Expression of cyclin-dependent kinase 9 is positively correlated with the autophagy level in colon cancer
Lei Zheng,Jia Lu,Da-Lu Kong
Abstract BACKGROUND Cyclin-dependent kinase 9 (CDK9) expression and autophagy in colorectal cancer (CRC) tissues has not been widely studied.CDK9,a key regulator of transcription,may influence the occurrence and progression of CRC.The expression of autophagy-related genes BECN1 and drug resistance factor ABCG2 may also play a role in CRC.Under normal physiological conditions,autophagy can inhibit tumorigenesis,but once a tumor forms,autophagy may promote tumor growth.Therefore,understanding the relationship between autophagy and cancer,particularly how autophagy promotes tumor growth after its formation,is a key motivation for this research.AIM To investigate the relationship between CDK9 expression and autophagy in CRC,assess differences in autophagy between left and right colon cancer,and analyze the associations of autophagy-related genes with clinical features and prognosis.METHODS We collected tumor tissues and paracarcinoma tissues from colon cancer patients with liver metastasis to observe the level of autophagy in tissues with high levels of CDK9 and low levels of CDK9.We also collected primary tissue from left and right colon cancer patients with liver metastasis to compare the autophagy levels and the expression of BECN1 and ABCG2 in the tumor and paracarcinoma tissues.RESULTS The incidence of autophagy and the expression of BECN1 and ABCG2 were different in left and right colon cancer,and autophagy might be involved in the occurrence of chemotherapy resistance.Further analysis of the relationship between the expression of autophagy-related genes CDK9,ABCG2,and BECN1 and the clinical features and prognosis of colorectal cancer showed that the high expression of CDK9 indicated a poor prognosis in colorectal cancer.CONCLUSION This study laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of patients with CRC.
Key Words: Autophagy;Tumorigenesis;Tumor tissue;Paracarcinoma tissue;Expression;Left/right colon cancer
lNTRODUCTlON
Colorectal cancer (CRC) is one of the most common malignant tumors in China,and its incidence rate is increasing every year.The latest research has confirmed that there are many differences between left-sided colon cancer and right-sided colon cancer in embryologic origin,pathogenesis,histopathology,epidemiology,epigenetics,clinical manifestations,and prognosis[1].Some scholars postulate that left-sided colon cancer and right-sided colon cancer are two distinct diseases[2].Therefore,the comparative study of left-sided and right-sided colon cancer is helpful to understand pathogenesis and the mechanism of progression.
Microscopically,the occurrence and development of tumors are mainly characterized by unlimited abnormal proliferation of cells.The proliferation and differentiation of cells in multicellular organisms[3,4] are regulated by the cell cycle.The main causes of tumor occurrence are cell cycle disorder and increased resistance to apoptosis.Cyclin-dependent kinases (CDKs) promote DNA synthesis and mitosis[5,6] through phosphorylation of key substrates and participate in cell cycle regulation.Abnormal activation of CDKs leads to the imbalance of the cell cycle,uncontrollable cell proliferation,and the occurrence and development of malignant tumors[7].For example,the CDK family includes 20 kinases that contribute to cell deterioration by promoting proliferation,migration,invasion,and anti-apoptosis of cancer cells[8].
CDK9 is a key regulator of transcription and is used to continuously produce short-lived proteins to maintain the survival of cancer cells[9].As a part of a functionally diverse group of enzymes responsible for cell cycle control and progression,CDK9 associates primarily with cyclin T1 and forms the positive transcription elongation factor b (p-TEFb) complex responsible for the regulation of transcription elongation and mRNA maturation.CDK9 can influence almost all cellular functions.Many studies have shown that the dysfunction of CDK9 signaling is obvious in various hematologic malignancies and solid tumor cell lines,and the inhibition of CDK9 can lead to growth suppression and apoptosis of cancer cells[10,11].Due to the extremely critical role of CDK9 in cancer cells,inhibiting its functions has been a research hotspot,with more and more studies focusing on small,molecular inhibitors,some of which are presently in clinical trials.The search for newer CDK9 inhibitors with higher specificity and lower toxicities continues[12].Targeting CDK9 with wogonin and related natural flavones could enhance the anticancer effect of theBCL-2family inhibitor ABT-263[13].
Macroautophagy,commonly known as autophagy,is the main form of cell autophagy.It sequesters proteins,lipids,or organelles that need to be degraded with a bimolecular membrane structure,forming autophagosomes.Autophagosomes can be integrated with lysosomes to form an autolysosome,which degrades the encapsulated cellular components through proteolytic enzymes and lipases in lysosomes and then recycles or excretes them.Autophagy in tumor cells is a double-edged sword.Autophagy can eliminate abnormally folded proteins and dysfunctional organelles,inhibit the stress response of cells,and ultimately prevent genome damage,thus inhibiting the occurrence of cancer[14].On the other hand,tumor cells can utilize autophagy to promote their survival under conditions of nutritional deficiency and hypoxia[15].
The relationship between autophagy and the progression of tumorigenesis is complicated.The promotion or suppression of autophagy depends on the tumor type and environment[16].The number of autophagosomes observed by transmission electron microscope (TEM) in cancer tissues can reflect the level of autophagy[17].In vitroandin vivodata support the hypothesis that autophagy could enhance cancer cell drug resistance to chemotherapy,and inhibition of autophagy may increase the sensitivity of cancer cells to chemotherapy[18,19].
Evidence suggests thatBECN1is a tumor suppressor.BECN1binds and activates VP34 through a conserved domain to induce autophagy[20].Through the positive regulators ofBECN1,UVRAG,and BIF-1,the combination ofBECN1and VP34 is enhanced,leading to an increase in autophagy and resulting in tumor inhibition[21].Overexpression of antiapoptotic protein BCL-2 promotes the survival of breast cancer,prostate cancer,small cell lung cancer,non-small cell lung cancer,and leukemia[22,23].The interaction between BCL-2 andBECN1could be exploited to prevent autophagy[24].
Mitochondrial autophagy (also known as mitophagy) is based on selective degradation dysfunction or damaged mitochondria and plays an important role in maintaining high-quality mitochondria[25].Regulation of this process is critical for maintaining cellular homeostasis and has been closely associated with acquired drug resistance.CDK9 may be closely associated with the regulation of mitochondrial function and mitophagy as well.It was reported that inhibition of CDK9 blocked PINK1-PRKN-mediated mitophagy in hepatocellular carcinoma by interrupting mitophagy initiation[26].However,the relationship between autophagy and CDK9 in CRC has not been fully investigated.Therefore,elucidating the interaction between autophagy and CDK9 would be helpful for improving the efficacy of anticancer drugs in the clinical treatment of CRC.
The ATP-binding cassette transmembrane transporter superfamily is involved in the transmembrane transport of anticancer drugs,which decreases the toxicity of anticancer drugs for tumor cells.TheABCG2transporter mediates multidrug resistance and regulates autophagy.Under normal growth conditions,cells with high levels ofABCG2have a higher basic autophagy rate than cells with low levels ofABCG2,which could be associated with the response to stressors[27].For example,high expression ofABCG2was associated with chemotherapy resistance and poor prognosis in patients with osteosarcoma[28].In childhood leukemia and chronic lymphocytic leukemia,drug resistance to adriamycin increased after overexpression ofABCG2[29].
In this study,the relationship between the expression of CDK9 and the incidence of autophagy in CRC was determined.We also compared the incidence of autophagy and prognosis withABCG2expression in left and right colon cancers.We found that the expression of CDK9 was positively correlated with autophagy in CRC.The incidence rate of autophagy and expression ofABCG2in patients with liver metastasis between left and right colon cancers was different,and autophagy may be involved in the resistance to chemotherapy.We further analyzed the relationship among the expression of autophagy-related genes CDK9,ABCG2,andBECN1,the clinical features,and the prognosis of CRC.
MATERlALS AND METHODS
CRC database source
Public gene expression data and corresponding clinical annotations for transcriptome analysis were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/;Date: March 31,2021;Version: V29.0).RNAseq data in level 3 HTSeq FPKM format in TCGA-COAD and TCGA-READ were used.The RNAseq data in FPKM format was converted into TPM format,and log2 conversion was performed.The clinicopathological features of public data are shown in Supplementary Table 1.
Clinical samples
Nineteen groups of fresh tumor and paracarcinoma colon tissue derived from 19 patients were collected.Twelve patients were diagnosed with left colon cancer,and seven patients were diagnosed with right colon cancer.All patients had liver metastasis.Twenty-four groups of fresh tumor tissue and paired tissue from 24 patients were collected.Fifteen patients were diagnosed with stage IV left colon cancer,and nine patients were diagnosed with right colon cancer.The basic information of the sample group is shown in Supplementary Table 2.Fresh tissues were defined as fresh surgical tissues,soybean size 1-2,and 40-50 mg.The proportion of tumor cells was greater than 40%,and the proportion of necrotic cells was not greater than 10%.Informed consent was obtained from all subjects and/or their legal guardians.
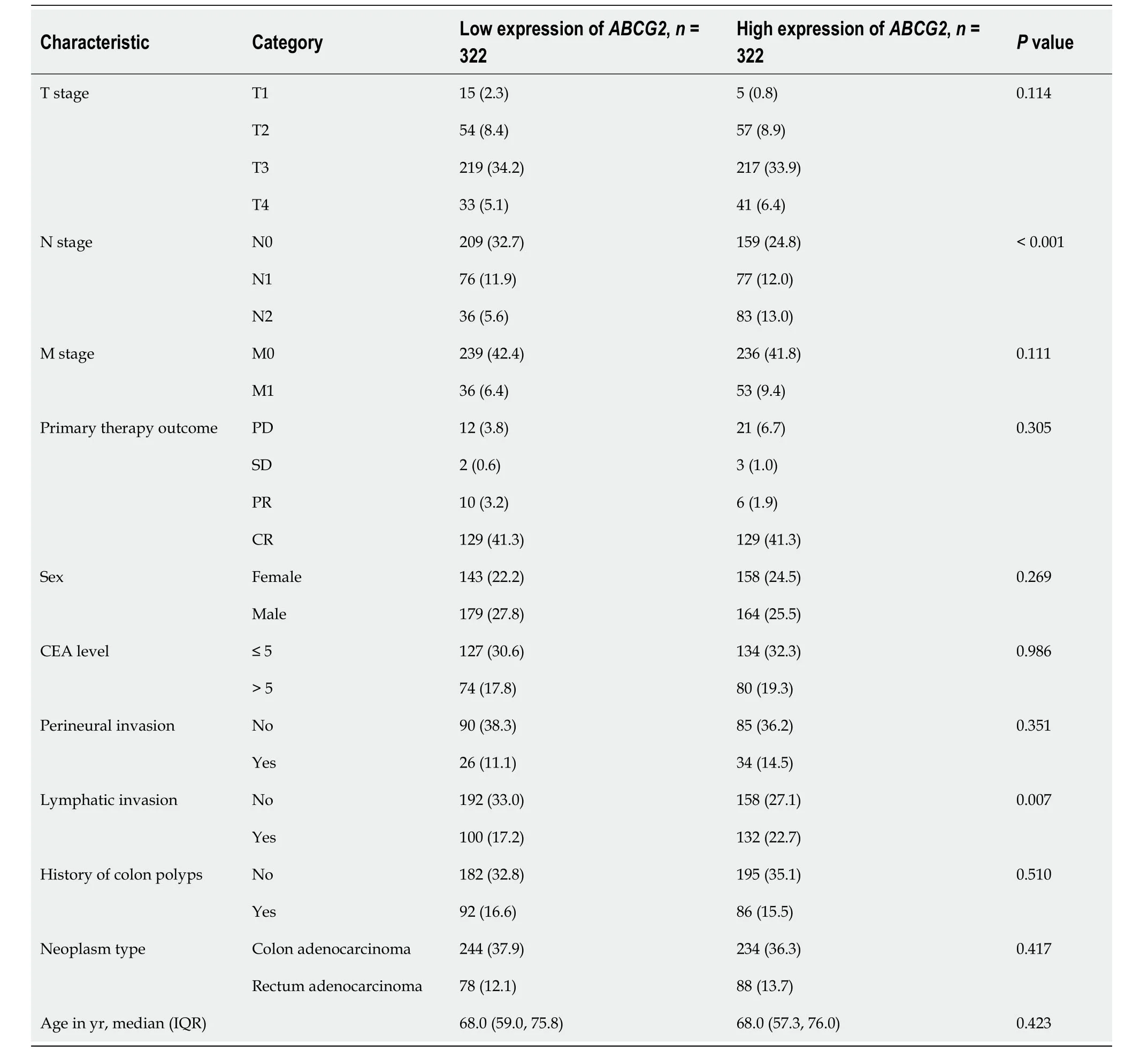
Table 2 Basic information of patients expressing ABCG2,n (%)
TEM and immunohistochemistry
Electron microscopy samples of 24 groups of fresh tumor tissue and paracarcinoma tissue from stage IV left and right colon cancer patients were prepared according to a previously published protocol[30].Nineteen groups of fresh tumor and paracarcinoma tissues derived from 19 patients with left and right colon cancer with liver metastasis were used for immunohistochemistry (IHC) based on protocols in our facility’s pathology department.
For TEM,tumor and paracarcinoma tissues from stage IV colon cancer patients were fixed in 2.5% glutaraldehyde,rinsed in 0.1 M phosphoric acid,and then immobilized with 1% osmic acid fixative.Dehydration included sequential steps in ethanol and acetone.Samples were embedded,solidified,and sliced into 70-nm sections.Staining was performed with 3% uranium acetate-lead citrate.Finally,observation and filming were carried out using the JEOL JEM-1230 TEM at 80 KV.
For IHC,paraffin section preparation involved tissue dehydration,transparency,wax immersion,block embedding,and sectioning.Dewaxing included xylene,100% alcohol,and subsequent alcohol concentrations with 10-min intervals.Duration varied with temperature.For antigen repair,the tissues were washed,treated with 3% hydrogen peroxide for 10 min and then citric acid buffer before being microwaved for antigen exposure.Serum blocking involved washing with phosphate buffered saline,drying,serum addition,and incubation at 37 °C.The serum was diluted 10 times.Primary antibody was applied,and slides were stored at 4 °C overnight.Secondary antibody was applied after phosphate buffered saline washing.StreptAvidin-Biotin Complex solution was applied for 30 min at 37 °C.The slides were exposed to developer solution,and staining with hematoxylin was completed in 0.5-5.0 min.Dehydration included various alcohol concentrations and xylene.Slides were finally sealed and dried.
CYTO-ID? autophagy detection kit
Autophagy was analyzed by flow cytometry using the CYTO-ID?Autophagy Detection kit (Enzo Life Sciences,Farmingdale,NY,United States).
Statistical analysis
Gene expression and clinical data were analyzed using R (version 3.6.3).Numerical variables were expressed as median and 25th-75thpercentile.Categorical variables were expressed as numbers and percentages.The Student’sttest was used for normally distributed data,and the Mann Whitney U test was used for comparing non-normally distributed data.Theχ2test was used to compare categorical data.For paired samples that met the normality test,the paired samplettest was used.For paired samples that did not meet the normality test,the Wilcoxon signed-rank test was used.
The median gene expression level was the cutoff point,and the expression was divided into high or low expression levels.Kaplan-Meier survival analysis was applied to evaluate the overall survival rate (OS),disease-specific survival rate (DSS),and progression-free interval (PFI) of CRC at high or low expression levels.To find the factors related to OS,DSS,and PFI,we used the Cox regression model for analysis.The area under the curve (AUC) of the time-dependent receiver operating characteristic (ROC) curve was used to evaluate the accuracy ofABCG2,CDK9,andBECN1expression levels in predicting the prognosis of patients.Autophagy pathway genes in the react database and CDK9 were imported into the string database to build protein-protein interaction (PPI) networks.CDK9 interaction genes were filtered and transferred to Cytoscape to make a PPI network.Pvalues < 0.05 were considered significant.
RESULTS
Expression of CDK9 in CRC tissues
TCGA-COAD and TCGA-READ analyses showed that the expression of CDK9 was increased in 50 paired CRC tissues (Figure 1,Supplementary Table 1).
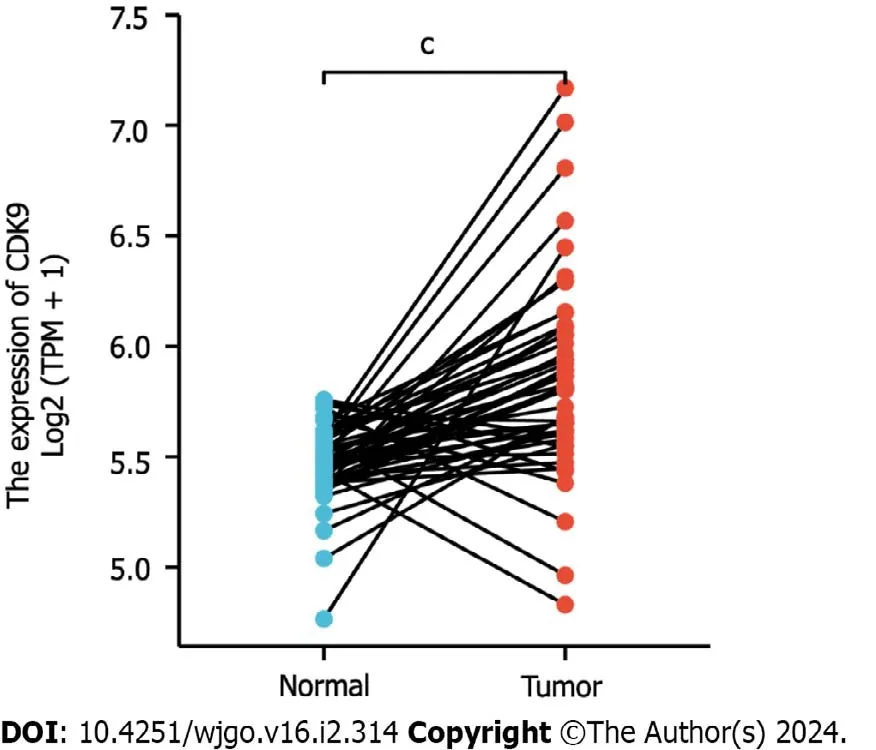
Figure 1 Expression of cyclin-dependent kinase 9 in colorectal cancer tissues. Expression of cyclin-dependent kinase 9 (CDK9) was extracted from The Cancer Genome Atlas.CDK9 was significantly increased in colorectal cancer tissues compared to paired paracarcinoma tissues.CDK9: Cyclin-dependent kinase 9.
Relationship between the expression of CDK9 and autophagy and its protein interactions
We collected tumor tissues and paracarcinoma tissues from colon cancer patients with liver metastasis.We observed by TEM that tissues with a high level of CDK9 had significantly higher levels of autophagy than the tissues with a low expression of CDK9 (Figure 2).We demonstrated that the expression of CDK9 in colon cancer was positively correlated with autophagy.PPI networks of CDK9 are shown in Supplementary Figure 1.CDK9 interacts with CSNK2B,HSP90-AA1,and RAPTOR,all of which have an association with the progression and prognosis of CRC[31-33].
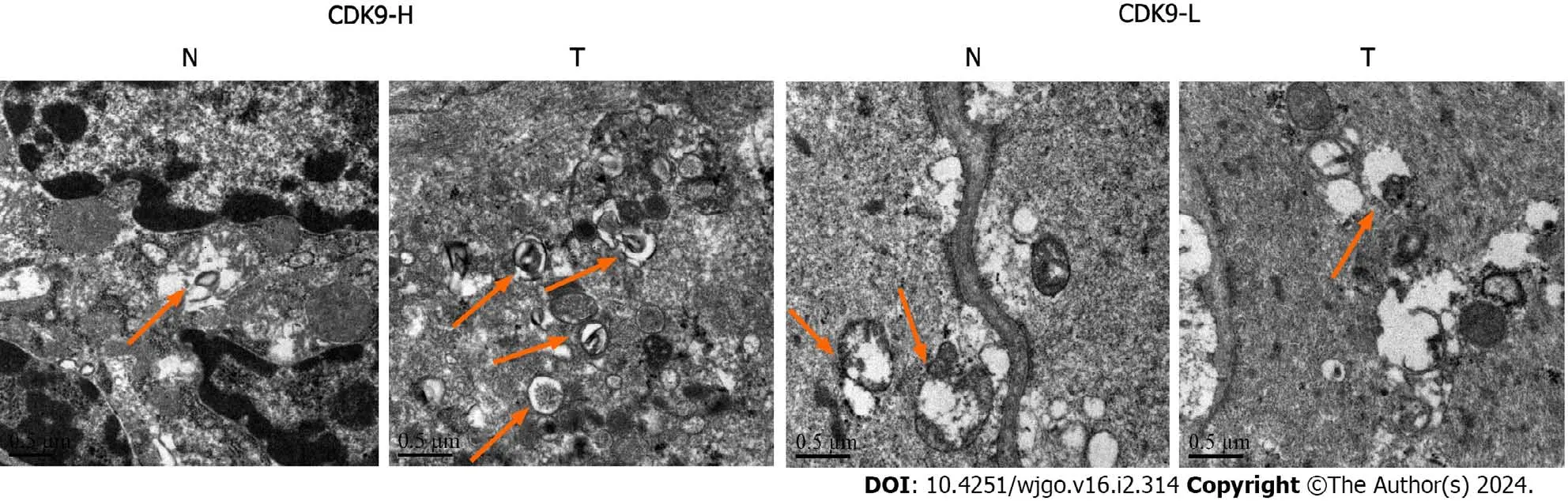
Figure 2 Transmission electron microscopy. Tissues with a high level of cyclin-dependent kinase 9 (CDK9) had significantly more autophagy than tissues with a low level of CDK9.Red arrows indicate the autophagosomes.CDK9-H: High level of cyclin-dependent kinase 9 expression;CDK9-L: Low level of cyclindependent kinase 9 expression;N: Paracarcinoma tissue;T: Tumor tissue.
CDK9 was an independent prognosis marker of CRC
We used Cox regression analysis to determine whether CDK9 expression levels could be used to evaluate the prognosis of patients with CRC.A high expression level of CDK9 was a risk factor for the prognosis of CRC that was associated with OS and DSS (allP< 0.050;Figure 3,Tables 1 and 2).Multivariate Cox regression analysis revealed that high expression of CDK9 was an independent prognosis marker associated with OS and DSS (allP< 0.050;Figure 3,Tables 1 and 2).
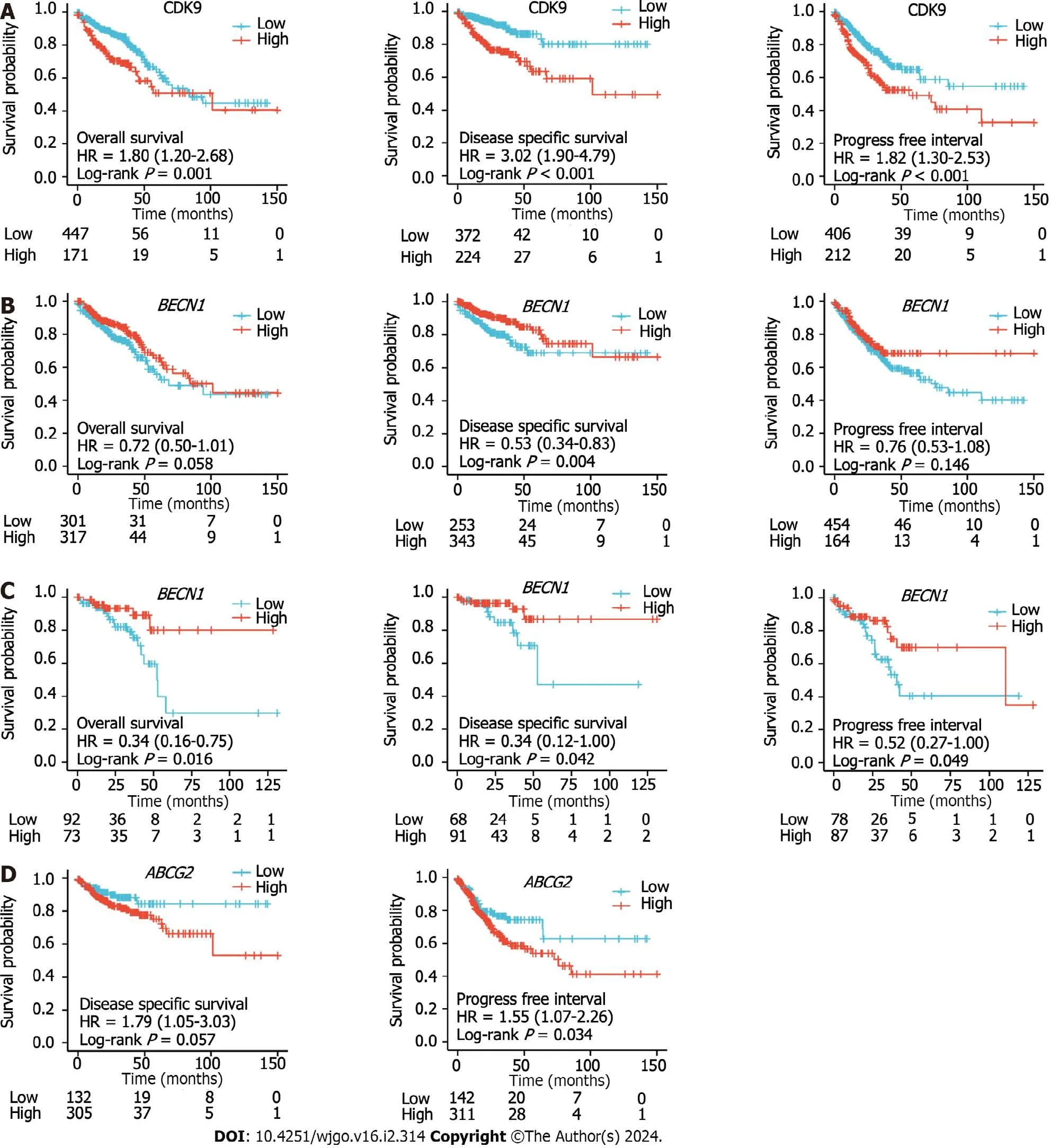
Figure 3 Kaplan-Meier curves of colorectal cancer patients. A: Kaplan-Meier curves of overall survival (OS),disease-specific survival (DSS),and progression-free interval (PFI) in colorectal cancer patients with high and low expression levels of cyclin-dependent kinase 9 (CDK9);B: Kaplan-Meier curves of OS,DSS,and PFI of colon cancer patients with high and low expression levels of BECN1;C: Kaplan-Meier curves of OS,DSS,and PFI of rectum cancer patients with high and low expression levels of BECN1;D: Kaplan-Meier curves of DSS and PFI of colon cancer patients with high and low expression levels of ABCG2.HR: Hazard ratio.
Autophagy incidence and expression levels of BECN1 and ABCG2 in left and right colon cancer tissue
We collected 24 groups of fresh tumor tissue and paired tissue from 24 patients with stage IV left and right colon cancer.We observed autophagy with a projection electron microscope and counted the positive rate of autophagy in multiple fields.The results showed that the positive rate (8/9) of autophagy in right colon cancer was significantly higher than that in left colon cancer (5/15;χ2=6.993,P=0.030) and paracarcinoma tissue (3/9;χ2=5.844,P=0.016) (Figure 4A).Primary cells were isolated from the fresh tumor tissues of colon cancer patients with liver metastasis.The CYTO-ID?autophagy detection reagent was used for staining.Flow cytometry was used to detect the formation of autophagy in the primary cancer cells.The results (Figure 4B) were consistent with the observations from electron microscopy.
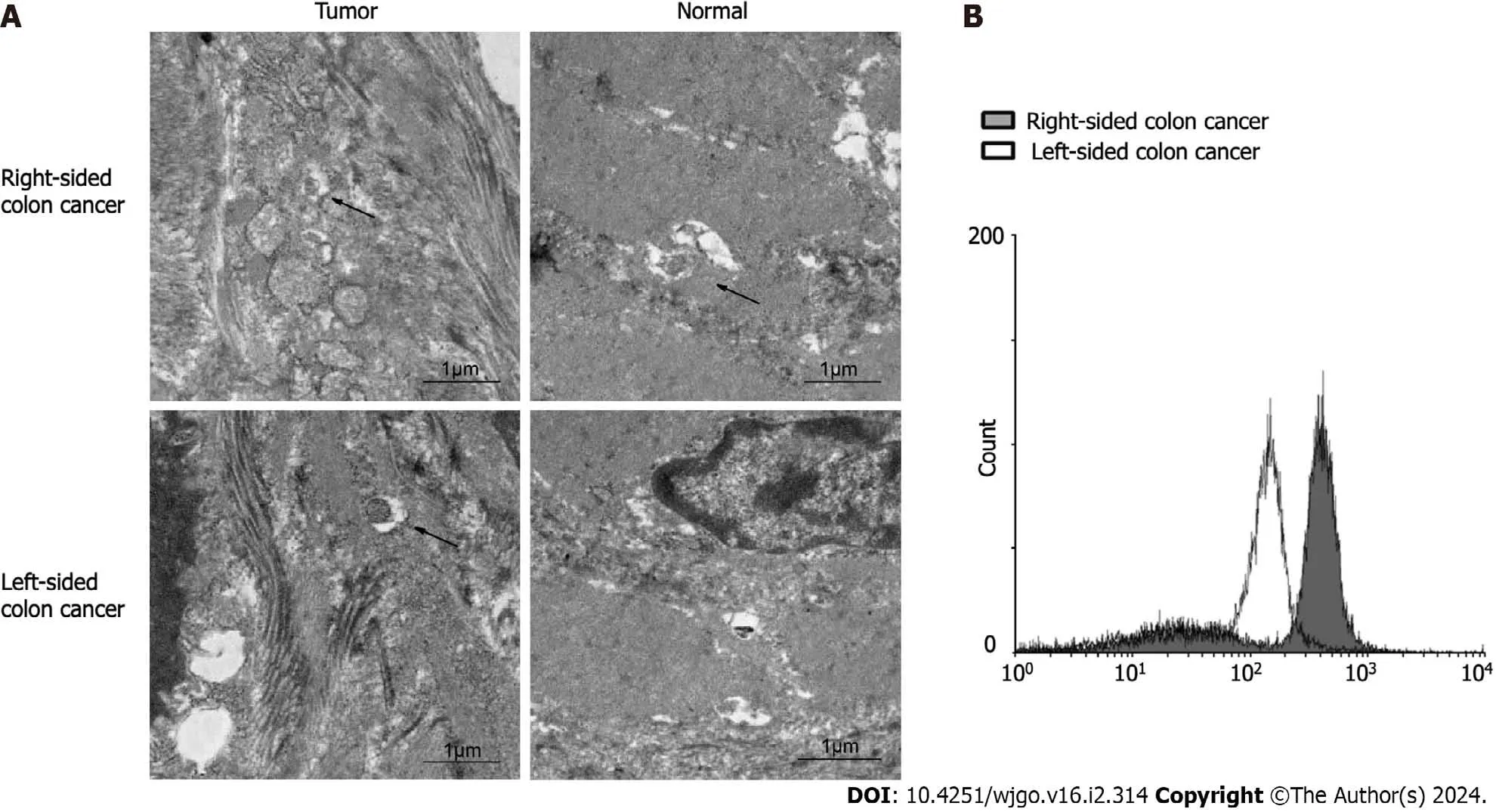
Figure 4 Level of autophagy in left and right colon cancer tissues.A: Autophagosomes (black arrows) in left and right colon cancer and adjacent tissues observed under electron microscopy;B: Expression levels of autophagic vesicles in left and right colon cancer primary cells.
We compared the expression ofBECN1andABCG2between 19 groups of fresh tumor tissue and paracarcinoma tissue from colon cancer patients with liver metastasis.We also compared the expression patterns ofBECN1andABCG2in left and right CRC.IHC results showed that there were no significant differences in the positive probability and expression level ofBECN1between advanced colon cancer and paracarcinoma tissue.However,the positive rate of right colon cancer (72.2%,6/7) was significantly higher than that of left colon cancer (3/12;χ2=6.537;P=0.011;Figure 5A).Inaddition,we found that compared with the paracarcinoma tissues,the right colon cancer tissue showed a high expression ofABCG2,while the expression level ofABCG2in the left colon cancer had no significant difference compared with the paracarcinoma tissues (Figure 5B).
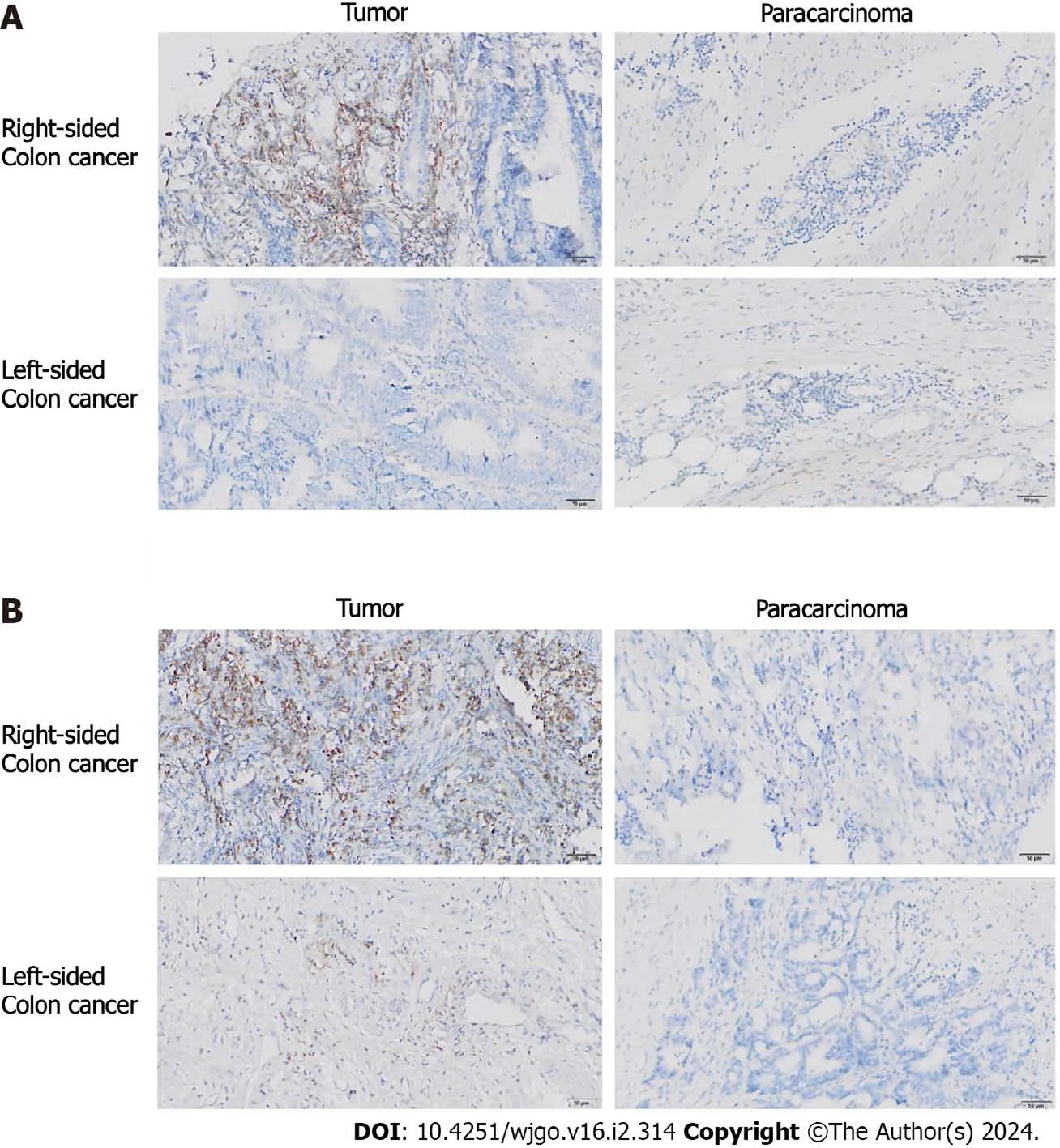
Figure 5 Expression of BECN1 (a key autophagy molecule) and ABCG2 (a drug resistance factor) in left and right colon cancer tissue. A: Expression of BECN1 in left and right colon cancer tissues and paracarcinoma tissue;B: Expression of ABCG2 in left and right colon cancer tissues and paracarcinoma tissue.
These results suggest that the incidence of autophagy in the primary liver metastasis of left and right colon cancers is different,and it may be involved in the occurrence of chemotherapy resistance.
Relationship between the expression levels of CDK9,ABCG2,and BECN1 and the clinical features and prognosis of CRC
Basic information ofBECN1,CDK9,andABCG2expression levels from 644 CRC samples in relation to clinical data were analyzed (Tables 1-3 and Supplementary Table 2).The expression levels of CDK9 were associated with tumor T stage and lymph node invasion (Table 1).The expression levels ofABCG2were associated with tumor N-stage and lymph node invasion (Table 2).The expression levels ofBECN1were related to age,history of colon polyps,and lymph node invasion (Table 3).There was no correlation between CDK9 and age andABCG2and age (P=0.821 andP=0.388,respectively).There was a weak correlation betweenBECN1and age (rho=-0.135,P=0.001).There was no correlation between CDK9 and tumor stage andBECN1and tumor stage (P=0.368 andP=0.884,respectively).There was a weak correlation betweenABCG2and tumor stage (rho=0.105,P=0.009).
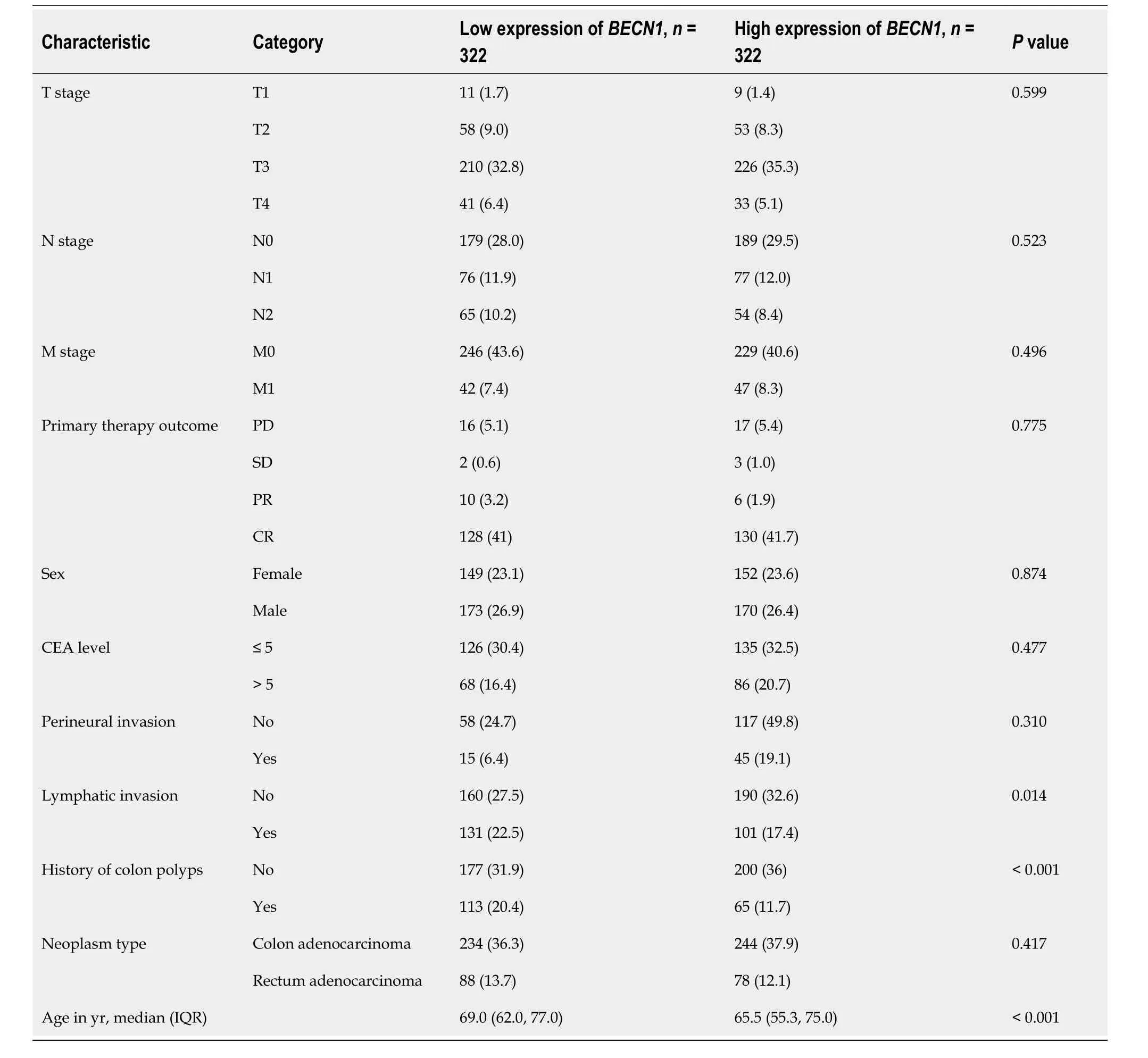
Table 3 Basic information of patients expressing BECN1,n (%)
The Kaplan-Meier survival analysis was used to evaluate the OS,DSS,and PFI of CRC patients with high or low expression levels ofBECN1,CDK9,andABCG2.Univariate and multivariate analysis of OS,disease-free survival,and DSS for CDK9,ABCG2,andBECN1are shown in Supplementary Table 3 and Supplementary Figure 2.The high expression level of CDK9 predicted the poor prognosis of CRC (OS,DSS,and PFI were consistent and statistically significant) (Figure 3A).The high expression level ofBECN1only predicted a poor prognosis of rectal cancer (Figure 3B and C).The high expression level ofABCG2only predicted a poor prognosis in colon cancer (Figure 3D).
According to the expression levels of CDK9 (Figure 6A),BECN1(Figure 6B),andABCG2(Figure 6C),the AUC of the time-dependent ROC curve was used to evaluate the prediction accuracy of prognosis in CRC patients.The larger the AUC value,the higher the accuracy of the prediction.ABCG2expression levels showed the highest level of accuracy (AUC=0.985 for rectal cancer;AUC=0.987 for colon cancer) for the prognosis of CRC.CDK9 showed the second-highest level of accuracy for the prognosis of CRC,whileBECN1showed a lower accuracy for the prognosis of CRC.
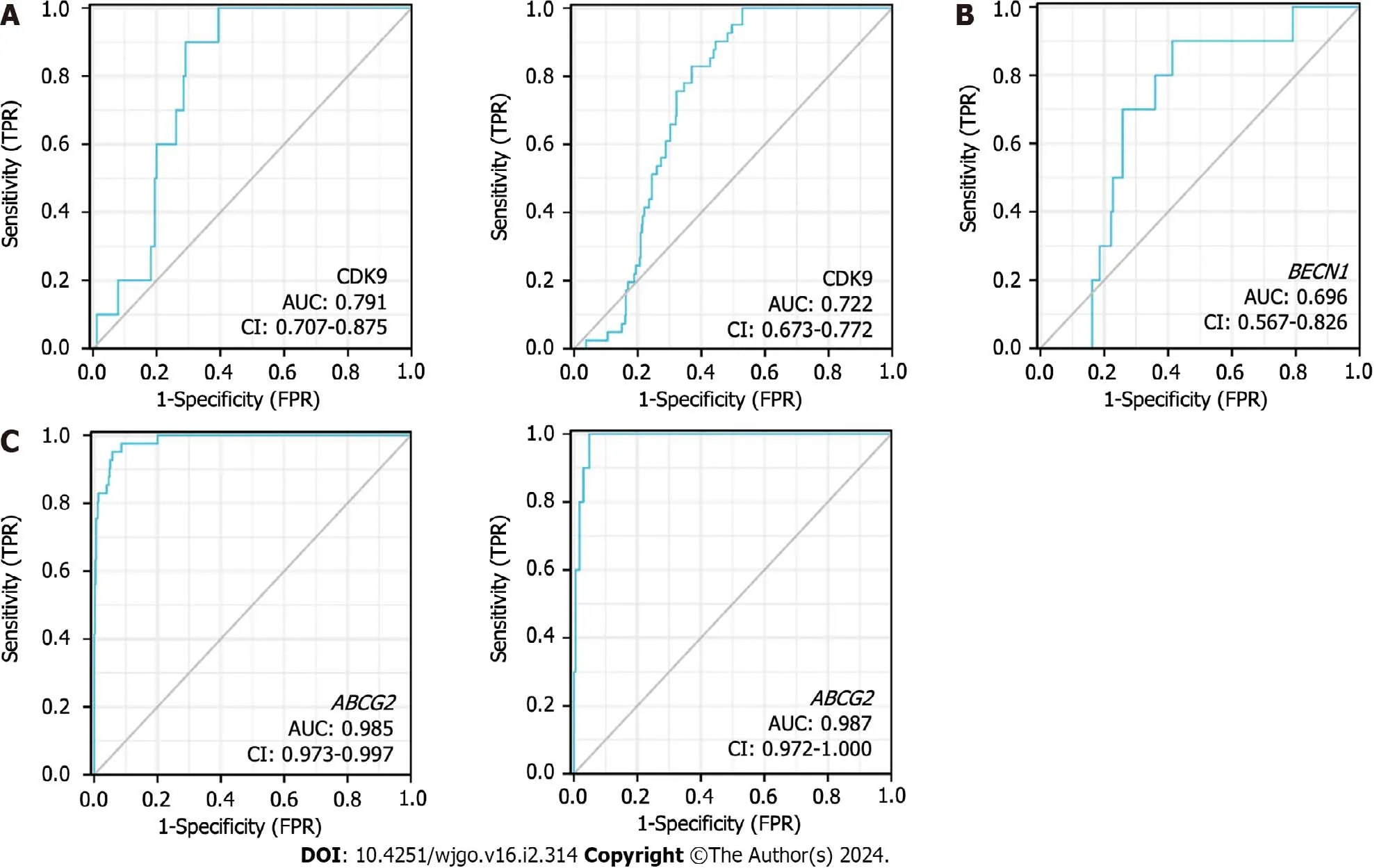
Figure 6 Receiver operating characteristic curves of colorectal cancer patients. A: Receiver operating characteristic (ROC) curves showed the accuracy of predicting the prognosis of rectal cancer (left figure) and colon cancer (right figure) by cyclin-dependent kinase 9 (CDK9) expression level;B: ROC curves showed the accuracy of the expression levels of BECN1 in predicting the prognosis of rectal cancer;C: ROC curves showed the accuracy of predicting the prognosis in colon cancer (left figure) and rectal cancer (right figure) by the expression levels of ABCG2.CDK9: Cyclin-dependent kinase 9;AUC: Area under the curve;FPR: False positive rate;TPR: True positive rate.
DlSCUSSlON
CDK9 is the catalytic kinase of p-TEFb,which is the key to the transcription elongation of most protein-coding genes[34].Studies have shown that CDK9 is highly expressed in CRC,breast cancer,prostate cancer,and lung cancer,and it isclosely related to the malignant degree of the tumors[35].In the early stage of tumorigenesis,autophagy serves as a survival approach and quality control mechanism to prevent the occurrence of tumors and inhibit the development of tumors.However,once the tumors are formed,autophagy can be subjected to environmental pressure,which contributes to the survival and growth of the formed tumors and promotes the invasion of the cancer by promoting metastasis[36].Few studies on the relationship between CDK9 and autophagy have been conducted.
CDK9 inhibitors promote dephosphorylation of SIRT1 and degradation of FOXO3,which is regulated by its acetylation,mediating the transcriptional repression ofFOXO3-drivenBNIP3and impairing theBNIP3-mediated stability of the PINK1 protein.Lysosomal degradation inhibitors cannot rescue mitophagy flux blocked by CDK9 inhibitors.Thus,CDK9 inhibitors inactivate theSIRT1-FOXO3-BNIP3axis andPINK1-PRKNpathway to subsequently block mitophagy initiation in hepatocellular carcinoma.CDK9 alongsidemTORcomplexes modulate translation.TORcomplexes have recently emerged as a potential target for autophagic regulation[37].This study confirmed that CDK9 is highly expressed in CRC and that expression of CDK9 is positively correlated with autophagy in colon cancer.
ABCG2plays a crucial role in drug transport and cellular defense mechanisms.In vitroexperiments have demonstrated that the expression ofABCG2can enhance cell survival and resistance to adverse conditions,such as amino acid starvation and radiation-associated cell death.These resistance mechanisms are closely associated withABCG2-dependent autophagy,a cellular process that helps maintain cell homeostasis and repair damage[27].The enhancement of autophagy depends on the overexpression ofABCG2[27].The positive rate of autophagy in right colon cancer was significantly higher than that in the left colon cancer.We also found that the expression level ofABCG2in right colon cancer was higher than that in the paracarcinoma tissue,while there was no significant difference between the expression level ofABCG2in left colon cancer and the paracarcinoma tissue (Figure 5B).The root cause of chemotherapy resistance in breast cancer is closely related to cancer stem cell subsets,and their activation depends on paclitaxel-promoted autophagy[38].It was found that rapamycin-induced autophagy could reduce the chemosensitivity of breast cancer cells to the XIAOPI formula[39].
High expression of the CDK9 marker represented a poor prognosis for patients with CRC.The expression CDK9 was upregulated in patients with papillary thyroid carcinoma,and it was associated with prognosis in these patients[40].High expression of CDK9 is an independent marker for the poor prognosis in osteosarcoma[41].CDK9 is also highly expressed in human ovarian cancer cell lines.Furthermore,an increase in CDK9 was significantly associated with poor prognosis in our CRC patients[10].
It was previously observed that there are differences between left and right colon cancers in molecular biological characteristics,morbidity,and response to targeted drug therapy[42].The disease-free survival rate of resected right colon cancer and left colon cancer after radical surgery is similar,but patients with metastatic left colon cancer have a longer survival rate than patients with right colon cancer after palliative chemotherapy[2].Some data indicate that the survival rate of right colon cancer patients has not improved since 1980 compared to left colon cancer patients,and the prognosis of these right colon cancer patients is worse[43].
It has been shown that autophagy plays a dual role in cancer: Autophagy activation can promote the survival of cancer cells (protective autophagy) or autophagy can lead to the death of cancer cells (cytotoxic/nonprotective autophagy)[43].The TEM results showed that the level of autophagy in the right colon cancer tissue was significantly higher than that in the left colon cancer tissue,which may be one of the reasons for the poor prognosis of right colon cancer compared with left colon cancer.The specific mechanism of the differences between left and right colon cancer needs further study.
CONCLUSlON
We found that the incidence of autophagy was different between left and right colon cancers with primary liver metastasis,and autophagy might be involved in the development of chemotherapy resistance.This study has laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of patients.The results from this study also show that tumor cell sensitivity to chemotherapy can be enhanced.
ARTlCLE HlGHLlGHTS
Research background
Cyclin-dependent kinase 9 (CDK9) expression and autophagy in colorectal cancer (CRC) tissues has not been widely studied.CDK9,a key regulator of transcription,may influence the occurrence and progression of CRC.The expression of autophagy-related genesBECN1and drug resistance factorABCG2may also play a role in CRC.
Research motivation
Under normal physiological conditions,autophagy can inhibit tumorigenesis,but once a tumor forms,autophagy may promote tumor growth.Therefore,understanding the relationship between autophagy and cancer,particularly how autophagy promotes tumor growth after its formation,is a key motivation for this research.
Research objectives
To investigate the relationship between CDK9 expression and autophagy in CRC,assess differences in autophagy between left and right colon cancers,and analyze the associations of autophagy-related genes with clinical features and prognosis.
Research methods
The research utilized a multi-faceted approach involving data analysis from The Cancer Genome Atlas database to study CDK9 expression in CRC.We also collected fresh tumor and paracarcinoma tissues for electron microscopy,immunohistochemistry,and flow cytometry to investigate autophagy.Additionally,the study analyzed the expression of autophagyrelated genesBECN1andABCG2and their correlation with clinical features and prognosis.
Research results
CDK9 expression was increased in CRC tissues.CDK9 expression positively correlated with the level of autophagy in colon cancer,indicating a potential link between CDK9 and autophagy in cancer progression.The incidence of autophagy was also found to be significantly higher in right colon cancer compared to left colon cancer with primary liver metastasis.Right colon cancer displayed a higher expression ofABCG2,suggesting a potential mechanism for the observed differences in prognosis between left and right colon cancers.High expression of CDK9 was found to be associated with a poor prognosis in CRC,affecting overall survival,disease-specific survival,and progression-free interval.The expression of CDK9 was linked to specific clinical features such as tumor T-stage and lymph node invasion.Finally,high expressions ofABCG2andBECN1were also associated with specific clinical features,and expression levels ofABCG2had a high predictive accuracy for CRC prognosis.
Research conclusions
CDK9 expression positively correlated with the level of autophagy in colon cancer.Thus,this study has laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of tumor patients.
Research perspectives
This study emphasized the significance of patient-specific treatment approaches in CRC cancer,highlighting the role of CDK9,autophagy,and drug resistance factors in prognosis.It underscored the need for combination therapies and further research to improve cancer treatment outcomes.
ACKNOWLEDGEMENTS
This work was supported by Cancer Biobank of Tianjin Medical University Cancer Institute and Hospital.
FOOTNOTES
Co-first authors:Lei Zheng and Jia Lu.
Author contributions:Kong DL conceived and designed the study;Zheng L provided study materials and collected and assembled data;Lu J analyzed data and prepared figures;Zheng L and Lu J wrote the manuscript and contributed equally to the study and as such are co-first authors;All authors read and approved the final manuscript.
Supported bythe Science and Technology Development Fund of Tianjin Education Commission for Higher Education,No.2020KJ133;and Tianjin Key Medical Discipline (Specialty) Construction Project,No.TJYXZDXK-009A.
lnstitutional review board statement:The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Tianjin,China) (Approval No.Ek2019085).All methods were performed in accordance with relevant guidelines and regulations.
Clinical trial registration statement:Our study is purely observational in nature and does not involve any intervention.As such,it falls under the category of observational research rather than interventional research.Consequently,there was no clinical trial registration undertaken for this study.
lnformed consent statement:Written informed consent for publication was obtained from all participants.
Conflict-of-interest statement:All the authors report no relevant conflicts of interest for this article.
Data sharing statement:The data that support the findings of this study are openly available in The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository?facetTab=cases&filters=%7B%22op%22%3A%22and%22%2C%22content%22%3A%5B%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.project.project_id%22%2C%22value%22%3A%5B%22TCGA-COAD%22%2C%22TCGA-READ%22%5D%7D%7D%5D%7D).Data from clinical samples were not uploaded to the database.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is non-commercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORClD number:Da-Lu Kong 0000-0002-1906-0981.
S-Editor:Li L
L-Editor:A
P-Editor:Zhang XD
 World Journal of Gastrointestinal Oncology2024年2期
World Journal of Gastrointestinal Oncology2024年2期
- World Journal of Gastrointestinal Oncology的其它文章
- Does enhanced recovery after surgery programs improve clinical outcomes in liver cancer surgery?
- Cardiotoxicity induced by fluoropyrimidine drugs in the treatment of gastrointestinal tumors
- Effect of screening colonoscopy frequency on colorectal cancer mortality in patients with a family history of colorectal cancer
- Preoperative controlling nutritional status as an optimal prognostic nutritional index to predict the outcome for colorectal cancer
- Tumour response following preoperative chemotherapy is affected by body mass index in patients with colorectal liver metastases
- Progress in the treatment of advanced hepatocellular carcinoma with immune combination therapy
