Transcriptome Sequencing Provides Evidence of Genetic Assimilation in a Toad-Headed Lizard at High Altitude
Weizhao YANG ,Tao ZHANG ,Zhongyi YAO ,Xiaolong TANG and Yin QI*
1Chengdu Institute of Biology,Chinese Academy of Sciences,Chengdu 610041,Sichuan,China
2Department of Biology,Lund University,Lund 22362,Sweden
3Institute of Biochemistry and Molecular Biology,School of Life Science,Lanzhou University,Lanzhou 730000,Gansu,China
Abstract Understanding how organisms adapt to the environment is a compelling question in modern evolutionary biology.Genetic assimilation provides an alternative hypothesis to explain adaptation,in which phenotypic plasticity is first triggered by environmental factors,followed by selection on genotypes that reduce the plastic expression of phenotypes.To investigate the evidence of genetic assimilation in a high-altitude dweller,the toad-headed agama Phrynocephalus vlangalii,we conducted a translocation experiment by moving individuals from high-to low-altitude environments.We then measured their gene expression profiles by transcriptome sequencing in heart,liver and muscle,and compared them to two low-altitude species P.axillaris and P.forsythii.The results showed that the general expression profile of P.vlangalii was similar to its viviparous relative P.forsythii,however,the differentially expressed genes in the liver of P.vlangalii showed a distinct pattern compared to both the lowaltitude species.In particular,several key genes (FASN,ACAA2 and ECI2) within fatty acid metabolic pathway were no longer differentially expressed in P.valgnalii,suggesting the loss of plasticity for this pathway after translocation.This study provides evidence of genetic assimilation in fatty acid metabolism that may have facilitated the adaptation to high-altitude for P.vlangalii.
Keywords genetic assimilation,gene expression,plasticity,Phrynocephalus vlangalii
1.Introduction
Elucidating the mechanisms of adaptation is a compelling question in modern evolutionary biology (Rose,2001;Smith and Eyre-Walker,2002).In the past decades,a “mutation-first evolution” model was widely accepted to explain the process of adaptation,in which mutations linked to fit phenotypes are under natural selection,resulting in changes in phenotype frequencies,and ultimately,adaptation (Carroll,2008).However,a “plasticity-first evolution” model has also been proposed in which phenotypic plasticity is first triggered by environmental factors,followed by selection on genotypes influencing the plastic expression of phenotypes through genetic accommodation (Moczeket al
.,2011;Jones and Robinson,2018).Genetic assimilation is a special case of such process,where the initially plastic phenotypes are gradually fixed,leading to reduced phenotypic plasticity and only adaptive phenotypes,even without environmental stimuli (Waddington,1953;Pigliucci,2006).Although a number of studies have shown evidence of genetic assimilation,such as cases inDrosophila melanogaster
(Dworkin,2005;Ghalamboret al
.,2010) andSpea bombifrons
(Levis and Pfennig,2019),the role of genetic assimilation in adaptive evolution remains unclear and controversial.Gene expression profiles provide an excellent tool to investigate genetic assimilation in evolution.In general,phenotypic plasticity springs from environmentally sensitive regulation of gene expression,which alters the profiles of gene expression (Colinet and Hoffmann,2012;Beamanet al
.,2016).As plastic phenotypes may evolve in response to selection with increased or decreased levels of plasticity,the gene expression associated with the phenotypes may evolve as well (Renn and Schumer,2013).Thus,variable patterns of gene expression mirror the evolution of phenotypic plasticity.In the case of genetic assimilation,plastically expressed genes in response to the environment may finally become fixed (Scoville and Pfrender,2010;Renn and Schumer,2013).Previous studies that have used gene expression profiles as a tool to investigate the pattern of genetic assimilation have mainly focused on eurociality in the honeybee (Tothet al
.,2007;Bloch and Grozinger,2011);host specialization inDrosophila
(Matzkinet al
.,2006;Matzkin,2012);and character displacement in spadefoot toads (Leviset al
.,2017).Toad-headed agamas (genusPhrynocephalus
) at the Qinghai-Tibetan Plateau (Huey,1982;Zhaoet al
.,1982) provide an excellent model system to study genetic assimilation.As true dwellers of high-altitude environments,as high as 5 300 m above sea level (a.s.l),this group has experienced a long-term adaptive evolution under several extreme stressors,including hypobaric hypoxia,low ambient temperatures,and strong UV radiation (Scheinfeldt and Tishkoff,2010;Cheviron and Brumfield,2011).Phylogenetic studies indicated that a total of six high-altitude species in this genus formed a monophyletic viviparous clade,including another low-altitude species,P.forsythii
(Figure 1A;Guoet al
.,2002;Guo and Wang,2007;Wanget al
.,2014).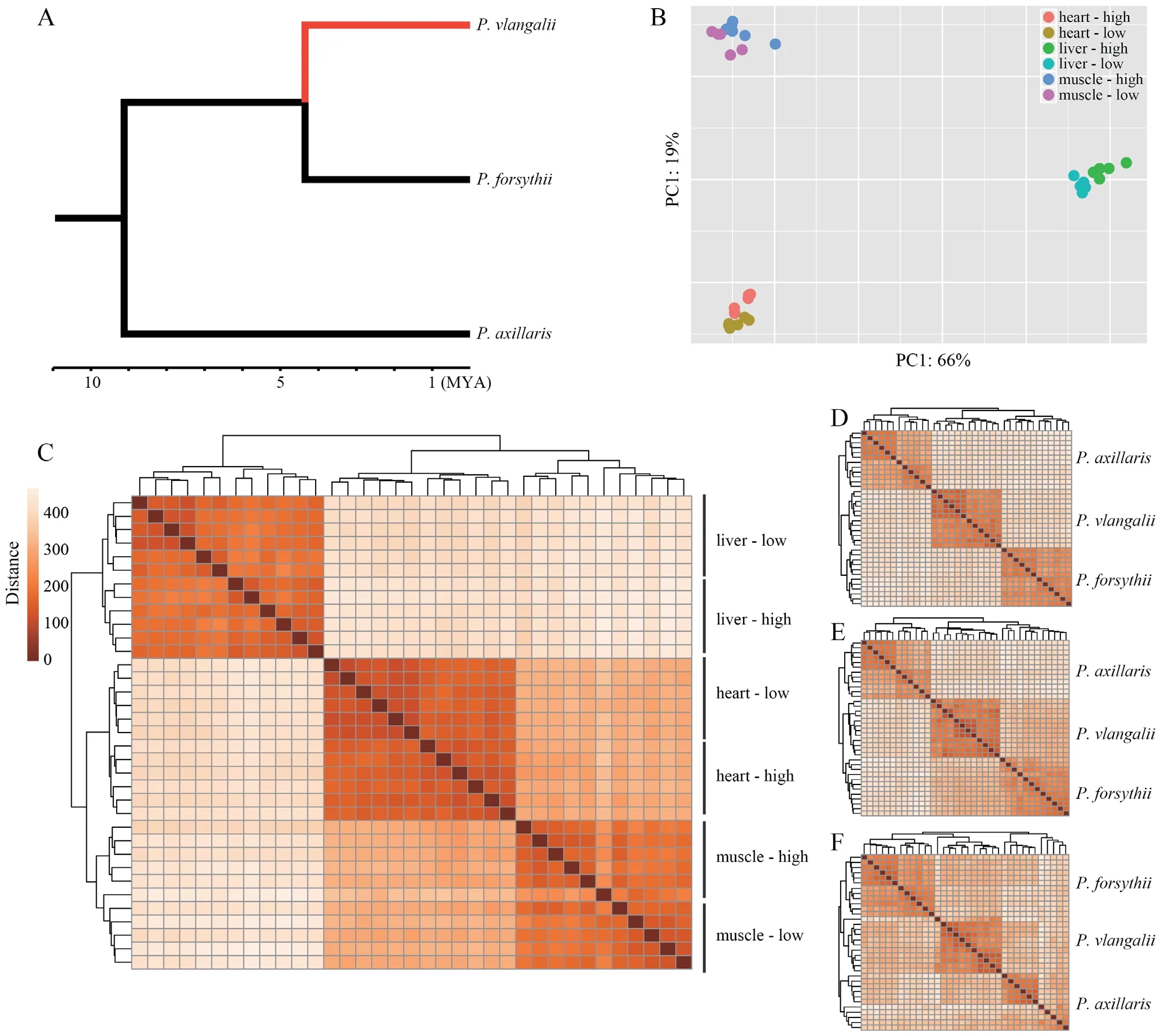
Figure 1 Translocation experiment and transcriptome sequencing.A:The phylogenetic relationship among Phrynocephalus axillaris,P.forsythii,and P.vlangalii.The red branch indicates the viviparous high-altitude clade leading to P.vlangalii.B:Principal component analysis (PCA) plot for gene expression profiles.C:Heatmap for gene expression profiles among P.vlangalii samples.D,E,and F:Heatmap for gene expression profiles among the three species for heart,liver,and muscle,respectively.In all the comparisons,P.vlangalii was clustered with P.forsythii,consistent with their phylogenetic relationship.
High-altitude toad-headed agamas have evolved a series of characteristics that underline their adaptations to extremely high-altitude environments.At DNA sequence level,a couple of genes have been identified with signature of positive selection associated with energy metabolism and DNA repair(Yanget al
.,2014;Yanget al
.,2015;Sunet al
.,2018).Accordingly,physiological studies revealed that,compared to low-altitude species,high-altitude toad-headed agamas may have decreased basal metabolic rate and increased the utilization of nutrients(e.g.fatty acid) to balance the energy budget (Tanget al
.,2013;Liet al
.,2016;Zhanget al
.,2018),which is a common strategy for high-altitude adaptation for ectothermic vertebrates (Cooperet al
.,2002;Liet al
.,2016).On the other hand,low-altitude toadheaded agamas exhibited the same direction of plastic response to highland environments (e.g.Tanget al
.,2013).Qiet al.
(unpublished) have conducted an experiment by translocating two low-altitude speciesP.axillaris
(oviparous) andP.forsythii
(viviparous) to highland environments and measured their transcriptomic,metabolomic,and behavioral responses.The two species showed a similar pattern,in which significantly plastic change occurred for core genes and metabolites within fatty acid metabolic pathway.However,whether the same plasticity still exists in high-altitude agamas remains unknown.For other taxa,several species have shown loss of plasticity while adapting to high-altitudes due to genetic assimilation,such asAlnum glutinosa
(Kortet al
.,2016),and montane butterflyColias eriphyle
(Kingsolver and Buckley,2017).Therefore,to investigate if toadheaded lizards have experienced the loss of plasticity at highaltitudes could provide new evidence of genetic assimilation in high-altitude adaptation for ectotherms.Here,we present a study to investigate the gene expression profiles and plasticity of a high-altitude species Qinghai toadheaded agama (P.vlangalii
).This species inhabits in Qinghai-Tibetan Plateau at altitudes ranging from 2000 to 4600 a.s.l.(Zhaoet al
.,1999).We implemented a translocation experiment by movingP.vlangalii
individuals from high-to low-altitude environment and measured the gene expression profiles for three organ tissues -heart,liver,and muscle,to investigate the plastic changes in gene expression.In addition,we comparedP.vlangalii
to two low-altitude species,P.axillaris
(oviparous) andP.forsythii
(viviparous),and revealed the pattern of expression plasticity forP.vlangalii
that may indicate whether genetic assimilation has contributed to theadaptation to high-altitude environmentfor this species.2.Materials and Methods
2.1.Ethical approval
All applicable international,national,and/or institutional guidelines for the care and use of animals were strictly followed.All animal sample collection protocols complied with the current laws of China.The sampling and experiment in this study were carried out with permission(Number 2017005) from the Ethical Committee for Animal Experiments in Chengdu Institute of Biology,Chinese Academy of Sciences.2.2.Translocation experiment
A total of 12 male adults ofP.vlangalii
were sampled from Zoige,Sichuan Province of China,at an altitude of 3 400 meters a.s.l.All samples were randomly and evenly assigned into an ‘origin group’ and ‘translocation group’.For the origin group,we measured their morphological traits and then performed euthanasia using decapitation and collected tissues of heart,liver,and muscle,by preserved in RNA later (Invitrogen,USA) in the field.For the translocation group,we moved the individuals to a workstation in Chengdu City,Sichuan Province of China,with an altitude of 650 meters a.s.l.All individuals had been kept in an outdoor enclosure similar to their native environment for six weeks to make the individuals acclimate to translocation environment.Then,the same procedure was conducted for the translocation group to collect tissue samples.To alleviate the potential bias stimulated by the environment,the field sites were standardized in three aspects:1) sand obtained from the origin sites ofP.vlangalii
to the translocation site as substrate;2) mealworms were provided as food every three days for both sites;and 3) fishing net above enclosures to reduce the risks of bird predation.2.3.Transcriptome sequencing
Total RNA was extracted from each tissue sample according to TRIzol protocols(Invitrogen,USA).Transcriptome sequencing was implemented on Illumina HiSeq 2500 platform with paired-end 150 base pair (bp) by Novogene (Beijing,China).Sequence data were deposited in NCBI Short Reads Archive (SRA) with BioProject accession number PRJNA718616.Raw sequence reads were first cleaned by removing the adapter sequences and low-quality base calls using a Novogene pipeline.Trimmomatic v0.35(Bolgeret al
.,2014) was used to trim the reads with LEADING:3,TRAILING:3,SLIDINGWINDOW:4:5,MINLEN:70,and default parameters.We checked for reads quality before and after filtering with FASTQC version 0.11.8 (Andrews,2010).One individual with all three types of tissues (E1,K1,and Q1;details in Table S1) was used forde novo
assembly of transcriptome via Trinity v2.8.4 afterin silico
read normalization (Grabherret al
.,2011;Haaset al
.,2013).We then used kallisto version 0.44.0 (Brayet al
.,2016) to quantify the abundance of the assembly and build the transcripts expression matrices.Assembled transcripts with ‘transcripts per million transcripts’ (TPM) less than three were filtered to generate the final assembly for each species.To compare to other two low-altitude speciesP.axillaris
andP.forsythii
(Qiet al
.,unpublished),a best reciprocal hit (BRH)method was applied to identify 1:1:1 orthologous sequences among the three species (Camachoet al
.,2009).2.4.Differential expression analysis
The clean reads for each sample were mapped against theP.vlangalii
transcriptome assembly by using STAR version 2.6 (Dobinet al
.,2013).The number of reads matched to the same transcripts was counted by HTSeq-count tool with the ‘union’ resolution mode (Anderset al
.,2015).The overall similarity among tissue samples was measured by Euclidean distance and visualized by clustering heatmap through ‘pheatmap’ package after regularized log transformation (rlog) of normalized counts via DESeq2 version 1.20 (Loveet al
.,2014).Principal component analysis (PCA) was also used to assess the relationship among different samples.Differentially expressed genes (DEGs) were estimated through generalized linear models in edgeR package version 3.22.5(Robinsonet al
.,2010;McCarthyet al
.,2012).We adopted a strict criterion to identify DEGs,with fold-value ≥ 2 and adjustedP
-value (FDR) < 0.05.Functional annotation was performed by mapping the transcripts against the UniProtKB/Swiss-Prot database (release“2018_08”) with blast hits E-value cut-off greater than 10.For those transcripts with annotation information,functional over-representations of DEGs were performed using the clusterProfiler package (Yuet al
.,2012) in R with annotation to GO category and KEGG pathway database.The minimum number of genes required for each test of a given category was 5.All tests were corrected by false discovery rate (FDR).To compare the plastic pattern withP.axillaris
andP.forsythii
for genes associated with fatty acid metabolism,we also checked the expression profiles of 12 diagnostic genes (Table S2),which showed differential expression in liver for the two low-altitude species,including key genes regulating the synthesis (Fatty Acid Synthase,FASN) and catabolic process (Acetyl-Coenzyme A Acyltransferase 2,ACAA2,and Enoyl-CoA Delta Isomerase 2,ECI2) of fatty acid.We calculated the absolute fold change of expression from low-to high-altitude for those genes among the three species,and tested the significance of differences by two-tailed Student’st
test.3.Results
3.1.Transcriptome sequencing
A total of 44 793 768-70 718 484 raw reads were generated forP.vlangalii
by Illumina sequencing.After filtering,42 722 956-67 578 714 reads were retained.One sample (R6) was excluded from the following analyses due to low quality of sequencing.Overall,36 336 transcripts were obtained with N50 size of 2 232 bp and mean length of 1 093 bp.By BRH method,8 892 orthologous transcripts were identified amongP.axillaris
,P.forsythii
,andP.vlangalii
.3.2.Gene expression profile
Both principal component analysis (PCA) and clustering analysis demonstrated that each tissue type presented a distinct expression signature and all samples were unambiguously grouped by tissue origin (Figure 1B).Within each tissue,samples were also separated by origin and translocation groups.In addition,samples from heart and muscle expressed closer than from liver,which may reflect that a large part of the heart is composed of cardiac muscle tissue(Figure 1C).By comparing the expression profiles ofP.vlangalii
to the other two low-altitude speciesP.axillaris
andP.forsythii
,we found that theP.vlangalii
was closer toP.forsythii
in all the three tissues,consistent with their phylogenetic relationship(Figure 1D,E,F).3.3.Differential expression analysis
Given low-altitude samples as reference,a total of 875 differentially expressed genes(DEGs) were identified in the heart forP.vlangalii
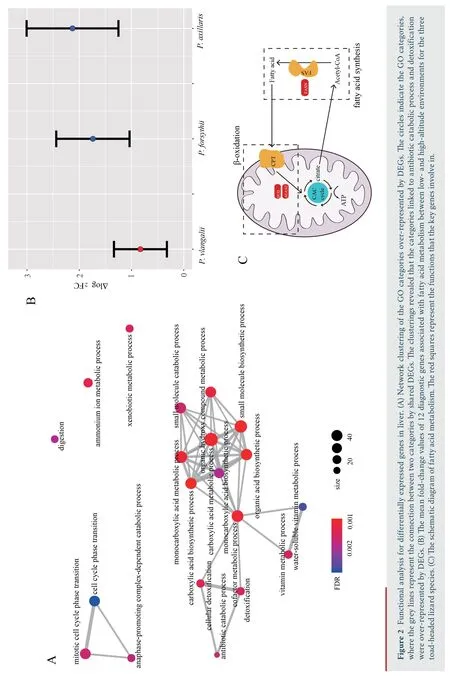
,with 400 down-regulated and 475 up-regulated.Similarly,688 DEGs were identified in muscle,with 345 down-regulated and 343 up-regulated.We identified 1 220 DEGs in liver,the greatest number of DEGs among all three tissues,with 612 downregulated and 608 up-regulated.Through functional annotation,we found that no GO category and KEGG pathway was over-represented by DEGs in heart.A total of 41 GO categories and 13 KEGG pathways were identified over-represented by DEGs in liver (Figure S1).Network clustering of the GO categories indicated that most of the categories were associated with monocarboxylic acid metabolic process,which could be linked to antibiotic catabolic process and detoxification (Figure 2A).In muscle,7 GO categories and 1 KEGG were over-represented by DEGs(Figure S2),which were associated with extracellular matrix organization and carbohydrate derivative catabolic process.
For the 12 diagnostic genes associated with fatty acid metabolism,the absolute fold change of expression in liver ofP.vlangalii
was significantly lower (ΔlogFC=0.8300) than that ofP.axillaris
(ΔlogFC=2.1298;P-value
=2.84×10) andP.forsythii
(ΔlogFC=1.7397;P-value
=1.31×10),along with no significant difference between the latter two species (P-value
=0.44) (Figure 2B).In addition,none of the key genes FASN,ACAA2 and ECI2 was differentially expressed between low-and high-altitude(Figure 2C).4.Discussion
P.vlangalii
from a high-to low-altitude environment and measured their expression profiles and plasticity via transcriptome sequencing.Our results clearly illustrated that each tissue type ofP.vlangalii
presented an unambiguous expression signature,with hundreds of DEGs up-regulated or down-regulated in each tissue,respectively.In comparison to the other two low-altitude speciesP.axillaris
(oviparous) andP.forsythii
(viviparous),although the expression profile ofP.vlangalii
was still closer toP.forsythii
,DEGs in liver exhibited a distinct pattern from the other two species,with reduced plasticity in expression of genes associated with fatty acid metabolism.A common strategy for ectothermic vertebrates in response to extreme environments at high-altitude is to suppress basal metabolism and increase utilization of nutrients to balance the energy budget (Cooperet al
.,2002;Liet al
.,2016;Zhanget al
.,2018).Tanget al.
(2013) revealed that a high-altitude toadheaded agama,P.erythrurus
,behaved similarly,with lower mitochondrial respiratory rate but higher fat utilization in liver compared to a low-altitude speciesP.przewalskii
.Qiet al.
(unpublished) further found that both oviparous (P.axillaris
)and viviparous (P.forsythii
) species in this genus showed a very similar plastic pattern of gene expression and metabolites associated with fatty acid metabolism in liver by translocating from low-to high-altitude.However,our study suggests that the true high-altitude speciesP.vlangalii
has a distinct pattern.Although the general expression profile ofP.vlangalii
was still closer toP.forsythii
,consistent with their phylogenetic relationship,DEGs in liver ofP.vlangalii
were mostly concentrated in functional categories like monocarboxylic acid metabolic process,antibiotic catabolic process and detoxification.Furthermore,the expression patterns of 12 diagnostic genes associated with fatty acid metabolism,which showed significantly differentially expression inP.axillaris
andP.forsythii
,illustrated reduced plastic expression inP.vlangalii
,including key genes FASN,ACAA2,and ECI2.FASN gene encodes an enzyme fatty acid synthase that plays a core role in fatty acid synthesis (Jayakumaret al
.,1995);ACAA2 and ECI2 encode proteins which are key mitochondrial enzymes involved in beta-oxidation,a step in the catabolic process of fatty acid(Abeet al
.,1993;Geisbrechtet al
.,1999).None of these genes were differentially expressed between low-and high-altitude groups forP.vlangalii
,suggesting no significant difference in fatty acid metabolism after translocation.The result indicated thatP.vlangalii
may have lost the capacity of plasticity in fatty acid metabolism due to genetic assimilation.Plasticity in fatty acid metabolism is thought to contribute to maintaining life activities for organisms moving to high altitude (Hammondet al
.,2001;Storzet al
.,2010;Refsnideret al
.,2018).As a true dweller adapted to high altitude,P.vlangalii
may have evolved other traits to trade-off the reduction of capacity for plasticity in fatty acid metabolism,similar to the case ofP.erythrurus
(Tanget al
.,2013).However,the detailed mechanism forP.vlangalii
still needs further research.Our study provided special evidence of genetic assimilation that may have facilitated the high-altitude adaptation forP.vlangalii
.Genetic assimilation refers to the process in which initially plastic phenotypes are gradually fixed,leading to reduced phenotypic plasticity and only adaptive phenotypes,even without environmental stimuli (Waddington,1953;Pigliucci,2006).At gene expression level,plastically expressed genes may finally become fixed,given that gene expression mirrors phenotypic plasticity (Scoville and Pfrender,2010;Renn and Schumer,2013).Genetic assimilation provides an alternative hypothesis to explain the mechanism of adaptive evolution,where phenotypic plasticity is first triggered by environmental factors,followed by selection on genotypes influencing the plastic expression of phenotypes (Moczeket al
.,2011;Jones and Robinson,2018).In fact,species such asAlnum glutinosa
(Kortet al
.,2016) andColias eriphyle
(Kingsolver and Buckley,2017) also showed similar loss of plasticity in adaptation to high-altitude environments,which indicated that the genetic assimilation might be an effective way for organisms to adapt to extreme environments.However,more studies especially on modeling of phenotypic plasticity are still required to elucidate the role of genetic assimilation in adaptation.Acknowledgements
We are grateful to Cuoke,Erga,X.Qiu,Y.Wu,and X.Zhu for logistic assistance in field station,and J.Ramos for English language editing.We have obtained the permits from Zoige National Wetland Nature Reserve,where we have a long-term field research station on ecology and evolution of lizards.This work was supported by National Natural Science Foundation of China (No.31501855) and the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) with Grant No.2019QZKK0402.Appendix
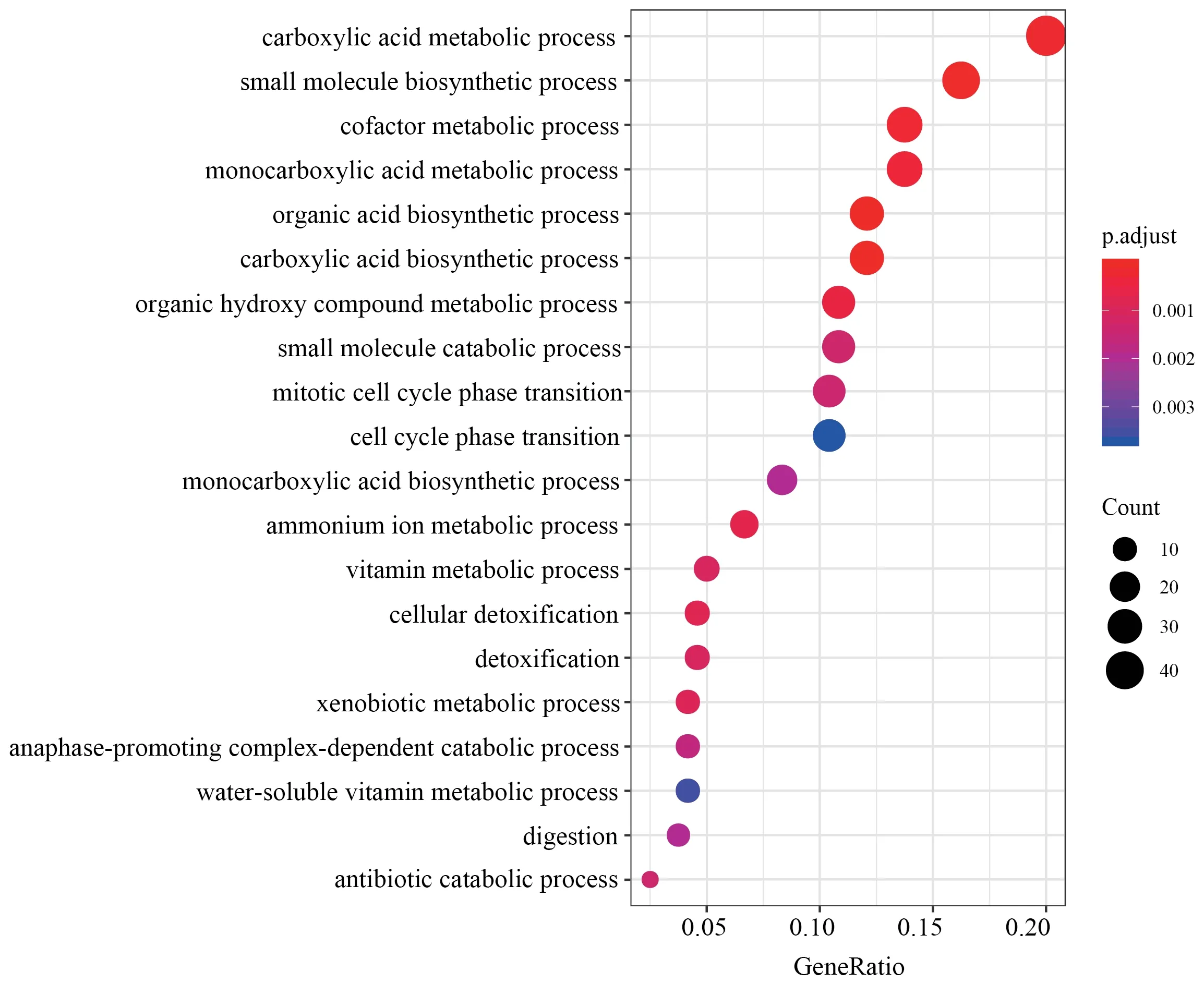
Figure S1 Top 20 GO categories over-represented by DEGs in liver.
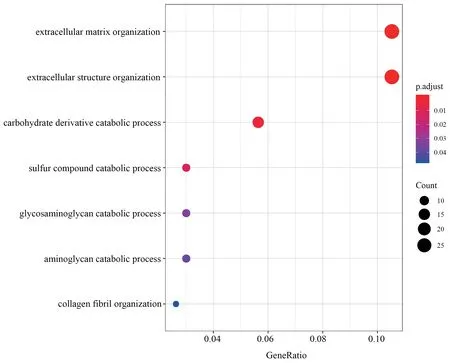
Figure S2 GO categories over-represented by DEGs in muscle.
Table S1 Sample information.
Table S2 Diagnostic genes associated with fatty acid metabolism.
 Asian Herpetological Research2021年3期
Asian Herpetological Research2021年3期
- Asian Herpetological Research的其它文章
- Diet of Juvenile Crocodylus porosus at Kuching Wetlands National Park,Sarawak,East Malaysia
- Complete Mitochondrial Genome of the Steppe Ribbon Racer (Psammophis lineolatus):The First Representative from the Snake Family Psammophiidae and its Phylogenetic Implications
- Age Structure of Two Species of Odorous Frogs (Odorrana margaretae andOdorrana grahami)
- Effects of Life Histories on Genome Size Variation in Squamata
- Molecular Evolution of Visual Opsin Genes during the Behavioral Shifts between Different Photic Environments in Geckos
- Disordered Translocation is Hastening Local Extinction of the Chinese Giant Salamander
