern,D.W.Applied Multivariate Statistical Analysis;Prentice Hall:New York,1982.
(85) Quinlan,J.R.C4.5,Programs for Machine Learning;Morgan Kaufmann:San Mateo,CA,1992.
(86) Baldi,P.;Brunak,S.;Chauvin,Y.;Andersen,C.A.;Nielsen,H. Bioinformatics 2000,16,412.
(87) Srivastava,A.K.;Pandey,A.;Srivastava,A.;Shukla,N.J.Sau. Chem.Soc.2011,15,25.
March 2,2011;Revised:March 29,2011;Published on Web:April 21,2011.
Prediction of Hepatitis C Virus Non-Structural Proteins 5B Polymerase Inhibitors Using Machine Learning Methods
Lü Wei1XUE Ying2,3,*
(1College of Life Sciences,State Key Laboratory of Crop Biology,Shandong Agricultural University,Tai′an 271018,Shandong Province,P.R.China;2College of Chemistry,Key Laboratory of Green Chemistry and Technology,Ministry of Education,Sichuan University,Chengdu 610064,P.R.China;3State Key Laboratory of Biotherapy,Sichuan University,Chengdu 610041,P.R.China)
Non-structural proteins 5B(NS5B)play an important role in protein maturation and gene replication as an RNA dependent RNA polymerase in the hepatitis C virus(HCV).Inhibiting NS5B polymerase will prevent RNA replication and,therefore,it is significant for the treatment of HCV.It is becoming increasingly important to screen and predict molecules that have NS5B inhibitory activity by computational methods.This work explores several machine learning(ML)methods(support vector machine(SVM),k-nearest neighbor(k-NN),and C4.5 decision tree(C4.5 DT))for the prediction of NS5B inhibitors(NS5BIs).This prediction system was tested using 1248 compounds(552 NS5BIs and 696 non-NS5BIs),which are significantly more diverse in chemical structure than those used in other studies.A feature selection method was used to improve the prediction accuracy and the selection of molecular descriptors responsible for distinguishing between NS5BIs and non-NS5BIs.The prediction accuracies were 81.4%-91.7%for the NS5BIs,78.2%-87.2%for the non-NS5BIs,and 84.1%-85.0%overall based on the three kinds of machine learning methods.SVM gave the best accuracy of 91.7%for the NS5BIs, C4.5 gave the best accuracy of 87.2%for the non-NS5BIs,and k-NN gave the best overall accuracy of 85.0%for all the compounds.This work suggests that machine learning methods can facilitate the prediction of the NS5BIs potential for unknown sets of compounds and to determine the molecular descriptors associated with NS5BIs.
Machine learning method;Molecular descriptor;Recursive feature elimination; Support vector machine;Hepatitis C virus
*Corresponding author.Email:xue@scu.edu.cn;Tel:+86-28-85418330.
The project was supported by the National Key Basic Research Program of China(2009CB118500)and Scientific Research Foundation for the Returned Overseas Chinese Scholars,Ministry of Education,China(20071108-18-15).
國(guó)家重點(diǎn)基礎(chǔ)研究發(fā)展規(guī)劃項(xiàng)目(2009CB118500)和教育部留學(xué)歸國(guó)人員科研啟動(dòng)基金(20071108-18-15)
O641

