Development of genomic resources for Wenchengia alternifolia(Lamiaceae) based on genome skimming data
Qi-Yue Zhou , Hui-Xi Ci , Zi-Hn Liu , Lng-Xing Yun , Lei Yng , Tuo Yng ,d,Bo Li , Pn Li ,*
a Laboratory of Systematic & Evolutionary Botany and Biodiversity, College of Life Sciences, Zhejiang University, Hangzhou, 310058, China
b Shanghai Key Laboratory of Plant Functional Genomics and Resources, Shanghai Chenshan Botanical Garden, Shanghai, 201602, China
c Haikou Duotan Wetlands Institute, Haikou, 570100, China
d Orchid Conservation & Research Center of Shenzhen, Shenzhen, 518114, China
e Research Centre of Ecological Sciences, College of Agronomy, Jiangxi Agricultural University, Nanchang, 330045, China
Keywords:Wenchengia Plastid hotspot Simple sequence repeat (SSR)Single nucleotide polymorphism (SNP)
A B S T R A C T Wenchengia alternifolia (Lamiaceae), the sole species of the genus Wenchengia is extremely rare and is currently listed as Critically Endangered(CR)on the IUCN Red List.The species had long been considered endemic to Hainan Island, China and was once believed to be extinct until a small remnant population was rediscovered at the type locality in 2010. Four more populations were later found on Hainan and in Vietnam. In order to develop genomic resources for further studies on population genetics and conservation biology of this rare species,we identified infraspecific molecular markers in the present study,using genome skimming data of five individuals collected from two populations on Hainan Island and three populations in Vietnam respectively. The length of plastome of the five individuals varied from 152,961 bp to 150,204 bp,and exhibited a typical angiosperm quadripartite structure.Six plastid hotspot regions with the Pi > 0.01 (trnH-psbA, psbA-trnK, rpl22, ndhE, ndhG-ndhI and rps15-ycf1), 1621 polymorphic gSSRs, as well as 1657 candidate SNPs in 237 variant nuclear genes were identified, thereby providing important information for further genetic studies.
1. Introduction
Wenchengia alternifolia C.Y. Wu & S. Chow (Lamiaceae) was described on the basis of two gatherings collected on Hainan Island,southern China, in the 1930s (Wu and Chow, 1965). It is characterized by having alternate leaves, racemose inflorescences, and a unique type of nutlet attachment described as vascular funicles with slender stalks (Wu and Chow,1965; Li et al., 2012). Because these traits are rare or unique in the Lamiaceae, Wu and Chow(1965) established the monotypic genus, Wenchengia C.Y. Wu & S.Chow, and the novel subfamily, Wenchengioideae C.Y. Wu & S.Chow,to accommodate it.W.alternifolia was believed to be extinct(Harley et al., 2004).
The phylogenetic position of Wenchengia has been controversial due to the unusual morphological traits and the lack of available materials for further studies. In 2010, a small population was rediscovered at the type locality, Shuangximu Valley,Wanning County (Fig.1), Hainan Province, China. Using materials from this population, Li et al. (2012) inferred the phylogenetic position of W. alternifolia using comprehensive evidence from molecular, morphological, anatomical and cytological data to confirm that Wenchengia is the first diverging lineage of the subfamily Scutellarioideae. The finding was further supported by phylogenomic analyses using plastome data (Zhao et al., 2020,2021).
Li et al. (2014) assessed the conservation status of W. alternifolia based on the rediscovered population and proposed that it be listed as Critically Endangered (CR) under the IUCN Red List Criteria. Fragmentation of habitats and persistent disturbance caused by the rapid development of urbanization, mining, plantations and tourism in Wanning County were thought to be a threat to the survival of W. alternifolia in the wild (Li et al., 2014).In 2017, a second larger population was discovered by Chunlei Xiang in Ding'an County(personal communication;Fig.2),Hainan Province. The individuals in this population differed morphologically from the population in Wanning County, but it was unclear whether the variance was due to genetic or environmental differences.

Fig.1. Wenchengia alternifolia population at Wanning, Hainan, China. A. habitat, B. plant, C. stem, D. leaves, E. flowers.
Wenchengia alternifolia has long been considered endemic to Hainan Island (Harley et al., 2004;Li et al., 2012).However,three old gatherings of W. alternifolia from Vietnam were found in the herbarium of the Mus′eum national d'Histoire naturelle in Paris(P) and in the herbarium at the Royal Botanic Gardens, Kew (K)(Paton et al., 2016). In the next year, three extant populations of W. alternifolia were discovered in Vietnam (Fig. 3) by Bo Li and his team. Thus, W. alternifolia probably had a wider distribution in the past. The species should be given high priority for conservation due to its distinct phylogenetic position and the limited number of known populations. Although previous studies have used plastomes to reconstruct phylogenetic relationships within the Scutellarioideae, including Wenchengia(Zhao et al., 2020), more information on the genomes and effective intraspecific molecular markers of W. alternifolia that could be used for population genetics and DNA barcoding were unavailable.
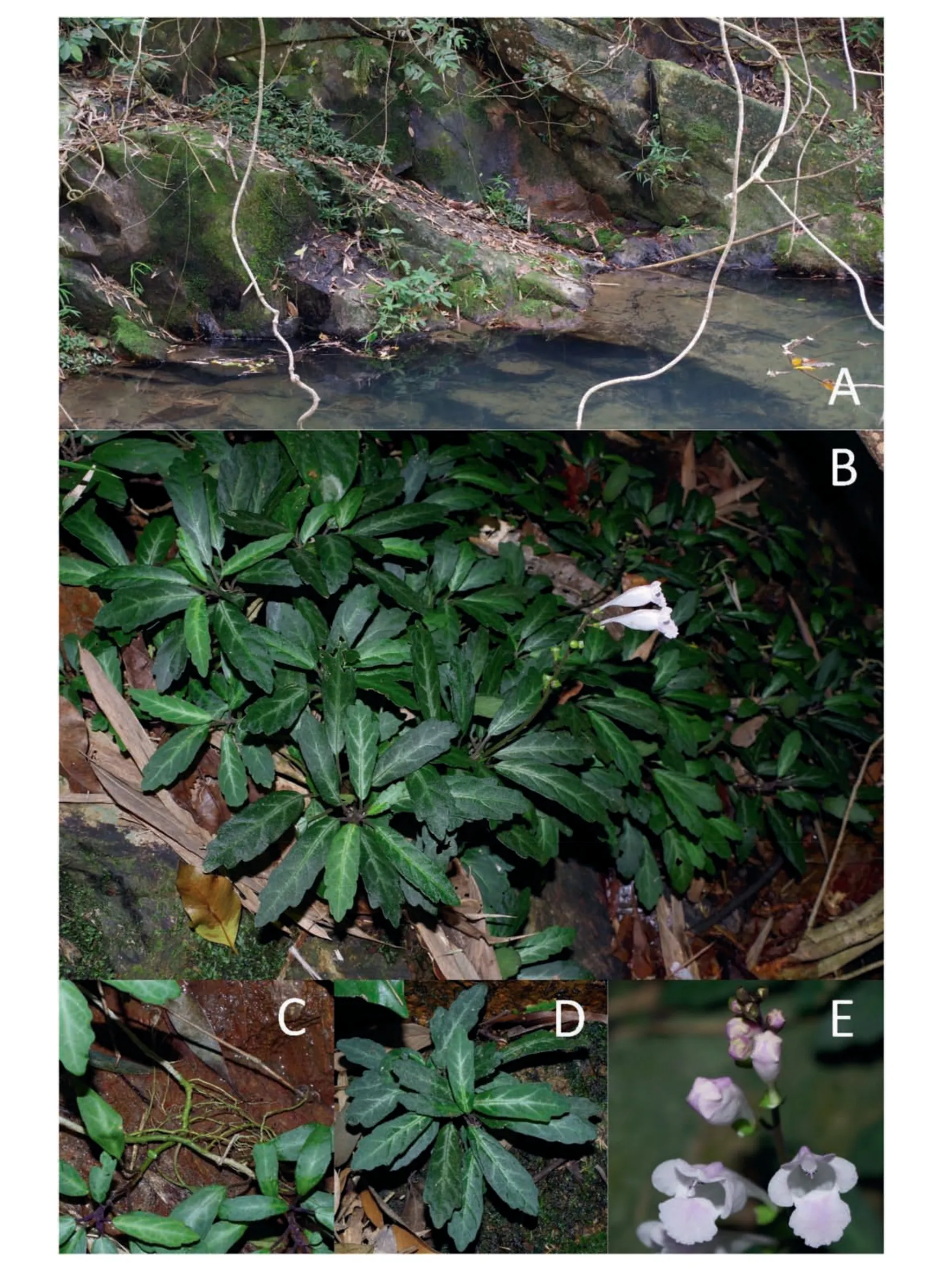
Fig. 2. Wenchengia alternifolia population at Ding'an, Hainan, China. A. habitat, B. plant, C. stem, D. leaves, E. flowers.

Fig. 3. Wenchengia alternifolia populations in Vietnam. Plants (A, C, E) and flowers (B, D, F, G). A-B. Hue Province, C-D. Da Nang Province, E-G. Quang Nam Province.
DNA barcoding and relative approaches can not only provide valuable tools for phylogenetic reconstruction and species identification(Calonje et al.,2009;Qin et al.,2020),but also offer insight into studies below the species level,including recognition of infraspecific variation, patterns of gene flow and habitat differentiation (Bockelmann et al., 2003; Kane et al., 2012;St?hlberg, 2007). Up to now, many fragments of intergenic spacers, coding regions and introns of plastomes have been widely used for DNA barcoding, but whether these regions are useful in closely related species or lower taxonomic levels is unclear(Dong et al., 2012;Hollingsworth et al.,2011;Kress et al.,2005). Therefore, searching for additional plastid hotspots with higher evolutionary rates and divergence is important. Simple sequence repeats (SSRs), containing 1-6 bp repetitive sequences,are widely used in genetic studies. To detect higher levels of polymorphism,genomic SSR(gSSR)markers have recently gained more attention comparing to EST-SSRs because intergenic spacers or intron contain more variations than extron sequences(Bae et al., 2015; Blair et al., 2006; Liu et al., 2021). In addition,targeted sequence capture is an efficient and cost-effective approach for generating phylogenomic data sets after the boost of high-throughput sequencing technology. A set of probes was designed from 353 putative single-copy nuclear genes (SCNGs)over 600 angiosperms to hybridize with template sequences and to capture target genes for subsequent analyses. It would be effective for phylogenetic studies at various taxonomic levels from species to the entire angiosperm clade (Johnson et al.,2019). The polymorphism detected within those genes, in combination with plastid hotspots and gSSR markers, will provide useful molecular tools for further studies on population genetics and evolutionary history.
In this study, we aim to identify plastid hotspots, polymorphic gSSRs,and SCNGs within Wenchengia alternifolia based on genome skimming data.Our findings will provide powerful tools for future studies on its conservation biology and population genetics.
2. Materials and methods
2.1. Plant material, DNA extraction and sequencing
Fresh leaf materials of five individuals of Wenchengia alternifolia from the two populations in Hainan Province (Pan Li & Langxing Yuan LP197753, Waning, Hainan, China; Pan Li & Langxing Yuan LP197754, Ding'an, Hainan, China) and three populations in Vietnam(Bo Li LB0647,Hue,Vietnam;Bo Li LB0824,Da Nang,Vietnam;Bo Li LB0938, Quang Nam, Vietnam) were dried in silica gel.Voucher specimens were deposited in the herbarium of Zhejiang University (HZU) and the herbarium of Jiangxi Agricultural University (JXAU). Total DNA was extracted from 3 mg of leaf tissue using the DNA Plantzol Reagent (Shanghai, China) and following the manufacturer's protocol. The quality and quantity of the genomic DNA was checked on an Agilent BioAnalyzer 2100(Agilent Technologies). After sample quality check, the genomic DNA was fragmented by ultrasound on a Covaris E220 (Covaris, Brighton,UK). Fragments 300~500 bp long were selected using Pippin Prep(Sage Science,Beverly,MA,USA).The selected DNA fragments were then repaired to obtain a blunt end and modified at the 3′end to obtain a dATP sticky end. The dTTP tailed adaptor was ligated to both ends of the DNA fragments. The ligation product was amplified by PCR and circularized to produce a single stranded circular(ssCir)library.The ssCir library was then amplified through rolling circle amplification(RCA)to obtain a DNA nanoball(DNB).The DNB was then loaded to a flowcell, and sequenced on a DNBSEQ Platform (Drmanac et al., 2010).
2.2. Genome assembly and annotation
The quality check of the raw reads was carried out in FastQC,and reads with Phred score <30 (0.001 probability error) were discarded.The resulting clean reads were used for de novo assembly of the plastomes through the GetOrganelle v1.6.2 pipeline (Jin et al.,2020). The complete plastomes were annotated according to reference genome downloaded from NCBI (GenBank accession number: MN128379; MN128378) in Geneious Prime (2020).0.5(Kearse et al.,2012).The putative starts,stops,and intron positions were identified by comparison with homologous genes of the reference genome (Zhao et al., 2020). The graphical map of the annotated circular plastomes was drawn using the OrganellarGenomeDRAW program(OGDRAW, Lohse et al., 2013).
To improve the efficiency of assembling, the overly large sequencing dataset of Wenchengia alternifolia, 8 Gb of WN and DN were sampled using Seqkit v0.14.0 and assembled into contigs in SPAdes v3.13.0 with default settings (Bankevich et al., 2012). All clean reads of each Vietnam individuals (~8 Gb) were directly processed in the pipeline to generate contigs.Contigs quality were assessed by QUAST v5.0.2 (Gurevich et al., 2013).
2.3. Genomic resources development based on Wenchengia alternifolia plastomes
Five plastomes were multiple aligned using Mafft Multiple Alignment plugin v1.4.0 (Katoh and Standley, 2013) in Geneious Prime. Variable regions more than 200 bp long were selected and generated in DnaSP 6 (Rozas et al., 2017) to calculate the total number of mutation (Eta) and average number of nucleotide differences (K), which were used to determine nucleotide diversity(Pi).
2.4. Genomic resources development based on Wenchengia alternifolia nuclear genomes
2.4.1. Polymorphic gSSRs identification
The contigs generated by SPAdes were aligned to the plastome sequence assembled by GetOrganelle and the mitochondrial DNA scaffolds of Scutellaria amoena C.H. Wright (GenBank accession number: MT277281.1, MT277264.1, MT277230.1, MT277181.1)downloaded from NCBI. Using BLAST search (BLAST v2.11.0), we were able to remove the plastome and mitogenome sequences.Then we discarded sequences less than 300 bp long. Candidate polymorphic gSSRs within W. alternifolia were identified based on the remanent multiple assembled sequences using the CandiSSR v20170602 software set at default parameters(Xia et al.,2016).For each target SSR,primers were automatically designed based on the Primer3 package built-in installation to the pipeline(Untergasser et al., 2012).
2.4.2. Putative single-copy genes and single nucleotide polymorphism identification
To capture targeted genes and analyze single nucleotide polymorphism(SNP)within them,we searched through the clean reads for each individual using HybPiper v1.3.1(Johnson et al.,2016)with default settings. 353 putative single-copy nuclear genes from 42 angiosperms (Johnson et al., 2019) were used as probes in the pipeline. Sequences captured for the same gene in at least two individuals were multiple aligned using the Mafft plugin in Geneious Prime to identify SNPs.To improve the accuracy and reliability of the results,we discarded sequences with a gene recovery rate of less than 25% and abandoned the polymorphism detected in sequence regions with an alignment identity of less than 80%.
3. Results
3.1. Genome assembly and features
Five complete plastome sequences were successfully constructed with no Ns or gaps in the GetOrganelle pipeline. Genome sizes varied from 150,204 bp in Wenchengia alternifolia DN to 152,961 bp in W.alternifolia DA.The plastome sequences have been submitted to GenBank (accession numbers shown in Table 1). All plastomes exhibited a quadripartite structure similar to other angiosperms(Li et al.,2017;Liu et al.,2017),comprising of two copies of IR regions separated by a LSC region and an SSC region (Fig. 4,Table 1).The GC content overall and in different regions was similar among five individuals.Plastomes of W.alternifolia WN,DA and QN contained 114 unique genes,including 80 protein-coding genes,30 tRNA genes, and 4 rRNA genes with 18 genes duplicated in the IR regions,while plastomes of W.alternifolia H and DN lost the protein coding gene ndhF and contained only 113 unique genes (Table 2).The difference in length of the five plastomes was mainly due to the loss of the ndhF gene in W. alternifolia DN and H.
A total of 129,404(W.alternifolia WN),126,341(DA),110,315(H)117,076(DN),128,838(QN)contigs with a length of more than 500 bp were generated after whole genome de novo assembly using SPAdes. The assembly results for each individual are shown in Table 3.

Fig.4. Plastome map of Wenchengia alternifolia.Genes inside circle are transcribed clockwise,gene outside are transcribed counter-clockwise.Light gray inner circle corresponds to AT content; dark gray to GC content. Genes belonging to different functional groups are shown in different colors; see legend for groups.
3.2. Development of genomic resources for Wenchengia alternifolia
3.2.1. Divergence hotspots in the plastomes of Wenchengia alternifolia
The divergence hotspots in the plastomes can provide effective phylogenetic information and serve as DNA barcodes.We screened 127 loci (48 inter-genic spacers, 62 coding genes, and 17 intron regions)with a length greater than 200 bp in the plastomes of five individuals of W.alternifolia(Fig.5).Nucleotide diversity(Pi)values for each locus ranged from 0.000213 (rrn23 gene) to 0.020896(ndhG-ndhI region).Six of the variable loci with Pi>0.01,including four inter-genic spacers (trnH-psbA, psbA-trnK, ndhG-ndhI and rps15-ycf1) and two coding genes (rpl22 and ndhE), showed relatively high nucleotide diversity values. Among the variable region above, Pi value of trnH-psbA and ndhG-ndhI was more than 0.015.All of them can be used as highly informative molecular markers for W. alternifolia.
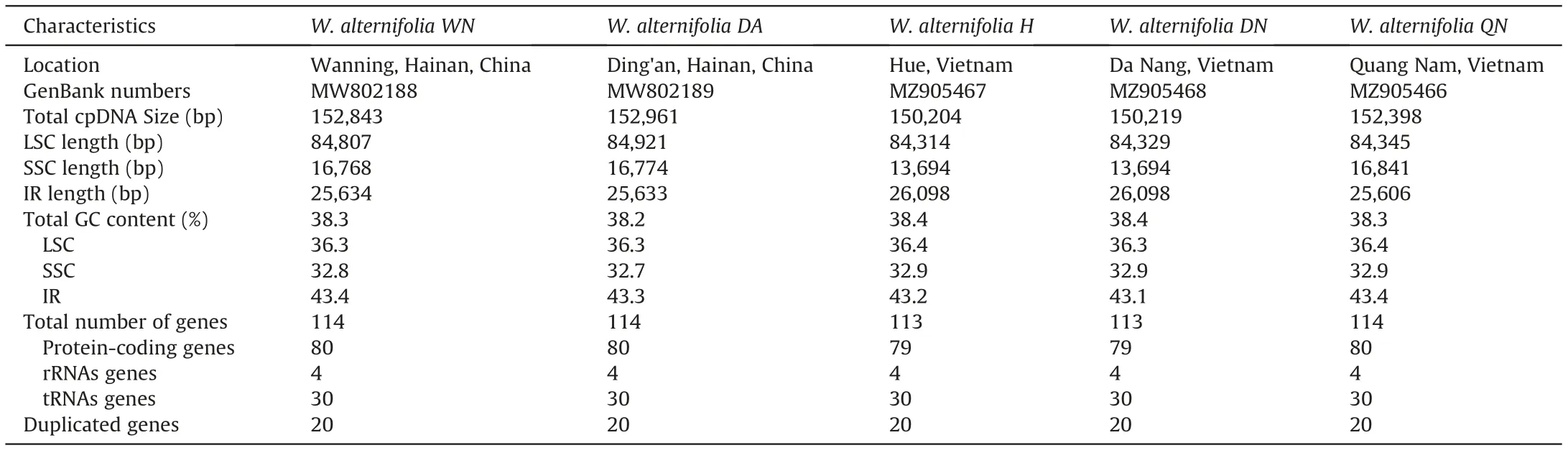
Table 1 Summary of five plastomes of Wenchengia alternifolia.
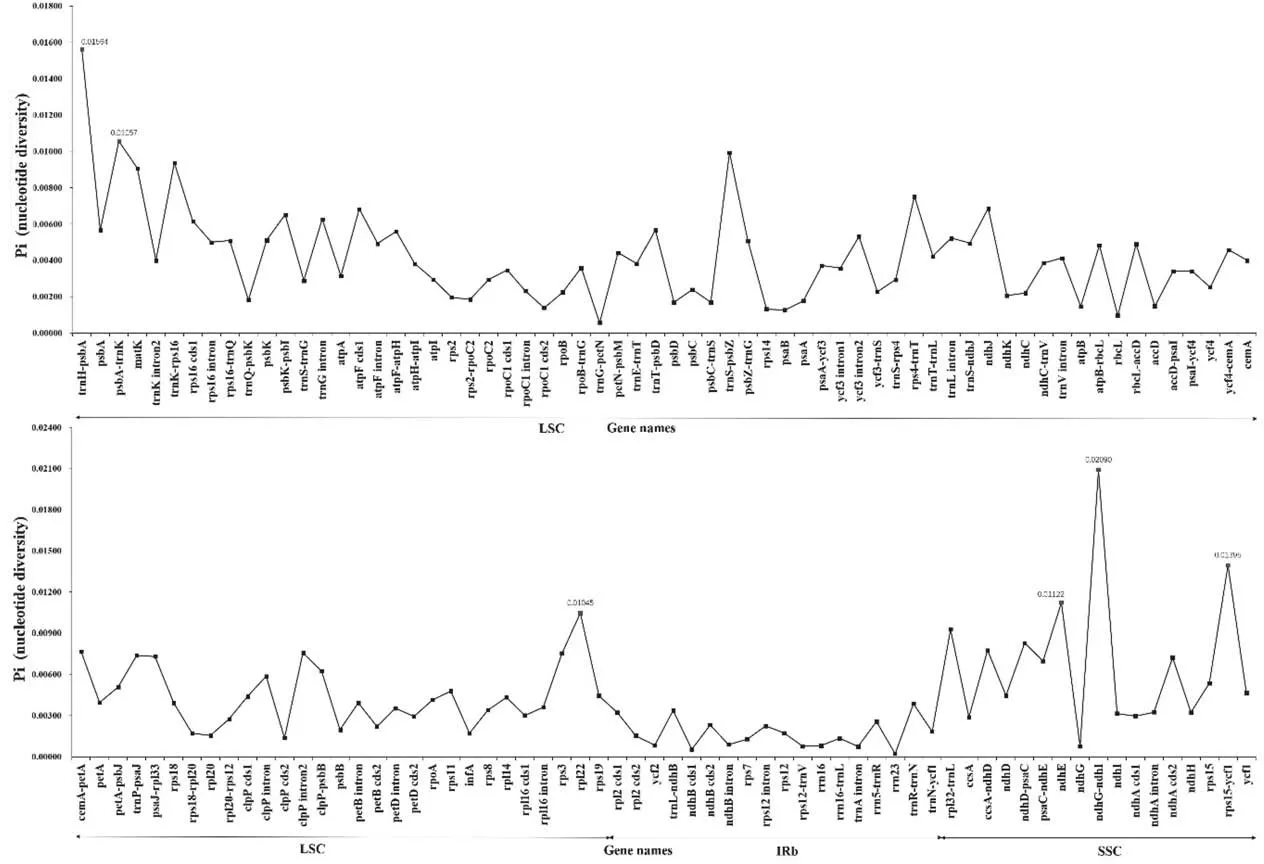
Fig. 5. Comparative analysis of nucleotide variability (Pi) values within Wenchengia alternifolia plastome. Regions surpassing the threshold (Pi > 0.01) are highlighted in red.
3.2.2. Genomic SSR (gSSR) markers
Within Wenchengia alternifolia,20,953 candidate polymorphic gSSRs were identified.After selecting the loci identified in all five individuals and discarding loci with either no available primers or sequence similarity <99%, we obtained 1621 polymorphic gSSRs with a standard deviation between 0.4 and 3.32 (Table S1;Table S2). Among the polymorphic gSSRs, di- (1197), tri- (362),tetra- (52), penta- (9) and hexanucleotides (1) accounted for 73.84%, 22.33%, 3.21%, 0.56% and 0.06%, respectively (Fig. 6). The polymorphism within W. alternifolia for each type of gSSRs is shown in the results of the standard deviation of the repeats(Table S3).
3.2.3. Putative single-copy genes and SNPs
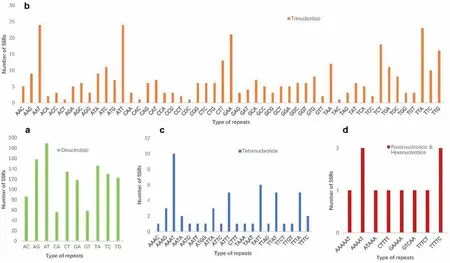
Fig. 6. Distribution of polymorphic genomic simple sequence repeats (gSSRs) for Wenchengia alternifolia. (a), (b), (c) and (d) represent di-, tri-, tetra-, penta- and hexanucleotide repeats, respectively.
Clean reads of five individuals were generated in the HybPiper pipeline to identify putative single-copy genes. Among all the genes recovered in two Hainan individuals (W. alternifolia WN &W. alternifolia DA), 220 and 229 sequences were at least 75% as long as the target gene. Only 9 and 17 sequences were less than 25%as long.In the individuals from Vietnam(W.alternifolia H,W.alternifolia DN and W. alternifolia QN), only 66, 51 and 39 sequences were more than 25% as long (Table S4). 237 genes were recovered in at least two of the five individuals, among which only 23 genes were recovered in all five individuals.Within those 237 genes, 1657 candidate SNPs were identified. Some of the single copy genes were specifically polymorphic, containing more than 35 SNPs (Table S5).
4. Discussion
4.1. Development of genomic resources in Wenchengia alternifolia
Plastid hotspots are plastome regions containing relatively more variations.The average Pi in all variable loci was 0.004.We selected the six most variable loci with a Pi>0.01,including four inter-genicspacers (trnH-psbA, psbA-trnK, ndhG-ndhI and rps15-ycf1) and two coding genes (rpl22 and ndhE), which can be used as molecular markers in population genetic studies. All of these loci exist in the LSC and SSC regions. As shown in Fig. 5, the average nucleotide diversity in IR is lower than in LSC and SSC. TrnH-psbA is highly divergent in many plant groups. Along with rps15-ycf1, it was also identified as a hyper-variable regions among 11 species of Scutellaria(Zhao et al.,2020).NdhG-ndhI has the highest Pi of 0.021,but it is rarely divergent in other plants. In addition, deletions and insertions are evident in the plastome,including the loss of the ndhF gene.

Table 3 Summary of whole genome de novo assembly in Wenchengia alternifolia.
The CandiSSR pipeline has been used in many studies to develop abundant intergeneric and intrageneric polymorphic gSSR markers that can be successfully amplified in target populations. We developed 1621 candidate polymorphic gSSRs markers in W. alternifolia, which not only have available primers but are also clear of their polymorphic status.Most of the polySSRs are di-and tri-nucleotides.The mean standard deviation of di-nucleotide SSRs is higher than in tri- and tetra-nucleotides, but they also vary greatly among different single SSRs.
The gene recovery rate in individuals of W. alternifolia from Hainan was higher than the average level and much higher than in individuals from Vietnam (Johnson et al., 2019; Li et al., 2014),perhaps because the sequencing depth was much higher, with 23 Gb clean reads of each individual in total. Although few genes were recovered in all five individuals, in at least two individuals,237 genes were recovered that contained adequate number of candidate SNPs that would be useful for genotyping and for other population genetics studies.
4.2. Polymorphism in genomes and morphology
W. alternifolia is a rheophyte, a riparian plants exposed to frequent submergence and strong currents during sporadic flooding events after heavy rain(Li et al.,2014;van Steenis,1987).Plastid hotspots in other plants seldom contain protein coding genes.353 putative single-copy nuclear genes used as targets in this study are suspected of being conserved in most angiosperms. However,polymorphisms in the plastome coding genes and in the recovered nuclear genes of W.alternifolia were relatively high,which might be due to long-term isolation and/or to environmental selection pressures(Mitsui and Setoguchi,2012).The sampled populations in Hainan and Vietnam show significant variation in morphology and habitat (Figs. 1-3), which calls for further studies on population genetics and adaptive evolution.
5. Conclusions
In this study, five individuals from distinct populations of W. alternifolia were sampled for genome skimming. Using these data, we assembled complete plastomes and characterized the plastid hotspots, polymorphic gSSRs, low or single copy gene fragments and candidate single nucleotide polymorphisms(SNPs).The rich genomic information presented here will be available for further studies on the population genetics, local adaptation and conservation biology of the critically endangered Wenchengia alternifolia.
Author contributions
PL,BL and LY conceived the ideas;PL and LXY contributed to the sampling; QYZ, HXC, ZHL and TY performed the experiments and analyzed the data.The manuscript was written by QYZ, BL and PL,then revised by all the other authors.
Data availability
The raw reads that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0001750.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (grant number 31970225 and 31900181), the Zhejiang Provincial Natural Science Foundation (grant number LY19C030007) and the Open Fund of Shanghai Key Laboratory of Plant Functional Genomics and Resources(PFGR202104).We thank Dr.Enhua Xia,Ms.Meizhen Wang and Dr.Xinjie Jin for helping with the analyses,Dr.David E.Boufford for revising the manuscript.We thank the Guangdong Provincial Academician Workstation of BGI Synthetic Genomics, BGI-Shenzhen, Guangdong, China and the China National GeneBank for producing the sequencing data. We are grateful to anonymous reviewers for the constructive comments.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.09.006.
- 植物多樣性的其它文章
- Plastome characteristics and species identification of Chinese medicinal wintergreens (Gaultheria, Ericaceae)
- Molecular cytogenetic study on the plants of Elymus nutans with varying fertility on the Qinghai-Tibet Plateau
- Seedling survival after simulating grazing and drought for two species from the Pamirs, northwestern China
- Borana rangeland of southern Ethiopia: Estimating biomass production and carrying capacity using field and remote sensing data
- An ethnobotanical study of medicinal plants in Güce district,north-eastern Turkey
- Does the critically endangered Rhododendron amesiae deserve top priority for conservation?

