Transcriptome Analysis Provides New Insights into Host Response to Hepatopancreatic Necrosis Disease in the Black Tiger Shrimp Penaeus monodon
ZHAO Jichen, CHEN Xieyan, HE Zihao, CHEN Guoliang, LIN Zhaojian,LIU Yongkui, SUN Chengbo, *, and WANG Wei, *
Transcriptome Analysis Provides New Insights into Host Response to Hepatopancreatic Necrosis Disease in the Black Tiger Shrimp
ZHAO Jichen1), CHEN Xieyan1), HE Zihao1), CHEN Guoliang2), LIN Zhaojian2),LIU Yongkui2), SUN Chengbo1), *, and WANG Wei1), *
1),,524088,2).,.,524088,
The hepatopancreatic necrosis disease (HPND) is a common shrimp disease. Aquaculture of the black tiger shrimphas suffered with the frequent outbreak of HPND. Understanding the molecular mechanisms of host response to HPND is of pivotal importance for improving its farming performance. In this study, the RNA-Seq platform was utilized to investigate transcriptomic changes of the hepatopancreas in pond-culturedwithHPND symptoms. A total of 62071 genes with an average length of 953bp were obtained, and a lot of simple sequence repeat (SSR) loci related to these genes were identified. Totally, 5204 differentially expressed genes (DEGs) were detected between healthy shrimp and HPND shrimp, with 3399 genes up- regulated and 1,805 genes down-regulated. These genes had a wide range of biological functions, and several well-known immune- related genes including integrin alpha 5, integrin beta 1, C type lectin and catalase were among the DEGs. Cell signaling pathways including the extracellular matrix (ECM) receptor interaction, phagosome and lysosome pathways were significantly upregulated in HPND-infected shrimps, indicating their involvement in the immune responses of shrimp against HPND. The data obtained in this study offers new insights into the molecular mechanisms of shrimp host response to HPND disease, and provides a resource for molecular marker development and genetic breeding studies of.
; shrimp; HPND; transcriptome; hepatopancreas
1 Introduction
The black tiger shrimp,, is one of the most important shrimp species cultured in the world. How- ever, the development ofaquaculture industry has encountered serious obstacles with the occurrence of various shrimp diseases in recent years, and the hepatopan- creatic necrosis disease (HPND) is one of the most harm- ful diseases (Thitamadee., 2016).
The hepatopancreatic necrosis disease can be caused by different factors, and one common form of HPND is the acute hepatopancreatic necrosis disease (AHPND). TheAHPND is also known as early mortality syndrome (EMS). Its first outbreak was observed in China around 2009, and then it spread sequentially to other countries such as Vietnam, Malaysia, Thailand, Philippine, and Mexico (Tran., 2013; Joshi., 2014; De la Pe?a., 2015; Soto-Rodriguez., 2015; Dabu., 2017). The post larvae mortality occurs after 35 to 45 days of culture in the pond, with the symptom of massive sloughing of he- patopancreatic tubule epithelial cells beginning in the cen- ter of the hepatopancreas and processing outward to the embryonic region (Thitamadee., 2016).
The AHPND has caused great economic losses. One of the most common causative agents of AHPND is the bac- teria straincarrying a large extra- chromosomal plasmid uniquely virulent to shrimp (Tran., 2013; Kondo., 2014;Han., 2015b). This AHPND-causing plasmid contains 2 toxin genes, namelyand, as they are homologous to theinsect-related (Pir) binary toxin (Lai., 2015; Si- rikharin., 2015). Theinitially co-lonizes the stomach of shrimp and then secrets toxins, spreads sequentially to hepatopancreas, and leads to he- patopancreatic necrosis. Ascarrying Pir toxin is more pathogenic than otherstrains with- out Pir toxin, Pir toxin is considered as the most important factor of AHPND pathogenesis (Lai., 2015; Zheng., 2018b). In addition, other pathogens including(Hidehiro., 2015),(Liu., 2018),(Dong., 2017a, 2017b),(Restrepo., 2018), and(Aranguren., 2017)can also cause HPND in shrimp. Therefore, various factors can result in HPND and the disease-causing mechanism of HPND may be different.
In order to control HPND, some detection methods have been established, such as the PCR/qPCR method(Han., 2015a), PCR and immunomagnetic separation ap- proach (Lun., 2018),and other methods (Arunrut., 2016). These methods are used for the detection of HPND pathogens in shrimp or water samples from environmen- tal source. On the other hand, some measures to control or prevent the disease have also been carried out. For ex- ample, probiotics, such as, pur- ple nonsulfur bacteria,spp, lactic acid bacteria, and proteins extracted from red seaweedandspare supplemented in food(Boonsri., 2016; Chumpol., 2017; Félix., 2017; Thammasorn., 2017; Chomwong., 2018; Wang., 2018);(Kongrueng., 2017) and phage (Jun., 2016; Stalin., 2016) are used to inhibit the growth of; Ozone na- nobubbles (Imaizumi., 2018) or phloroglucinol (Ku- mar., 2018) are applied to treat aquaculture water. Nonetheless, these measures are not very effective in pre- venting the bacteria infection during shrimp cultivation. Furthermore, with the increase of antibiotic-resistance in pathogenic bacteria, the therapeutic effect of antibiotics is weakened, and the disease is getting harder to control(Dong., 2017a). Hence, more attention needs to be focused on understanding the molecular mechanisms of host response to HPND.
In recent years, with the development of high-through- put sequencing technologies including RNA Sequencing (RNA-Seq), new methods have been applied to the study of crustacean immunology (Wang., 2017;Zhang., 2018). Studies on the transcriptomic responses to HPND can provide insights into the host physiological response and immune mechanism (Qin., 2018; Zheng., 2018a). Some well-known immune-related genes, such as crustins, serine proteinases, serine proteinase inhibitors, C type lectins, alkaline phosphatases, and heat shock pro- teins, have been studied in shrimp with HPND disease (Ge., 2017). Furthermore, several novel isoforms of anti- lipopolysaccharide factors were found involved in HPND(Soonthornchai., 2016). Another study reported that the Rho signaling pathway helps to mediate AHPND pa- thogenesis in shrimp (Ng., 2018). Moreover, cyto- skeletal reorganization occurred in prawns after infection of(Zheng., 2018b). These results have deepened our understanding of HPND resistance. However, most studies on shrimp in this area have focus- ed on the Pacific white shrimp(Ma- ralit., 2018; Ng., 2018), while very few studies have explored the scenario in other shrimp species including(Soonthornchai., 2016).
Aquaculture ofhas been seriously affected by HPND in recent years (Thitamadee,., 2016). Al- though there are some studies onwith AHPND(Soonthornchai., 2016), our understanding of host response mechanism to the disease is still fragmentary, es- pecially for artificially culturedwith natural- ly occurring HPND symptoms. Hepatopancreas is an im- portant organ of shrimp, fulfilling multiple functions in- cluding digestion, immune defense and reproduction. It is also the major target organ of HPND. Nonetheless, infor- mation about genes and pathways involved in hepatopan- creas response to HPND inis still limited (Ge., 2017). In this study, we utilized the RNA-Seq plat- form to perform a comparative transcriptomic analysis be- tween a naturally HPND-infected group and a healthy groupof, aiming to identify genes and biological pathways related to HPND response in the hepatopancreas of
2 Materials and Methods
2.1 Shrimp and HPND Experiments
In August 2018, shrimp HPND disease was detected in somefarms in Fangchenggang city, Guangxi province, China, after 35 to 45 days of post larvae culti- vation in the ponds. Sick shrimp with HPND was deter- mined according to morphological characteristics and ab- normality of the gastrointestinal tract. Compared with heal- thy shrimp, HPND infected shrimp showed empty sto- mach and intestine and atrophic hepatopancreas. Shrimp with typical symptoms of HPND were collected from the pond, and the hepatopancreas tissues of nine individuals were separately dissected and immediately frozen in li- quid nitrogen. These animals were used as the experimen- tal group. Another group of nine healthy shrimp without the symptoms of HPND in other ponds were taken as the control group. The nine shrimps selected in each grouphad similar sizes to avoid potential effects of different body size. The tissues were then stored at ?80℃ until RNA ex- traction. For each group, tissues from three shrimp were collected to prepare an RNA sample, and there were three parallel samples in each control or experimental group.
2.2 RNA Extraction, Library Construction and Sequencing
Total RNA was extracted from the hepatopancreas of nine individuals using Trizol Reagent (Invitrogen) accord- ing to the manufacturer’s instructions. The RNA quality of each sample was monitored on 1.0% agarose gels, with electrophoresis at 120V for 30min. RNA concentration and integrity of each sample were further measured using an Agilent 2100 Bioanalyzer and Nanodrop. For each group, there were three parallel samples for sequencing. Equal amount of RNA from every three individuals were pooled as one sample for cDNA library construction,whichwas carried out at the Guangzhou Sagene Biotech Co., Ltd., Briefly, mRNA was isolated from the pooled RNA using poly-T oligo-attached magnetic beads, and then the puri- fied mRNA was cleaved into small fragments, followed byfirst-strand cDNA synthesis. Subsequently, the second strand cDNA was synthesized and sequencing adapters were con- nected to the products. cDNA fragments were selected on a 2% agarose gel, then the products were subjected to PCR amplification. The resultant cDNA libraries were then sequenced on the Illumina HiSeqTM platform with 125bp paired-end sequencing reads.
2.3 Transcriptome Assembly and Gene Annotation
Before the assembly, raw reads with adaptor sequences, ambiguous nucleotides or contaminations were filtered from the total reads, and only reads with quality scores above 20 were used in downstream analysis. Sequentially, theclean reads were used forassembly to produce assembled transcripts (unigenes) using the software Tri- nity (Grabherr., 2011). Then, the assembled unigenes were Blast annotated against major gene databases includ- ing the Nr, SwissProt, KEGG and COG/KOG databases (Camacho., 2009). The cutoff E-value was set at 1e?5and only the top hit was used to annotate each unigene. Next, according to the Nr annotation, the Blast2GO soft- ware was used to obtain the GO annotation for these uni- genes (Conesa., 2005).
2.4 Analysis of Differentially Expressed Genes (DEGs) and Functional Enrichment
The clean reads from different samples were mapped to the assembled unigenes, and the expression level was cal- culated using the software RSEM (Li., 2011) (RNASeq by Expectation Maximization) based on the FPKM (Frag- ment Per Kilo bases per Million Reads) value. Differen- tial gene expression analysis was conducted using the edgeR package with default parameters (Robinson., 2010). To ensure the high quality of DEG analysis, False Discovery Rate (FDR) control method was used. Unigenes with an adjusted<0.05 and |log2(FC)|≥1 were deter- mined as the DEGs (Reiner., 2003). To identify sig- nificantly enriched GO terms and KEGG pathways among the differentially expressed genes, enrichment analyses were conducted using the software Goseq V1.16.2 (Young., 2010) and KOBAS (Xie., 2011), respectively, with avalue cutoff of 0.05.
2.5 Validation by Real-Time PCR
To validate the gene expression profiles from RNA-seq analysis, a total of thirteen DEGs were selected to per- form quantitative real-time polymerase chain reaction (qRT- PCR) analysis. The thirteen genes included five up-re- gulated and eight down-regulated genes showing different levels of transcriptional changes, and these genes were related to several significantly enriched biological pro- cesses, including cytoskeletal reorganization and immune defense. The primers were designed using the Primers 5 software. Sequences of the primers were provided in Ta- ble 1. The elongation factor-1α gene (EF-1α) of(GenBank: DQ021452.1) was used as internal control. qRT-PCR was performed using the SYBR?PremixEX- TaqTMII (TaKaRa, Japan) kit. The PCR reaction was car- ried out in a total volume of 12.5μL with 6.25μL of 2× SYBR Green PCR buffer, 0.5μL of each primer (10μmolL?1), ddH2O and diluted cDNA. The thermal cycling pro- gram included initial denaturation at 95℃ for 30s, then 40 cycles of 95℃ for 5s and holding at 60℃ for 30s, followed by 95℃ for 10s. A dissociation curve analysis was further performed to check if only one PCR product was amplified. The samples had three biological repli- cates and each reaction had three technical replicates. The fluorescence intensities of the samples for each gene were measured by threshold cycle (Ct) values, and the expres- sion levels of all genes examined were normalized to the levels of EF-1α gene in the same sample. Gene expres- sion level was compared and converted to fold differences by the comparative CT method (2???CTmethod).
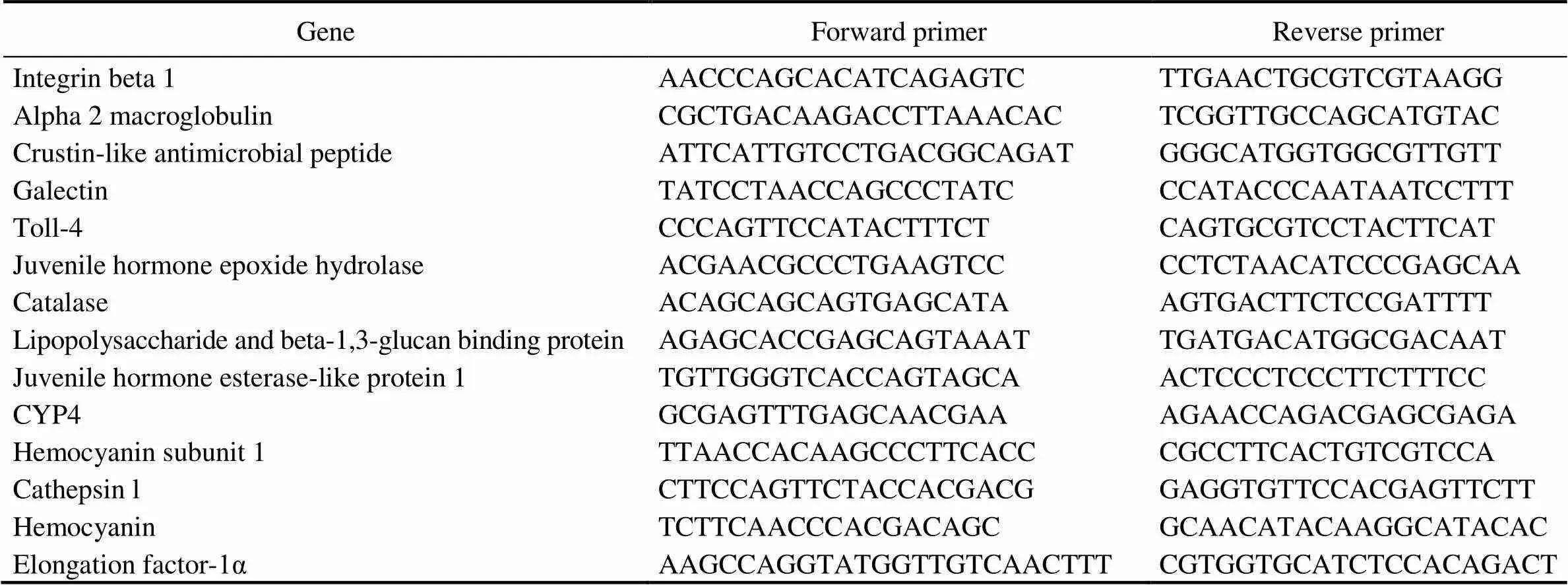
Table 1 Primers used for gene expression validation
2.6 Identi?cation of Simple Sequence Repeats (SSRs) Loci in the Transcripts
SSRs are wildly used molecular markers for geneticbreeding studies. To identify potential SSR loci from the transcriptome, sequences with various types of repeat unit including di-, tri-, tetra-, penta-, and hexa-nucleotide mo- tifs were detected using the software MicroSatellite (MISA,http://pgrc.ipk-gatersleben.de/misa/). The minimum num- bers of repetitive units for di-, tri-, tetra-, penta-, and hexa- nucleotide were set as 6, 5, 4, 4 and 4, respectively, based on parameters used in other similar studies with some customizations to allow identification of a larger number of potential SSR loci.
3 Results
3.1 De novo Assembly
Three cDNA libraries (H1, H2, H3) representing the HPND group and another three libraries (D1, D2, D3) re- presenting the control group were constructed and sub- jected to Illumina sequencing. The HPND group had 212985322 raw reads in total, and the control group had 225101296 raw reads in total (Table 2). After data filter- ing, 62071 unique transcripts (unigenes) with an average length of 953bp were obtained (Table 3). As shown in Fig.1, the unigene length ranged from 200nt to≥3000nt, and a large proportion of the unigenes were in the range of 200–400nt. Among them, 53491 (86.18%) genes and 46489 (74.9%) genes were detected in the experimentalgroup and control group, respectively. Raw sequence reads are available at the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with the acces- sion No. SRP271721.
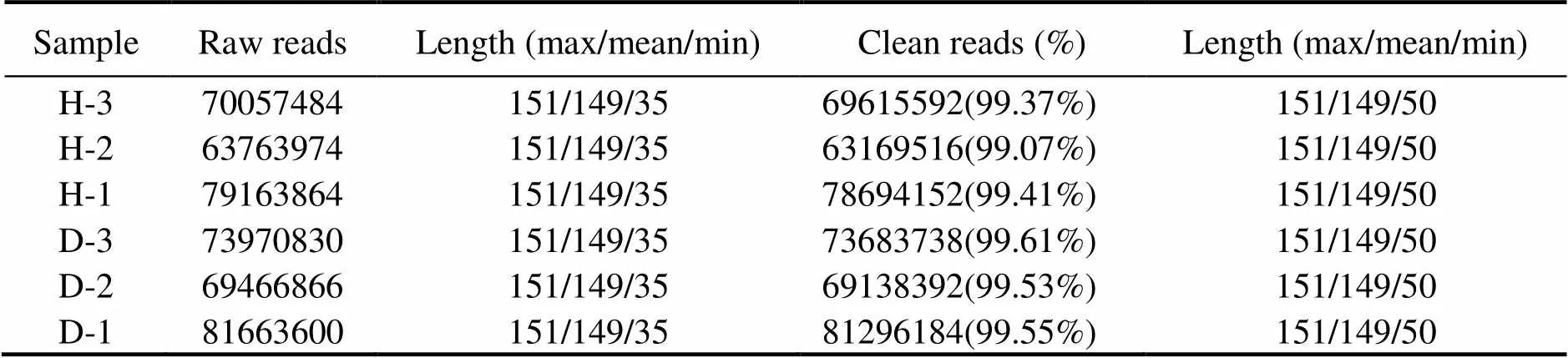
Table 2 Summary of transcriptome sequencing data

Table 3 De novo assembly of P. monodon hepatopancreas transcriptome
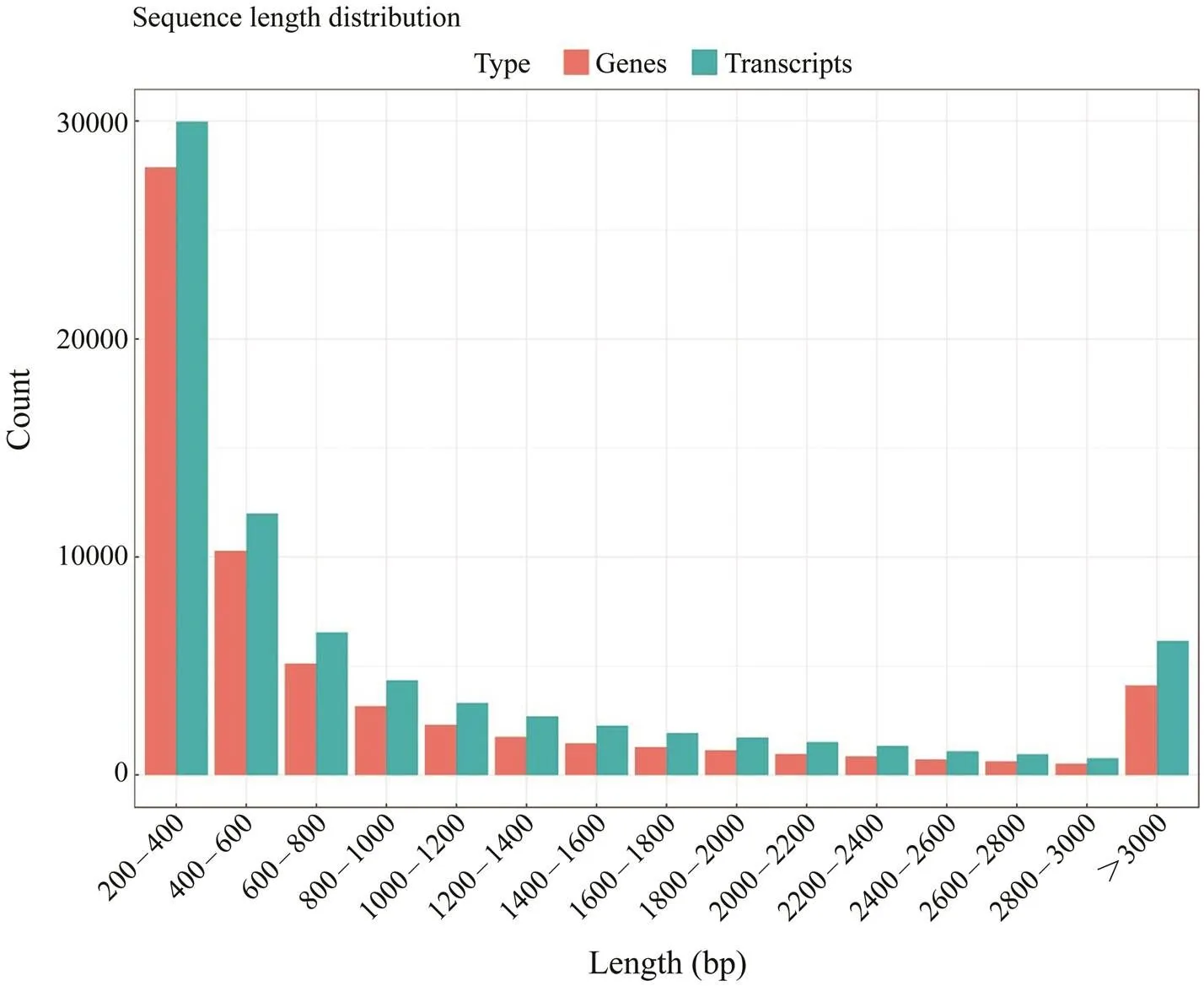
Fig.1 Length distribution of the assembled sequences. The gene length ranges from 200nt to >3000nt, and a large pro- portion of the genes were in the range of 200–400bp.
3.2 Gene Annotation
All assembled unigenes were annotated to databases including the Nr, KEGG, SwissProt, and KOG database. A total of 17044 unigenes can find significant hits when they were compared to known sequences in the databases, corresponding to 27.46% of all the unigenes. Nr database had the largest number of homologous sequences to the assembled unigenes, with 16952 unigenes annotated againstthis database, or 27.31% of the unigenes (Table 4). As shown in Fig.2, 13 species possessed homologous genes to more than 1% of the unigenes based on the NR an- notation, constituting about 55.99% of the total number. These species included crustaceans, insects, shellfish, fish, and coral. Thewas the largest species, accounting for 26.25% of all the unigenes. The shrimp(3.66%),(2.16%) and(1.28%) accounted for 7.10% of the total number.
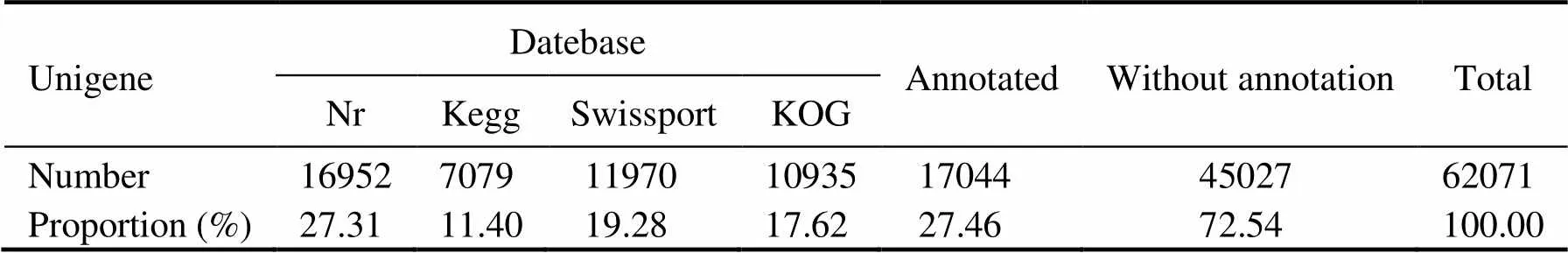
Table 4 Number of unigenes annotated to different databases
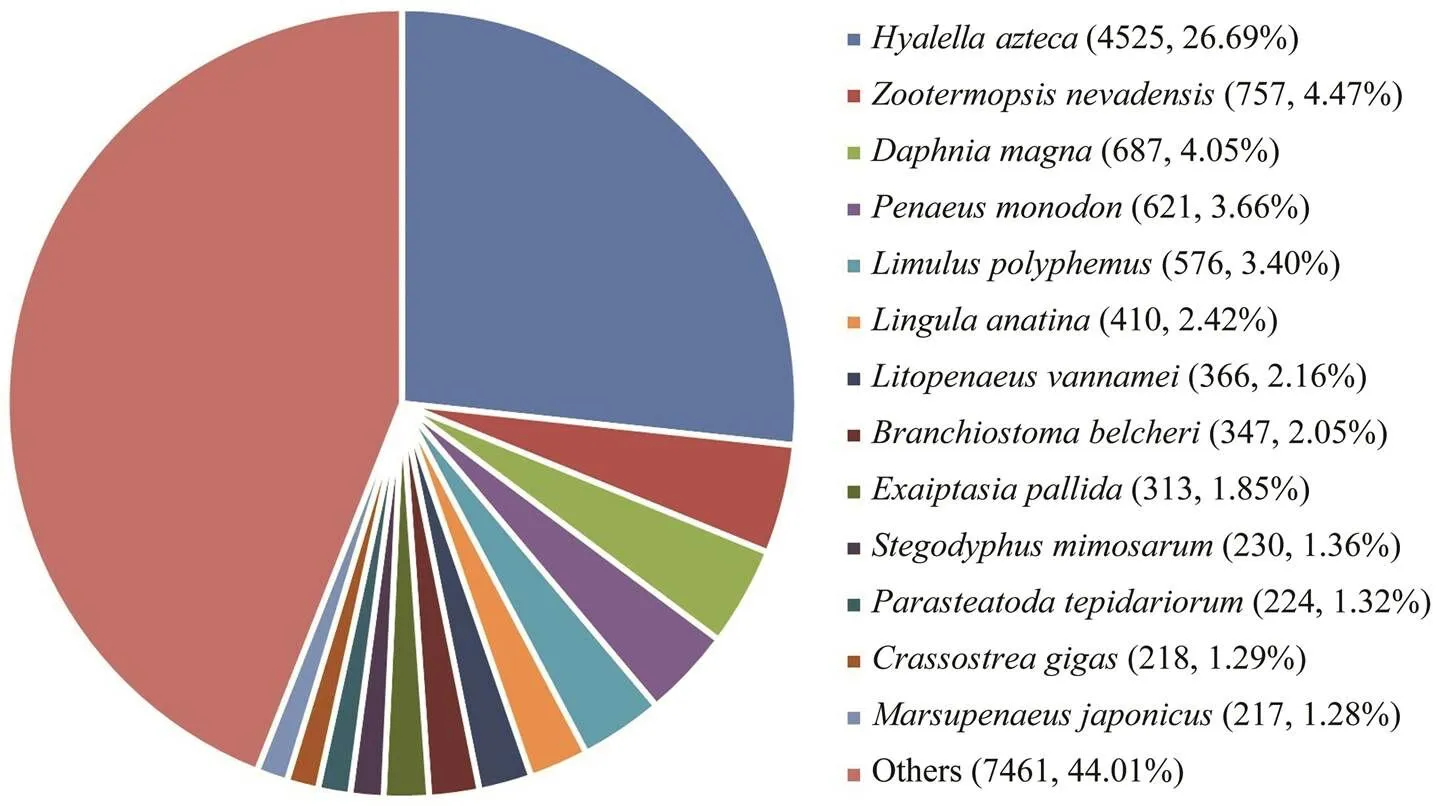
Fig.2 Species distribution of the Blast hits. The largest number of unigenes were matched to homologous genes from Hyalella azteca.
The unigenes were further compared with sequences in the GO database, and they were assigned to different classes in three main categories of GO, namely biological process (20 subcategories), cellular component (16 subcategories) and molecular function (11 subcategories) (Fig.3A). In the category of biological process, the largest number of uni- genes was assigned to the term ‘cellular process’, while ‘cell’and ‘catalytic activity’ contained the largest number of uni- genes in the category of cellular component and molecu- lar function, respectively.
Moreover, a total of 10935 unigenes were successfully annotated to 26 specific protein function definitions ororthologous categories of the KOG database (Fig.3B). Among the KOG terms, the largest three protein function categories were ‘general function prediction only’ (2935), ‘signal transduction mechanism’ (1631) and ‘posttransla- tional modification, protein turnover, chaperones’ (1008) (Fig.3B).
We also identified associated gene pathways of the as- sembled unigenes according to the KEGG database, and 7049 unigenes were assigned to 6 major KEGG catego- ries, including metabolism, human disease, cellular pro- cesses, organismal systems, genetic information process- ing, and environmental information processing. These an- notated unigenes were further divided into 39 level 2 sub- category pathways. Among them, the largest subcategory group was carbohydrate metabolism containing 614 an- notated unigenes, followed by transport and catabolism (540) and signal transduction (523) unigenes (Fig.3C). In total, 272 level 3 KEGG subcategory pathways were fur- ther annotated.
3.3 Analysis of Differentially Expressed Genes
To analyze the differentially expressed genes (DEGs), transcript abundance for each gene was compared be- tweenthe two groups, and 5204 unigenes were identified as DEGs. Among them, 3399 unigenes were identified as up-regulated and 1805 unigenes were identified as down- regulated (Fig.4). Moreover, 2356 of these DEGs could be annotated to the Nr database, and 1586 of them were significantly induced, while 770 unigenes were repressed.
Some immune effector genes, such as the anti-lipopoly- saccharide factors, crustins, prophenoloxidases, tumor ne- crosis factor ligand superfamily members, and chitinases, have been identified from. Most of these im- mune effector genes were induced in HPND-infected shrimp. These genes were likely to participate in the phy- siological response to HPND, and they provide some im- portant candidates to perform functional validation in fol- low up studies.
To further understand the functions of DEGs, these DEGs were also analyzed for GO enrichment. There were 12 GO terms significantly enriched with a-value cutoff of 0.05, including single-organism process, membrane and wide pore channel activity (Table 5).
KEGG enrichment analysis further revealed that there were 18 pathways significantly enriched with a-value cutoff of 0.05. The most significantly enriched pathways were several pathways related to immune defense, inclu- ding ECM-receptor interaction, amoebiasis, lysosome, fo- cal adhesion, peroxisome and phagosome (Fig.5). Some common immune genes were enriched in these pathways, such as integrin alpha 5, integrin beta 1, actin, C type lec- tin, catalase, cathepsin C, and cathepsin L.
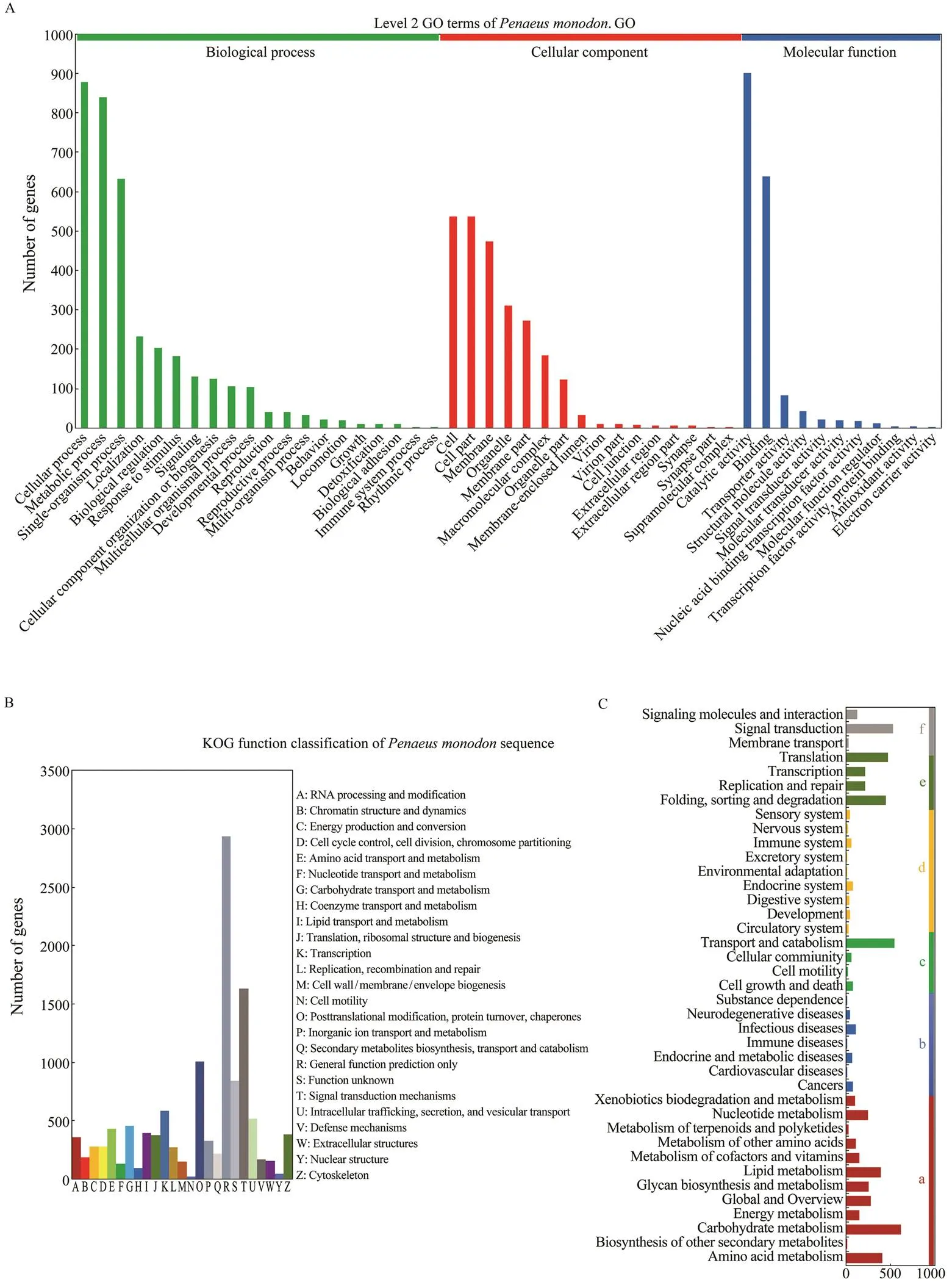
Fig.3 Functional annotation of the assembled unigenes. A, Gene ontology (GO) classi?cations of theunigenes. The uni- genes are assigned to various GO terms in 3 categories including biological process, cellular component and molecular function. B, The cluster of orthologous groups (COG) classi?cation of putative proteins. A total of 10935 unigenes are successfully annotated to 25 categories. C, KEGG annotation of the unigenes. The unigenes are assigned to different sig- naling pathways in six major categories of the KEGG database, including metabolism (a), human disease (b), cellular pro- cesses (c), organismal systems (d), genetic information processing (e), and environmental information processing (f).
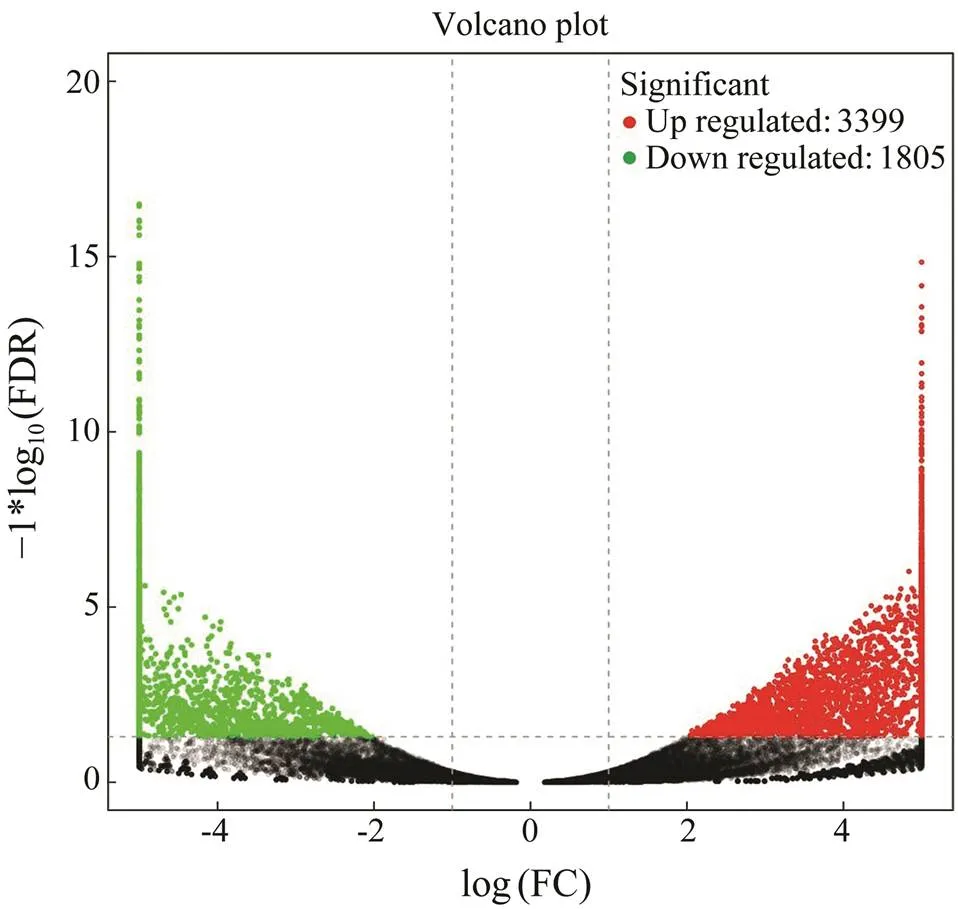
Fig.4 Volcano diagram of differentially expressed genes (DEGs). The x-axis indicates fold change and the y-axis indicates statistical significance of the differences. Red dots represent up-regulated DEGs, while green dots represent down-re- gulated DEGs (FDR≤0.05).
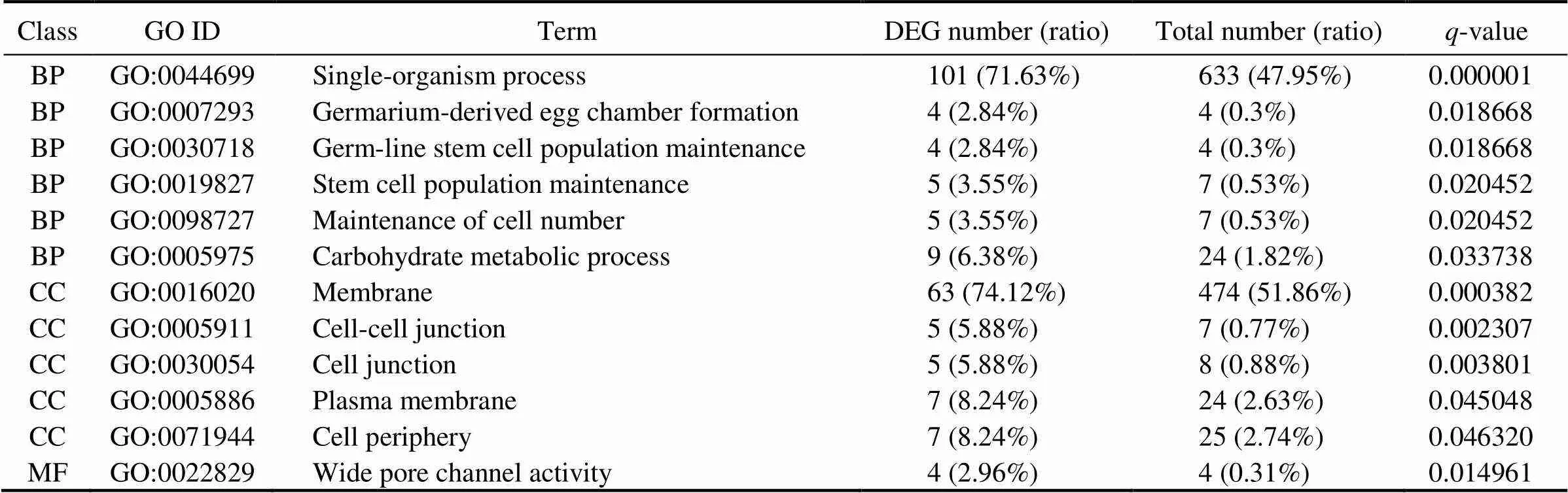
Table 5 Significantly enriched GO terms
Notes: BP, biological process; MF, molecular function; CC, cellular component. Numbers of DEGs and all genes in each item were shown.
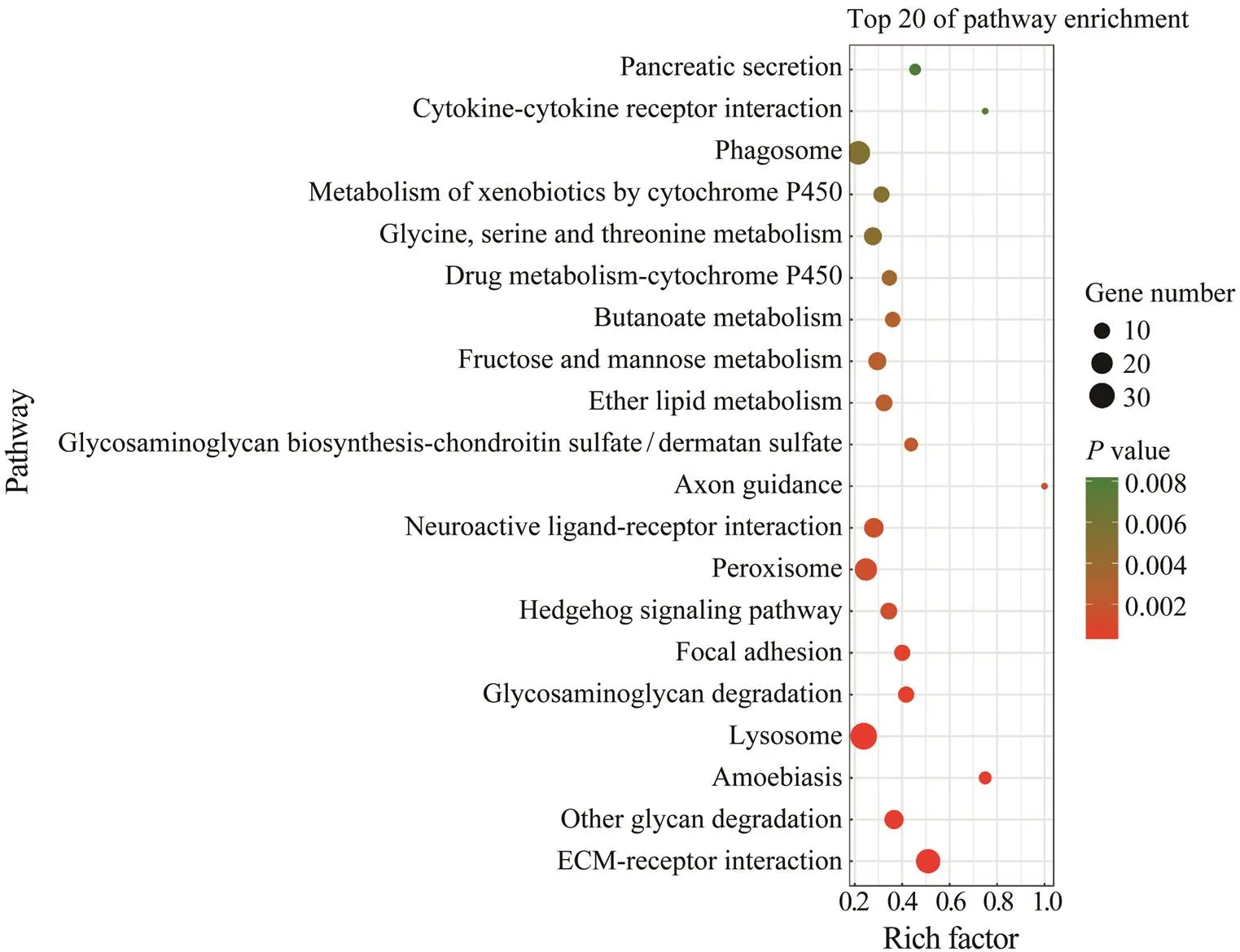
Fig.5 Top 20 enriched KEGG pathways. The most significantly enriched pathways included ECM-receptor interaction, glycan degradation, amoebiasis, lysosome, glycosaminoglycan degradation, and focal adhesion.
3.4 Validation of RNA-Seq Data via Real-Time PCR
In order to validate the differential expression data from RNA-Seq analysis, quantitative real-time PCR was further conducted for thirteen selected genes, including five up- regulated genes (, integrin beta 1, alpha 2 macroglobu- lin, crustin-like antimicrobial peptide, galectin, and toll-4) and eight down-regulated genes (., juvenile hormone epoxide hydrolase, catalase, lipopolysaccharide and beta- 1,3-glucan binding protein, juvenile hormone esterase- like protein 1, cytochrome P450 family 4, hemocyanin sub- unit 1, cathepsin l, and hemocyanin). As shown in Fig.6, expression patterns of these tested genes were consistent when the two different methods were used. Thus, the re- sult showed that gene expression data from transcriptome analysis was reliable.
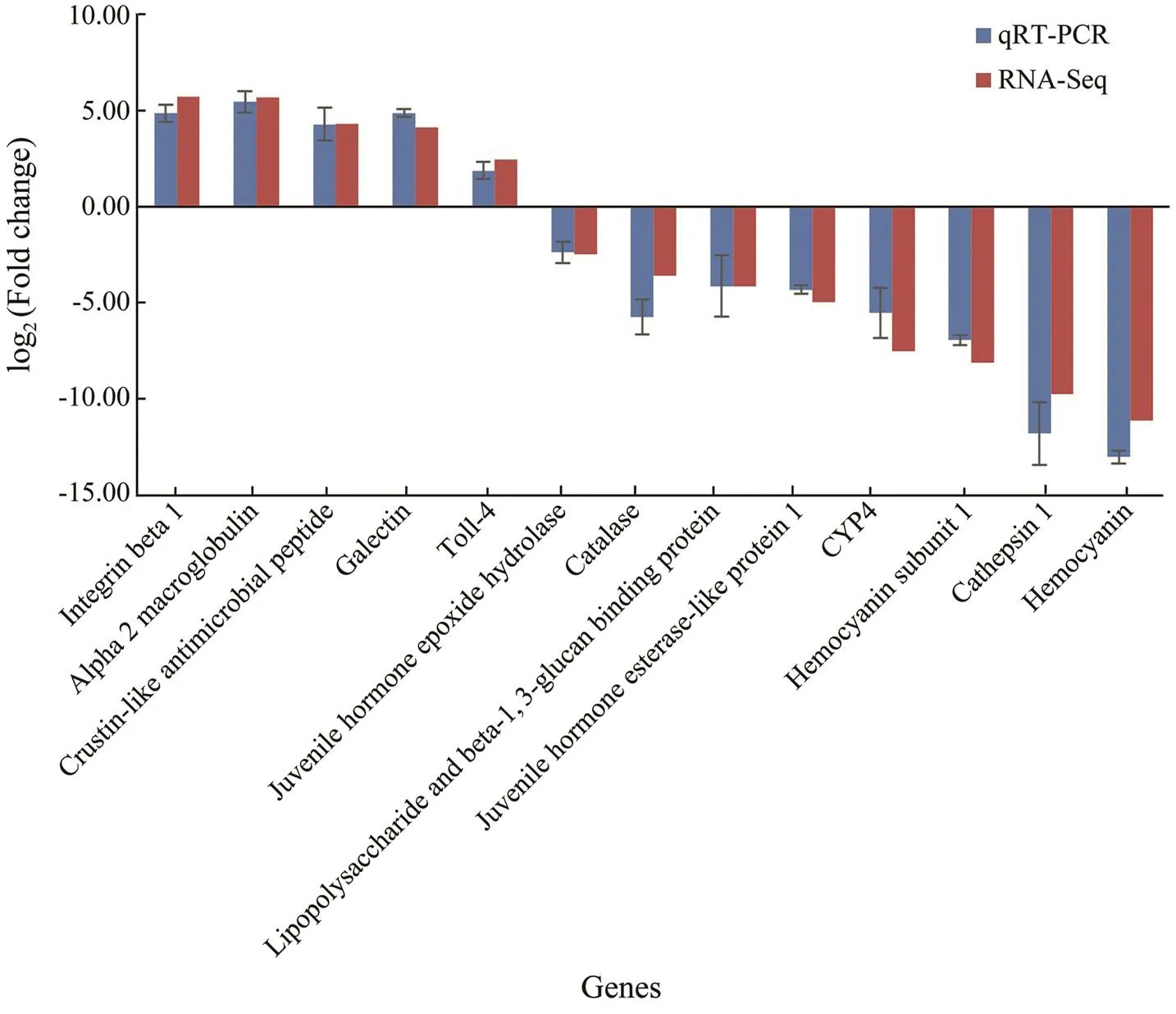
Fig.6 Comparison of gene expression data between RNA-Seq and Real-time PCR. The thirteen selected genes showed consistent expression patterns when the two different methods were used.
3.5 Identification of SSR Loci in the Transcriptome
As an important type of molecular marker, SSR is wide- ly used in genetic breeding studies. In this study, a total of 22894 potential SSRs were identified from the assembled transcripts. Among these SSR loci, the di-nucleotide re- peat was the most common motif (11264) in the tran-scripts, followed by tri-nucleotide repeats (7042), tetra-nu- cleotide repeats (3427), hexa-nucleotide repeats (655) and penta-nucleotide repeats (506). Among the di-nucleotide repeats, AG/CT was the most abundant motif, comprising 19.7% of the total, followed by AC/GT (18.8%) and AT/ AT (10.3%) (Fig.7). AAT/ATT (5.5%) was the most abun- dant motif in the tri-nucleotide repeats, followed by AGG/ CCT (5.5%) and AAG/CTT (3.7%). In total, 15609 tran- scripts were found to have at least one SSR locus and 4711 transcripts harbored more SSR loci.
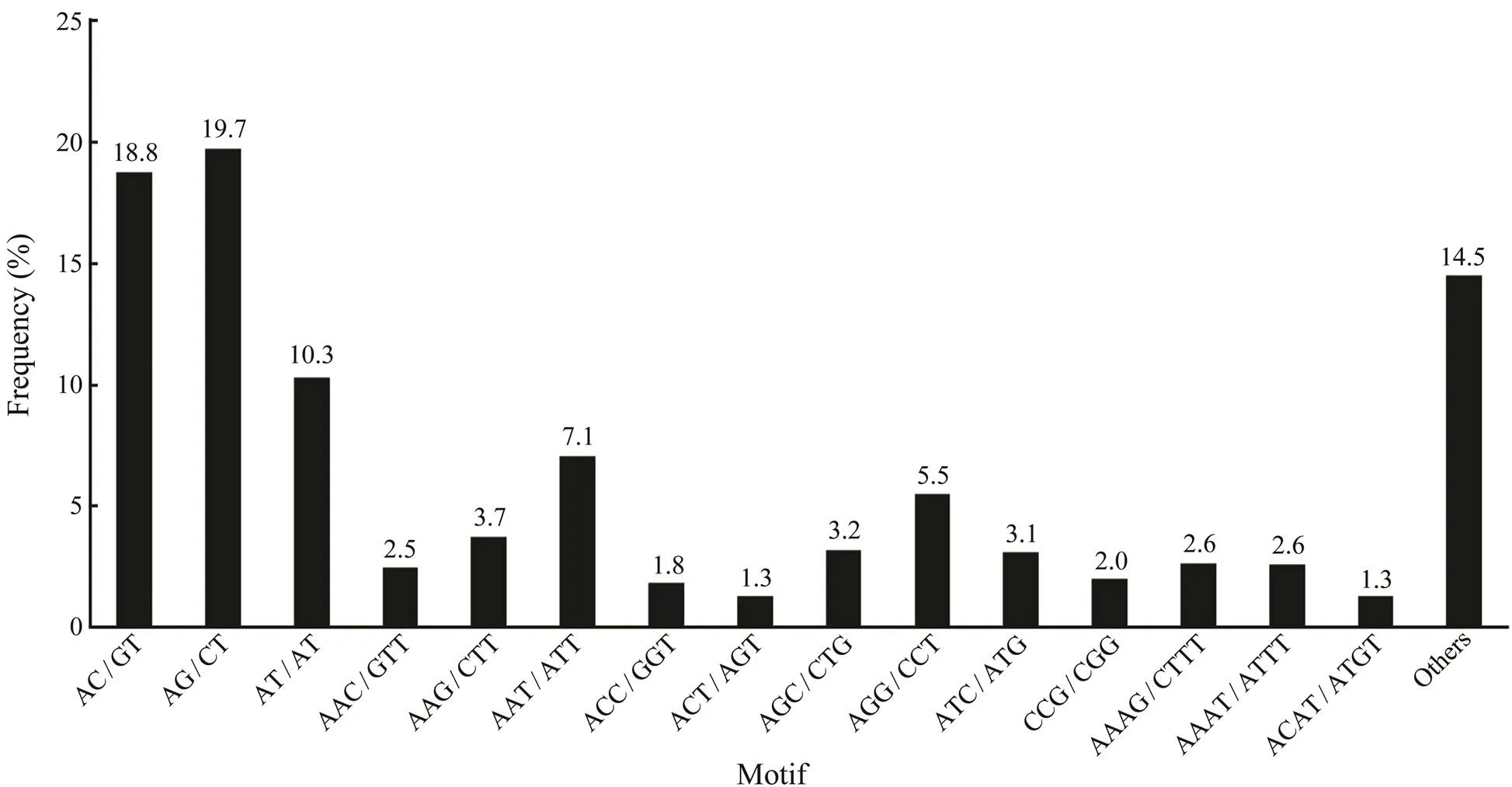
Fig.7 Statistics of different SSR types in the transcriptome. AG/CT and AAT/ATT are the most common motif for di-nucleotide repeats and tri-nucleotide repeats, respectively.
4 Discussion
The black tiger shrimp,, is one of the most important shrimp species cultured in the world. With the development of shrimp farming industry, more and more shrimp diseases have emerged and caused catastrophic eco- nomic loss (Thitamadee., 2016). HPND is a serious shrimp disease, but studies on the host response mecha- nism to HPND are still limited (Ge., 2017; Ng., 2018). In this study, the RNA-Seq platform was utilized to investigate the transcriptomic modulation of commer- cially culturedwith naturally occurring HPND symptoms, providing new insights into the molecular me- chanism of shrimp resistance to HPND.
Shrimp with HPND may need to activate more genes re- lated to immunity or stress resistance to counteract thedisease challenge. In previous studies,was found to have more up-regulated unigenes than down-regulated unigenes after WSSV infection (Rao., 2016; Cao., 2017; Ding., 2018). A similar phenomenon occurred in(Zheng., 2018b)and(Soonthornchai., 2016) infected withAHPND. In our study, a total of 62071 genes were ob-tained using Trinity software. Among them, more genes were identified from the HPND group (53491) than con- trol group (46489). Meanwhile, through DEG analysis, a larger number of up-regulated unigenes were detected than down-regulated unigenes. It can be hypothesized that the accumulation of novel transcripts was associated with HPND infection in shrimp, and the induced genes mayplay important roles in the elimination of pathogens (Ding., 2018).
Among the above-mentioned DEGs, integrins were in- volved in cytoskeletal reorganization, and cytoskeletal re- organization may be an important strategy for shrimp to respond to HPND. Integrins are α-β heterodimeric cell sur- face receptors that mediate cell-cell and cell-matrix inter- actions and serve essential cell adhesion functions (Gian- cotti., 1999; Huhtala., 2005; Johnson., 2009). In our study, the dysregulation of integrin alpha 5 and in- tegrin beta 1 in ECM-receptor interaction pathway maylead to cytoskeletal reorganization of HPND-infected shrimp. In lepidopteran pests, cytoskeletal reorganization is a means to respond to the exotoxin from(Castagnola., 2014). In fish, remodeling of gill microstructure is an important strategy to protect against ammonia stress (Liew., 2013; Sinha., 2014). In prawns, cytoskeleton regulation mediated by the Rho sig- naling pathway helps to attenuate AHPND pathogenesis (Ng., 2018). It can also lead to apoptosis of hemo- cytes and immune response in shrimp (Zheng., 2018a).These data suggest that cytoskeletal reorganization may represent an important strategy forto respond to HPND infection.
It is speculated that cytoskeletal reorganization may con- tribute to the immune defense process of shrimp (Wang., 2017). Phagocytosis is a conserved cellular immune response in eukaryotes (Koiwai., 2018). Phagocyto- sis is an important process that phagocytic cells engulf pathogens or antigens into phagosomes. As a multifunc- tional gene, integrins have been found to mediate phago- cytosis in oyster (Jia., 2015), drosophila (Nonaka., 2013), crab (Huang., 2015) and prawns (Koi- wai., 2018) in previous studies. Thus, integrin-in- volved phagocytosis may play an important role in animal immunity. In this study, the differentially expressed inte- grin alpha 5 and integrin beta 1 genes were present in boththe ECM-receptor interaction pathway and phagosome pathway. These data indicated that there may be active in- teraction between cytoskeletal reorganization and immune defense. Cytoskeletal reorganization may play a role in phagocytosis of shrimp infected with HPND.
Lysosome is involved in the clearance of pathogens. Phagosomes combine with lysosomes to form phagolyso- somes, where the pathogens are degraded due to respira- tory or oxidative burst, low pH, activity of lysosomal acid hydrolases, and secretion of microbicidal substances (Wu., 2019). In this study, a series of immune-related genes in the pathways of phagosome and lysosome were severe- ly affected in shrimp infected with HPND. Previously, ly- sosomes have been found to be involved in immune de- fense ofafter infection with(Yao., 2008). These data indicated that phagosome and lysosome may play important roles ineliminating pathogens after shrimpsare infected with HPND.However, the definite molecular mechanism of host re- sponse to the pathogen is still unclear. The molecular re- gulation of the pathways of ECM-receptor interaction, pha- gosome and lysosome need more studies in the future.
In addition to the above results, we also identified a large number of SSRs as potential molecular markers in our study. Similar studies have been reported in several crustaceans, including(Yu., 2014),(Wang., 2017),(Shen., 2014), and(Ding., 2018). In the shrimp, a high-density linkage map was built for the In- dia population based on SNP markers (Baranski., 2014). However, few data were available about SSR mar- kers ofinfected with HPND. The potential SSR markers identified in this study can provide impor- tant genetic resource for the selective breeding of disease- resistant shrimp.
5 Conclusions
In this study, RNA sequencing and comparative tran- scriptomics analysis were performed to identify genes and signaling pathways involved in hepatopancreas response to HPND in cultured. A large number of func- tional genes and potential SSR loci were identified from the transcriptome, adding a substantial contribution to the genetic resource available for. Totally 5204DEGs were detected in HPND-infected shrimps, with 3399up-regulated genes and 1805 down-regulated ones. Se- veral well-known immune-related genes including a list of immune effector genes were differentially regulated, andpathways related to ECM-receptor interaction, phagosome and lysosome were significantly enriched, implying their crucial roles in shrimp resistance to HPND. The results obtained in this study have offered new insights into host response mechanism to HPND. The study also establishes a foundation for future research on molecular marker de- velopment and genetic breeding of HPND-resistant shrimp.
Acknowledgements
This study was funded in part by the Natural Science Foundation of Guangdong Province (No. 2018A030310049), 2019 Annual Guangdong Provincial Special fund of Nan- hai Economic Shrimp Breeding and Culture Laboratory (No.2319412525), the Nanhai Scholar Project of Guangdong Ocean University, Fangchenggang Science and Technol- ogy Plan Project (No. AD19008017), the R & D Program of Key Areas in Guangdong Province (No. 2020B020201 0009), and 2019 Provincial Financial Special Fund Con- struction Project of Guangdong Ocean University (No. 231619003).
Aranguren, L.F., Han, J.E., and Tang, K.F.J., 2017.(EHP) is a risk factor for acute hepato-pancreatic necrosis disease (AHPND) and septic hepatopan- creatic necrosis (SHPN) in the Pacific white shrimp., 471: 37-42, https://doi.org/10.1016/j.aquaculture.2016.12.038.
Arunrut, N., Kampeera, J., Sirithammajak, S., Sanguanrut, P., Pro-espraiwong, P., Suebsing, R.,., 2016. Sensitive visual de- tection of AHPND bacteria using loop-mediated isothermal am-plification combined with DNA-functionalized gold nanopar- ticles as probes., 11: e0151769, https://doi.org/10.1371/journal.pone.0151769.
Baranski, M., Gopikrishna, G., Robinson, N.A., Katneni, V.K., Shekhar, M.S., Shanmugakarthik, J.,., 2014. The develop-ment of a high density linkage map for black tiger shrimp () based on cSNPs., 9 (1):e85413, https://doi.org/10.1371/journal.pone.0085413.
Boonsri, N., Rudtanatip, T., Withyachumnarnkul, B., and Wong- prasert, K., 2016. Protein extract from red seaweedprevents acute hepatopancreatic necrosis disease (AHPND) infection in shrimp., 29: 1597-1608, https://doi.org/10.1007/s10811-016-0969-2.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K.,., 2009. BLAST plus: Architecture and app-lications., 10: 421, https://doi.org/10.1186/1471-2105-10-421.
Cao, J., Wu, L., Jin, M., Li, T.T., Hui, K.M., and Ren, Q., 2017. Transcriptome profiling of thelymphoid organ under the white spot syndrome virus chal- lenge., 67: 27-39, https://doi.org/10.1016/j.fsi.2017.05.059.
Castagnola, A., and Stock, S.P., 2014. Common virulence fac- tors and tissue targets of entomopathogenic bacteria for biolo- gical control of lepidopteran pests., 5: 139-166, https://doi.org/10.3390/insects5010139.
Chomwong, S., Charoensapsri, W., Amparyup, P., and Tassa- nakajon, A., 2018. Two host gut-derived lactic acid bacteria activate the proPO system and increase resistance to an AHPND-causing strain ofin the shrimp., 89: 54-65, https://doi.org/10.1016/j.dci.2018.08.002.
Chumpol, S., Kantachote, D., Nitoda, T., and Kanzaki, H., 2017. The roles of probiotic purple nonsulfur bacteria to control wa- ter quality and prevent acute hepatopancreatic necrosis disease(AHPND) for enhancement growth with higher survival in white shrimp () during cultivation., 473: 327-336, https://doi.org/10.1016/j.aquaculture.2017.02.033.
Conesa, A., Gotz, S., Garcia-Gomez, J.M., Terol, J., Talon, M., and Robles, M., 2005. Blast2GO: A universal tool for annota- tion, visualization and analysis in functional genomics research., 21: 3674-3676, https://doi.org/10.1093/bioinformatics/bti610.
Dabu, I.M., Lim, J.J., Arabit, P.M.T., Orense, S.J.A.B., Tabar-dillo, J.A., Corre, V.L.,., 2017. The first record of acute hepatopancreatic necrosis disease in the Philippines., 48: 792-799, https://doi.org/10.1111/are.12923.
De la Pe?a, L. D., Cabillon, N.A., Catedral, D.D., Amar, E.C., Usero, R.C., Monotilla, W.D.,., 2015. Acute hepatopan- creatic necrosis disease (AHPND) outbreaks inandcultured in the Philippines., 116: 251-254, https://doi.org/10.3354/dao02919.
Ding, Z.F., Jin, M., and Ren, Q., 2018. Transcriptome analysis ofintestines under the white spot syndrome virus and poly (I:C) challenges., 13 (9): e0204626, https://doi.org/10.1371/journal.pone.0204626.
Dong, X., Bi, D., Wang, H., Zou, P., Xie, G., Wan, X.,., 2017a. pirABvp-bearingandpathogens isolated from the same AHPND-af- fected pond possess highly similar pathogenic plasmids., 8: 1859, https://doi.org/10.3389/fmicb.2017.01859.
Dong, X., Wang, H., Xie, G., Zou, P., Guo, C., Liang, Y.,., 2017b. An isolate ofcarrying the pir(VP) gene causes acute hepatopancreatic necrosis disease., 6: e2. https://doi.org/10.1038/emi.2016.131.
Félix, D. M., Elías, J. A. L., Córdova, A. I. C., Córdova, L. R. M., González, A. L., Jacinto, E. C.,.,2017. Survival ofshrimp fed on diets supplemented withsp. is improved after challenges by., 148: 118-123, https://doi.org/10.1016/j.jip.2017.06.003.
Ge, Q., Li, J., Wang, J., Li, J., Ge, H., and Zhai, Q., 2017. Tran- scriptome analysis of the hepatopancreas ininfected with an AHPND-causing strain of., 67: 620-633, https://doi.org/10.1016/j.fsi.2017.06.047.
Giancotti, F.G., and Ruoslahti, E., 1999. Integrin signaling., 285: 1028-1032, https://doi.org/10.1126/science.285.5430.1028.
Grabherr, M.G., Haas, B.J., Yassour, M., Levin, J.Z., Thomp- son, D.A., Amit, I.,., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome., 29: 644-652, https://doi.org/10.1038/nbt.1883.
Han, J.E., Tang, K.F.J., Pantoja, C.R., White, B.L., and Light-ner, D.V., 2015a. qPCR assay for detecting and quantifying a virulence plasmid in acute hepatopancreatic necrosis disease (AHPND) due to pathogenic., 442: 12-15, https://doi.org/10.1016/j.aquaculture.2015.02.024.
Han, J.E., Tang, K.F.J., Tran, L.H., and Lightner, D.V., 2015b. Photorhabdus insect-related (Pir) toxin-like genes in a plas- mid of, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp., 113: 33, https://doi.org/10.3354/dao02830.
Hidehiro, K., Phan Thi, V., Dang, L.T., and Ikuo, H., 2015. Draftgenome sequence of non-acute hepa-topancreatic necrosis disease strain kc13.17.5, isolated from diseased shrimp in Vietnam., 3: e00978-00915, https://doi.org/10.1128/genomeA.00978-15.
Huang, Y., Zhao, L.L., Feng, J.L., Zhu, H.X., Huang, X., Ren, Q.,., 2015. A novel integrin function in innate immunity from Chinese mitten crab ()., 52: 155-165, https://doi.org/10.1016/j.dci.2015.05.005.
Huhtala, M., Heino, J., Casciari, D., de Luise, A., and Johnson, M.S., 2005. Integrin evolution: Insights from ascidian and teleost fish genomes., 24: 83-95, https://doi.org/10.1016/j.matbio.2005.01.003.
Imaizumi, K., Tinwongger, S., Kondo, H., and Hirono, I., 2018. Disinfection of an EMS/AHPND strain ofusing ozone nanobubbles., 41: 725-727, https://doi.org/10.1111/jfd.12783.
Jia, Z.H., Zhang, T., Jiang, S., Wang, M.Q., Cheng, Q., Sun, M.Z.,., 2015. An integrin from oystermediates the phagocytosis towardthrough LPS binding activity., 53: 253-264, https://doi.org/10.1016/j.dci.2015.07.014.
Johnson, M.S., Lu, N., Denessiouk, K., Heino, J., and Gullberg, D., 2009. Integrins during evolution: Evolutionary trees and model organisms.–, 1788: 779-789, https://doi.org/10.1016/j.bbamem.2008.12.013.
Joshi, J., Srisala, J., Truong, V.H., Chen, I., Nuangsaeng, B., and Suthienkul, O.,., 2014. Variation inisolates from a single Thai shrimp farm experien- cing an outbreak of acute hepatopancreatic necrosis disease (AHPND)., 428-429: 297-302, https://doi.org/10.1016/j.aquaculture.2014.03.030.
Jun, J.W., Han, J.E., Tang, K.F.J., Lightner, D.V., Kim, J., Seo,S.W.,., 2016. Potential application of bacteriophage pVp-1: Agent combatingstrains associated with acute hepatopancreatic necrosis disease (AHPND) in shrimp., 457: 100-103, https://doi.org/10.1016/j.aquaculture.2016.02.018.
Koiwai, K., Kondo, H., and Hirono, I., 2018. RNA-seq identi- fies integrin alpha of kuruma shrimpas a candidate molecular marker for phagocytic hemo-cytes., 81: 271-278, https://doi.org/10.1016/j.dci.2017.12.014.
Kondo, H., Tinwongger, S., Proespraiwong, P., Mavichak, R.,Unajak, S., Nozaki, R.,., 2014. Draft genome sequences of six strains ofisolated from early mor- tality syndrome/acute hepatopancreatic necrosis disease shrimp in Thailand., 2:e00221-14, https://doi.org/10.1128/genomeA.00221-14.
Kongrueng, J., Mitraparp-Arthorn, P., Bangpanwimon, K., Ro- bins, W., Vuddhakul, V., and Mekalanos, J., 2017. Isolation ofand like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by., 124: 223-232, https://doi.org/10.3354/dao03120.
Kumar, V., Baruah, K., Nguyen, D.V., Smagghe, G., Vossen, E., and Bossier, P., 2018. Phloroglucinol-mediated Hsp70 produc- tion in crustaceans: Protection againstinand., 9: 1091, https://doi.org/10.3389/fimmu.2018.01091.
Lai, H.C., Ng, T.H., Ando, M., Lee, C.T., Chen, I.T., Chuang, J.C.,., 2015. Pathogenesis of acute hepatopancreatic ne- crosis disease (AHPND) in shrimp., 47: 1006-1014, https://doi.org/10.1016/j.fsi.2015.11.008.
Li, B., and Dewey, C.N., 2011. RSEM: Accurate transcript quan- tification from RNA-Seq data with or without a reference ge- nome., 12: 323, https://doi.org/10.1186/1471-2105-12-323.
Liew, H.J., Sinha, A.K., Nawata, C.M., Blust, R., Wood, C.M., and De Boeck, G., 2013. Differential responses in ammonia excretion, sodium fluxes and gill permeability explain diffe- rent sensitivities to acute high environmental ammonia in threefreshwater teleosts., 126: 63-76, https://doi.org/10.1016/j.aquatox.2012.10.012.
Liu, L., Xiao, J., Zhang, M., Zhu, W., Xia, X., Dai, X.,., 2018. Astrain as the causative agent of AHPND in cultured shrimp,., 153: 156-164, https://doi.org/10.1016/j.jip.2018.02.005.
Lun, J., Liu, D., Liu, T., Zhang, S., Dong, Y., Li, C.,., 2018. Evaluation of outer membrane protein U (OmpU) as a novel capture target ofand rapid detection of acute hepatopancreatic necrosis disease (AHPND) using PCRcombined with immunomagnetic separation., 485: 225-232, https://doi.org/10.1016/j.aquaculture.2017.11.046.
Maralit, B.A., Jaree, P., Boonchuen, P., Tassanakajon, A., and Somboonwiwat, K., 2018. Differentially expressed genes in he- mocytes ofchallenged withAHPND (VPAHPND) and VPAHPNDtoxin., 81: 284-296, https://doi.org/10.1016/j.fsi.2018.06.054.
Ng, T.H., Lu, C.W., Lin, S.S., Chang, C.C., Tran, L.H., Chang, W.C.,., 2018. The Rho signalling pathway mediates the pathogenicity of AHPND-causingin shrimp., 20: e12849, https://doi.org/10.1111/cmi.12849.
Nonaka, S., Nagaosa, K., Mori, T., Shiratsuchi, A., and Nakani- shi, Y., 2013. Integrin alpha PS3/beta nu-mediated phagocy- tosis of apoptotic cells and bacteria in., 288: 10374-10380, https://doi.org/10.1074/jbc.M113.451427.
Qin, Z., Babu, V.S., Wan, Q., Zhou, M., Liang, R., Muhammad, A.,., 2018. Transcriptome analysis of Pacific white shrimp () challenged byreveals unique immune-related genes., 77: 164-174, https://doi.org/10.1016/j.fsi.2018.03.030.
Rao, R., Bhassu, S., Bing, R.Z.Y., Alinejad, T., Hassan, S.S., and Wang, J., 2016. A transcriptome study onhepatopancreas experimentally challenged with white spot syndrome virus (WSSV)., 136: 10-22, https://doi.org/10.1016/j.jip.2016.01.002.
Reiner, A., Yekutieli, D., and Benjamini, Y., 2003. Identifying dif- ferentially expressed genes using false discovery rate control- ling procedures., 19: 368-375, https://doi.org/10.1093/bioinformatics/btf877.
Restrepo, L., Bayot, B., Arciniegas, S., Bajana, L., Betancourt, I., Panchana, F.,., 2018. PirVP genes causing AHPND iden- tified in a newspecies () within the com-mensalclade., 8: 13080, https://doi.org/10.1038/s41598-018-30903-x.
Robinson, M.D., and Oshlack, A., 2010. A scaling normaliza- tion method for differential expression analysis of RNA-seq data., 11: R25, https://doi.org/10.1186/gb-2010-11-3-r25.
Shen, H.S., Hu, Y.C., Ma, Y.C., Zhou, X., Xu, Z.H., Shui, Y.,., 2014. In-depth transcriptome analysis of the red swamp crayfish.,9 (10): e110548, https://doi.org/10.1371/journal.pone.0110548.
Sinha, A.K., Matey, V., Giblen, T., Blust, R., and De Boeck, G., 2014. Gill remodeling in three freshwater teleosts in response to high environmental ammonia., 155: 166-180, https://doi.org/10.1016/j.aquatox.2014.06.018.
Sirikharin, R., Taengchaiyaphum, S., Sanguanrut, P., Chi, T.D., Mavichak, R., Proespraiwong, P.,., 2015. Characteriza- tion and PCR detection of binary, Pir-Like toxins fromisolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp., 10: e0126987,https://doi.org/10.1371/journal.pone.0126987.
Soonthornchai, W., Chaiyapechara, S., Klinbunga, S., Thongda, W., Tangphatsornruang, S., Yoocha, T.,., 2016. Differen- tially expressed transcripts in stomach ofin response to AHPND infection., 65: 53-63, https://doi.org/10.1016/j.dci.2016.06.013.
Soto-Rodriguez, S.A., Gomez-Gil, B., Lozano-Olvera, R., Betan-court-Lozano, M., and Morales-Covarrubias, M.S., 2015. Field and experimental evidence ofas the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp () in Northwestern Me- xico., 81: 1689-1699, https://doi.org/10.1128/AEM.03610-14.
Stalin, N., and Srinivasan, P., 2016. Characterization ofand its specific phage from shrimp pond in Palk Strait, South East coast of India., 44: 526-533, https://doi.org/10.1016/j.biologicals.2016.08.003.
Thammasorn, T., Jitrakorn, S., Charoonnart, P., Sirimanakul, S., Rattanarojpong, T., Chaturongakul, S.,., 2017. Probiotic bacteria () expressing specific double-stranded RNA and its potential for controlling shrimp viral and bacterial diseases., 25: 1679-1692, https://doi.org/10.1007/s10499-017-0144-z.
Thitamadee, S., Prachumwat, A., Srisala, J., Jaroenlak, P., Sala- chan, P.V., Sritunyalucksana, K.,., 2016. Review of cur- rent disease threats for cultivated penaeid shrimp in Asia., 452: 69-87, https://doi.org/10.1016/j.aquaculture.2015.10.028.
Tran, L., Nunan, L., Redman, R.M., Mohney, L.L., Pantoja, C.R., Fitzsimmons, K.,., 2013. Determination of the infec- tious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp., 105: 45-55, https://doi.org/10.3354/dao02621.
Wang, H., Wang, C., Tang, Y., Sun, B., Huang, J., and Song, X., 2018.probiotics as potential biocontrol agents improve the survival ofchallenged with acute hepatopancreatic necrosis disease (AHPND)-cau- sing., 494: 30-36, https://doi.org/10.1016/j.aquaculture.2018.05.020.
Wang, W., Yang, S., Wang, C., Shi, L., Guo, H., and Chan, S., 2017. Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp., 71: 319-328, https://doi.org/10.1016/j.fsi.2017.10.028.
Wu, W., Dai, C., Duan, X., Wang, C., Lin, X., Ke, J.,., 2019. miRNAs induced by white spot syndrome virus involve in im-munity pathways in shrimp, 93: 743-751, https://doi.org/10.1016/j.fsi.2019.08.009.
Xie, C., Mao, X.Z., Huang, J.J., Ding, Y., Wu, J.M., Dong, S.,., 2011. KOBAS 2.0: A web server for annotation and iden-tification of enriched pathways and diseases., 39: W316-W322, https://doi.org/10.1093/nar/gkr483.
Yao, C.L., Wu, C.G., Xiang, J.H., Li, F.H., Wang, Z.Y., and Han, X. Z., 2008. The lysosome and lysozyme response in Chi- nese shrimptoand laminarin stimulation., 363: 124-129, https://doi.org/10.1016/j.jembe.2008.06.035.
Young, M.D., Wakefield, M.J., Smyth, G.K., and Oshlack, A., 2010. Gene ontology analysis for RNA-seq: Accounting for selection bias.,11: R14, https://doi.org/10.1186/gb-2010-11-2-r14.
Yu, Y., Wei, J.K., Zhang, X.J., Liu, J.W., Liu, C.Z., Li, F.H.,., 2014. SNP discovery in the transcriptome of white Pa- cific shrimpby next generation se- quencing., 9(1): e87218, https://doi.org/10.1371/journal.pone.0087218.
Zhang, Y., Gao, G., Lin, R., Aweya, J.J., Tao, M., and Wang, F., 2018. Transcriptome analyses revealhemocytes response to lipopolysaccharide., 76: 187-195, https://doi.org/10.1016/j.fsi.2018.03.002.
Zheng, Z., Aweya, J.J., Wang, F., Yao, D., Lun, J., Li, S.,., 2018a. Acute hepatopancreatic necrosis disease (AHPND) related microRNAs ininfected with AHPND-causing strain of., 19: 335, https://doi.org/10.1186/s12864-018-4728-4.
Zheng, Z., Wang, F., Aweya, J.J., Li, R., Yao, D., Zhong, M.,., 2018b. Comparative transcriptomic analysis of shrimp hemo- cytes in response to acute hepatopancreas necrosis disease (AHPND) causinginfection., 74: 10-18,https://doi.org/10.1016/j.fsi. 2017.12.032.
. E-mail: wangwei.hg@outlook.com E-mail: scb248@126.com
August 30, 2020;
October 26, 2020;
January 13, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
(Edited by Qiu Yantao)
 Journal of Ocean University of China2021年5期
Journal of Ocean University of China2021年5期
- Journal of Ocean University of China的其它文章
- Numerical Modelling for Dynamic Instability Process of Submarine Soft Clay Slopes Under Seismic Loading
- DcNet: Dilated Convolutional Neural Networks for Side-Scan Sonar Image Semantic Segmentation
- Bleaching with the Mixed Adsorbents of Activated Earth and Activated Alumina to Reduce Color and Oxidation Products of Anchovy Oil
- The Brown Algae Saccharina japonica and Sargassum horneri Exhibit Species-Specific Responses to Synergistic Stress of Ocean Acidification and Eutrophication
- Effects of Dietary Protein and Lipid Levels on Growth Performance, Muscle Composition, Immunity Index and Biochemical Index of the Greenfin Horse-Faced Filefish (Thamnaconus septentrionalis) Juvenile
- Genome-Wide Patterns of Codon Usage in the Pacific Oyster Genome
