Molecular mapping of YrTZ2, a stripe rust resistance gene in wild emmer accession TZ-2 and its comparative analyses with Aegilops tauschii
WANG Zhen-zhong , XIE Jing-zhong, GUO Li, ZHANG De-yun, LI Gen-qiao, FANG Ti-lin, CHEN Yong-xing, LI Jun, WU Qiu-hong, LU Ping, LI Miao-miao, WU Hai-bin, , ZHANG Huai-zhi, ZHANG Yan, YANG Wu-yun, LUO Ming-cheng, Fahima Tzion, LIU Zhi-yong
1 China Rural Technology Development Center, Beijing 100045, P.R.China
2 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, P.R.China
3 College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, P.R.China
4 Department of Plant and Soil Sciences, Oklahoma State University, Stillwater, OK 74078, USA
5 Crop Research Institute, Sichuan Academy of Agriculture Sciences, Chengdu 610066, P.R.China
6 China National Seed Group Co., Ltd., Beijing 100045, P.R.China
7 Department of Plant Sciences, University of California, Davis, CA 95616, USA
8 Institute of Evolution, University of Haifa, Mount Carmel, Haifa 31905, Israel
1. Introduction
Bread wheat (Triticum aestivumL.) is one of the top three most important food crops which affects worldwide food security. Stripe rust, caused byPuccinia striiformisf. sp.tritici(Pst), is one of the severest wheat diseases worldwide.Growing resistant cultivars is the most cost-effective and environmentally friendly method to control this disease.Up to date, more than 70 formally named and many other provisionally designated stripe rust resistance genes or quantitative trait loci (QTLs) have been reported (McIntoshet al. 2013, 2014, 2016). Out of these, two stripe rust resistance genes,Yr36andYr18, have been isolated through map-based cloning (Fuet al. 2009; Krattingeret al. 2009). However, stripe rust resistance genes tend to become ineffective due to the continuous evolution ofPstraces and monoculture of cultivars carrying same resistance gene in wide area (Wanet al. 2004, 2007; de Vallavieille-Popeet al. 2012). So there is a continued need for identification and utilization of diversified resistance genes from various wheat germplasm resources.
Wild emmer (T.turgidumssp.dicoccoides, 2n=4x=28,AABB) is allotetraploid with A and B genomes, which was derived from a spontaneous hybridization of two diploid wild grassesT.urartu(2n=14, AA) and an as yet unidentifiedAegilopsspecies related toAe.speltoides(2n=14, SS)(Dvorak and Zhang 1990; Dvoraket al. 1993). Wild emmer is the progenitor of cultivated tetraploid durum wheat(T.turgidumssp.durum, AABB) and hexaploid bread wheat (T.aestivum, AABBDD) (Feldman 2001), harboring abundant genetic resources for wheat improvement,including tolerance to abiotic stresses (salt, drought and heat), resistance to biotic stresses (powdery mildew, rusts and Fusarium head blight), grain protein quality and quantity,and micronutrient concentrations (Zn, Fe, and Mn) (Xie and Nevo 2008). Artificial selection during wheat domestication resulted in an inadvertent loss of genes and QTL beneficial for improving wheat agronomic and economic traits.Although they could be introgressed into modern wheat cultivars through traditional breeding methods, molecular breeding provides an improved strategy which can greatly shorten the breeding period.
Previously, molecular markers used for genetic linkage maps are mainly comprised of restriction fragment length polymorphisms (RFLPs) (Blancoet al. 1998), amplified fragment length polymorphisms (AFLPs) (Nachitet al.2001), simple sequence repeats (SSRs) (Somerset al.2004; Songet al. 2005), and diversity arrays technology(DArT) (Akbariet al. 2006; Peleget al. 2008). Recently,single nucleotide polymorphisms (SNPs) are increasingly applied for high-density genetic mapping, physical map construction, comparative genomics analysis, genome-wide association studies (GWAS), and genomic selection in rice(Oryza sativa) (Zhaoet al. 2011), maize (Zea mays) (Ganalet al. 2011; Riedelsheimeret al. 2012), and wheat (Akhunovet al. 2009; Luoet al. 2009; Cavanaghet al. 2013; Wanget al. 2014).
Fine mapping and map-based cloning of resistance genes in wheat is a tedious process because of the allopolyploid (AABBDD), large genome size (17 gigabase),and numerous repeat DNA (90%). The availability of draft genome sequences and International Wheat Genome Sequencing Consortium (IWGSC) survey sequences ofT.aestivum cv. Chinese Spring,T.urartuaccession G1812,andAe.tauschiiaccession AL8/78 (Brenchleyet al. 2012;Jiaet al. 2013; Linget al. 2013; IWGSC 2014) facilitates gene mapping. In particular, the released high-resolution SNP genetic linkage map and physical map ofAe.tauschiiaccession AL8/78 provide closely wheat-related target for comparative genomics analyses (Luoet al. 2013).
In present study, we report: (1) the identification and genetic mapping a near-immunity stripe rust resistance geneYrTZ2derived from wild emmer with microsatellite markers and 90K iSelect SNP genotyping assay, and (2) comparative genomics analysis of the genomic regions ofYrTZ2with the genetic linkage map and physical map ofAe.tauschii.
2. Materials and methods
2.1. Plant materials
The wild emmer accession TZ-2 resistant to stripe rust was crossed with a highly susceptible durum wheat cultivar Langdon. A set of 200 F6:7recombinant inbred lines (RILs)advanced by single-seed descent approach and the parents Langdon and TZ-2 were evaluated for stripe rust resistance with the prevailingPstrace CYR34. A highly susceptible wheat variety Mingxian 169 was used as control.
2.2. Stripe rust evaluations
The parental lines Langdon and TZ-2, Langdon/TZ-2 F1,200 F6:7RILs, and the susceptible control Mingxian 169 were sowed in two-meter rows (each row with 30 seeds),which inoculated withPstrace CYR34 at the jointing stage in Chengdu of Sichuan Province, China. At 18–20 days post inoculation when the susceptible control Mingxian 169 had become severely infected, the infection type (IT) was recorded with a scale of 0–4, with 0 (immune reaction), 0;(hypersensitive reaction), 1 (highly resistant), 2 (moderately resistant), 3 (moderately susceptible), and 4 (highly susceptible), the values of 0–2 were rated as resistant, and those of 3–4 were rated as susceptible (Zhanget al. 2001).ITs were recorded after 10 days.
2.3. Genomic DNA isolation and SSR marker analysis
Genomic DNAs of the parental lines and the F6:7RILs population were extracted from seedings using the Plant Genomic DNA Kit (Tiangen Biotech, Co., Ltd., Beijing, China). DNA concentration was quantified using NanoPhotometer?P360(Implem GmbH, Munich, Germany) and normalized to 100 ng μL–1. Resistant and susceptible DNA bulks were prepared by mixing equal amounts of DNA from 10 homozygous resistant and 10 homozygous susceptible F6:7families, respectively, for bulked segregant analysis (Michelmoreet al. 1991). SSR markers (Xgwm,Xwmc,Xbarc,Xcfa, andXcfdseries, https://wheat.pw.usda.gov) were tested for polymorphism and the polymorphic SSR markers were subsequently genotyped in the RIL mapping populations.
PCR reactions were carried out in a 10-μL reaction volume with the following conditions: one denaturation cycle at 94°C for 5 min, followed by 35 cycles at 94°C for 45 s,55–65°C (depending on specific primers) for 45 s, and 72°C for 1 min, followed by an extension step of 72°C for 10 min.Fragment analysis of PCR products were carried out on 8% non-denaturing polyacrylamide gels (39 acrylamide:1 bisacrylamide). After electrophoresis, the gels were silver stained and photographed.
2.4. lnfinium 90K iSelect SNP genotyping
To saturate the genomic region harboring the stripe rust resistance gene, the 200 F6:7RILs were genotyped using wheat 90K iSelect SNP genotyping assay platform at the Genome Center of University of California, Davis according to the manufacturer’s protocol. SNP allele clustering was conducted with two population-based detection algorithms:Density Based Spatial Clustering of Applications with Noise (DBSCAN) and Ordering Points to Identify the Clustering Structure (OPTICS) using the polyploid version of GenomeStudio software as described in Wanget al.(2014). Subsequently, the cluster matrix of polymorphic SNP markers was output from the polyploid version of GenomeStudio, and the genotypes of samples assigned in TZ-2 cluster were marked ‘1’, the genotypes of sample located in Langdon cluster were marked ‘2’, and the others were marked ‘0’.
2.5. Genetic mapping of the stripe rust resistance gene
The polymorphic SNP markers, SSR markers, and stripe rust resistance data were used for linkage analysis with the MultiPoint mapping software as described in Peleget al.(2008) and Luoet al. (2013). Co-segregating SNP markers were regarded as a polymorphic locus. The linkage map was constructed with the software Mapdraw V2.1 (Liu and Meng 2003).
3. Results
3.1. Inheritance of the stripe rust resistance gene in TZ-2
The wild emmer accession TZ-2 and durum wheat cultivar Langdon showed nearly immune and highly susceptible reaction to stripe rust, respectively. The F1plants are highly resistant to CYR34, indicating the dominant nature of the stripe rust resistance in TZ-2. Of the 200 F6:7RILs, 103 were resistant (IT 0–2) and 97 were susceptible (3–4), which fits the expected 1:1 ratio (χ21:1=0.18,P<0.05), indicating that a single locus, provisionally designatedYrTZ2, in TZ-2 is responsible for the stripe rust resistance.
3.2. ldentification of microsatellite markers linked to YrTZ2
Initially, 194 SSR primers distributed randomly throughout the whole genome were screened for polymorphisms between the parents as well as the resistant and susceptible DNA bulks. SSR markers,Xwmc406,Xwmc230,Xgwm413,Xwmc128, andXcfd65revealed polymorphisms between the parents and linkage with the bulks. These markers were tested on the F6:7population and a linkage map for stripe rust disease resistance geneYrTZ2was constructed. The geneYrTZ2was located into a 1.1-cM genetic interval between SSR markersXwmc230andXgwm413(Fig. 1).
3.3. Chromosome arm assignment and physical bin mapping
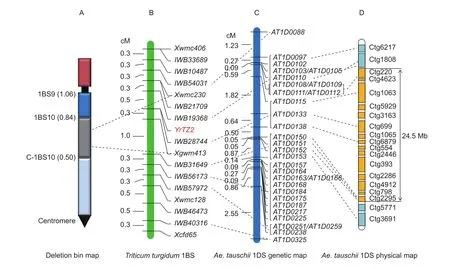
Fig. 1 Genetic linkage map of the stripe rust resistance gene YrTZ2. Ae. tauschii, Aegilops tauschii.
In order to locate theYrTZ2in the deletion bins on chromosome 1BS, Chinese Spring homoeologous group 1 nullisomic-tetrasomics, ditelosomics and deletion lines were used to assign the chromosome and physical bin location of theYrTZ2-linked SSR markers. Both SSR markersXgwm413andXwmc230were present in N1A-T1B, N1DT1A, Dt1BS, and 1BS-9, but absent in N1B-T1A, Dt1BL,and 1BS-10 (Fig. 2), indicating thatYrTZ2is located on chromosome 1BS bin 0.50–0.84 (Fig. 1).
3.4. ldentification of SNP markers linked to YrTZ2
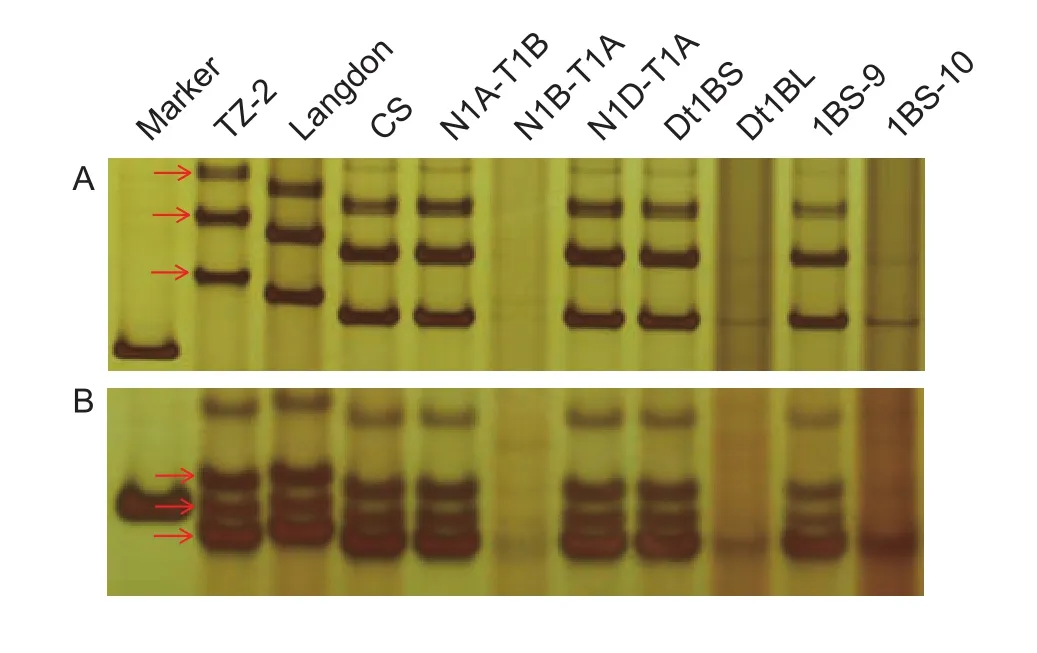
Fig. 2 Amplification patterns of markers Xwmc230 (2A)and Xgwm413 (2B) in the parental lines TZ-2 and Langdon,Chinese Spring (CS) and its homoeologous group 1 nullisomictetrasomics, ditelosomics, and deletion lines.
The 200 F6:7RILs were genotyped with 90K iSelect SNP genotyping assay. Based on the maximum likelihood estimation with LOD threshold of 3.0, linkage groups were constructed with cluster threshold (recombination rate) value of 0.1. After ordering, the matrix data of each linkage group were output and the markers with low confidence order were rechecked with clustering graph in polyploid version of GenomeStudio, especially makers with double crossover.Linkage analysis and recheck between the matrix data and GenomeStudio clustering graph were performed alternately to adjust or delete the markers with low confidence order.The jackknife resampling procedure was used to evaluate the reliability of the ordered linkage groups with the parameter setting: Jackknife value 90, number of iteration 10, time to Es 0.3, and control of monotony threshold of 1.0, which can remove the markers causing unstable neighborhoods in each linkage group. Then the cluster threshold (recombination rate) value was increased to merge different pairs of linkage groups that maybe belong to one chromosome. Finally, 15 625 polymorphic SNP markers were clustered into 14 linkage groups. Polymorphic SNP and SSR markers linked toYrTZ2were used to construct a high-resolution linkage map ofYrTZ2. Due to the limitation of population size, multiple co-segregating SNP markers were mapped at one locus and used as skeleton marker.All together, 11 polymorphic loci (consisting of 250 SNP markers),IWB33689,IWB10487,IWB54031,IWB21709,IWB19368,IWB28744,IWB31649,IWB56173,IWB57972,IWB46473, andIWB40316, were integrated into the genetic linkage map ofYrTZ2(Fig. 1).YrTZ2was finally delimited into a 0.8-cM interval between SNP locusIWB19368and SSR markerXgwm413, and co-segregated with SNP locusIWB28744(co-segregated with 28 SNP markers)(Fig. 1).
3.5. ldentification of collinearity genomic region of YrTZ2 in Ae. tauschii and comparative genomics analysis
The sequences of the 250 SNP markers clustered into 11 polymorphic loci were used as queries to search theAe.tauschiiSNP marker extended sequence database to identify the orthologous gene pairs betweenT.dicoccoides1BS andAe.tauschii1DS. Out of the 11 polymorphic loci, 7 loci,IWB54031,IWB19368, IWB28744,IWB31649,IWB56173,IWB57972,andIWB40316, identified 31 orthologous SNP marker extended sequence inAe.tauschii. Compararive genomics analysis revealed high levels collinearity betweenYrTZ2genomic region and its orthologous genomic regions inAe.tauschii1DS (Fig. 1;Appendix A).
YrTZ2was mapped between SNP markersIWB19368andIWB31649, and co-segregated withIWB28744.IWB19368andIWB31649correspond to the extended sequences of markers AT1D0112 (distal) and AT1D0150(proximal), respectively, on chromosome 1DS that were anchored to the assembled BAC contigs ctg220 and ctg2295 in the physical map ofAe.tauschii. Therefore, the genomic region betweenIWB19368andIWB31649was orthologous to a 24.5-Mb containing 15 BAC contigs, ctg220, ctg4623,ctg1063, ctg5929, ctg3163, ctg699, ctg1065, ctg6879,ctg554, ctg2446, ctg393, ctg2286, ctg4912, ctg798, and ctg2295 on chromosome 1DS (Fig. 1).
4. Discussion
4.1. Comparison of YrTZ2 with other stripe rust resistance genes on chromosome 1BS
Bread wheat is serving as an important global food crop all the time. The maximizing wheat production is becoming a big challenge for researchers, breeders, and growers.
Wild relatives of wheat harbor rich genetic resource for wheat improvement (Schneideret al. 2008; Xie and Nevo 2008). Wild emmer is the ancestor of modern cultivated wheat and mainly distributed in central-eastern (Turkey,Iran and Iraq) and western areas (Syria, Lebanon, Jordan and Israel) of the Fertile Crescent (Avniet al. 2014). Wild emmer harbors abundant beneficial traits that can be introgressed into tetraploid and hexaploid wheat in modern wheat breeding programs. However, wild emmer has not been explored thoroughly and its potential in wheat breeding programs remains to be further characterized (Xie and Nevo 2008).
Wild emmer accession TZ-2 was collected from Mount Hermon, Israel, and showed highly stripe rust resistance to manyPstraces (CYR29, CYR30, CYR31, CYR32, CYR33,and CYR34) in the greenhouse at the seedling stage and in field at the adult plant stage. In this study, genetic analysis showed that the stripe rust resistance to CYR34 in TZ-2 is controlled by a single dominant geneYrTZ2that was flanked byIWB19368andXgwm413in a 0.8-cM genetic interval on chromosome 1BS in deletion Bin 1BS10-0.50-0.84. Two stripe rust resistance genes,Yr15andYrH52, were derived from Israeli wild emmer wheat and located on chromosome 1BS.Yr15was identified from wild emmer accession G25 and mapped on chromosome 1BS using cytogenetic analysis (McIntoshet al. 1996) and molecular markers (Sunet al. 1997; Chaguéet al. 1999; Ramirez-Gonzalezet al.2015). Stripe rust resistance geneYrH52inT.dicoccoidesaccession Hermon 52 (Penget al. 1999, 2000) was linked to SSR markerXgwm413with a genetic distance of 1.3 cM(proximal).YrH52-linked polymorphic microsatellite markers analysis revealed thatYr15(Xgwm413/UBC212a-Yr15-Nor1) is different fromYrH52(Xgwm413/UBC212a/Nor1-YrH52-Xgwm273) on 1BS (Penget al. 2000). In the current study,YrTZ2(Xgwm413-YrTZ2-IWB19368) was located at similar portion of chromosome 1BS as that ofYr15andYrH52. Allelism tests need to be conducted in the future to clarify ifYrTZ2is allelic or closely linked toYr15orYrH52.
In addition toYr15,YrH52, andYrTZ2, several other stripe rust resistance genes have been identified on chromosome 1BS.Yr10was identified from Turkish hexaploid wheat accession PI 178383 and mapped at the telomeric region of chromosome 1BS (Wanget al. 2002).Yr24was derived fromT.turgidumsubsp.durumaccession K733 (McIntosh and Lagudah 2000).Yr26was assumed to be from durum line γ80-1, a γ-radiated mutant (Maet al.2001).YrCH42was identified from Chinese wheat cultivar Chuanmai 42 (Liet al. 2006). Evidences showed thatYr24,Yr26, andYrCH42were the same gene (Maet al. 2001; Liet al. 2006; McIntoshet al. 2013) and are ineffective against the new virulentPstrace CYR34 in China (Hanet al. 2012).YrAlpwas derived from spring wheat cultivar Alpowa with race-specific all-stage resistance (Lin and Chen 2007).Chenget al. (2014) identified broad-spectrum all-stage stripe rust resistance genesYr64andYr65in different bins of chromosome 1BS from durum wheat accessions PI 331260 and PI 480016, respectively. Converting these co-segregating SNPs into KASP assay and validate in the Chinese breeding materials would benefit the pyramiding of these genes on chromosome 1BS and development of durable and broad-spectrum stripe rust resistance varieties in wheat breeding program.
4.2. A SNP-based genetic linkage map of stripe rust resistance gene YrTZ2
The characteristics of large genome size, hexaploid nature and numerous repetitive DNA sequences presented a formidable challenge to fine mapping and map-based cloning of wheat genes. The nature of biallelic, costeffective, and high-throughput genotyping makes SNPs more suitable for genetic studies. The advent of wheat 90K iSelect SNP genotyping assay increased the number of gene-based markers which was applied for wheat genetic linkage map construction, genome-wide association studies,and comparative genomics analysis (Cavanaghet al. 2013;Wanget al. 2014; Wuet al. 2015). In this study,YrTZ2was initially mapped into a 1.1-cM genetic interval between SSR markersXwmc230andXgwm413. SNP genotyping assay was applied to saturate the genomic region ofYrTZ2.Finally,YrTZ2was delimited within a 0.8-cM genetic interval between locusIWB19368and markerXgwm413, and cosegregated with locusIWB28744(consisting of 28 attaching SNP markers) that could be served as a starting point for chromosome landing and map-based cloning as well as marker-assisted selection (MAS) of theYrTZ2gene.
4.3. Comparative genomics analyses of YrTZ2 with Aegilops tauschiii
Comparative genomics analyses provided an effective way for wheat gene mapping. By applying comparative genomics analysis using genome sequences ofBrachypodium,rice or sorghum, high-density genetic linkage maps of vernalization (VRN) genes (Yanet al. 2003, 2004, 2006),pairing homologous 1 (Ph1) (Griffithset al. 2006), grain protein content-B1 (Gpc-B1) (Uauyet al. 2006), yellow rust resistance geneYr36(Fuet al. 2009), wax production geneW1(Luet al. 2015), and powdery mildew resistance genePm6(Qinet al. 2011),Pm41 (Wanget al. 2014),Ml3D232(Zhanget al. 2010),MlIW170(Liuet al. 2012; Lianget al.2015), andMlIW172(Ouyanget al. 2014) were constructed.The draft genome sequences ofT.aestivum cv. Chinese Spring,T.urartuaccession G1812, andAe.tauschiiaccession AL8/78 enriched the available sequence resource and accelerated the wheat genomics research(Brenchleyet al. 2012; Jiaet al. 2013; Linget al. 2013).The physical map ofAe.tauschii, anchored with 7185 SNP marker-extended sequences, provided an efficient tool for comparative genomics analyses among grass families,and marker development for fine mapping and map-based cloning of genes in wheat (Luoet al. 2013). Comparative genomics analysis indicated highly collinearity betweenYrTZ2genomic region (IWB19368–IWB31649) of 1BS and a 24.5-Mb orthologous genomic region spanning 15 BAC-contigs ofAe.tauschii1DS. The recently finished BAC-contig sequence ofAe.tauschiiand Chinese Spring IWGSC whole genome assembly ver. 1.0 would further contribute to fine mapping, map-based cloning, and MAS ofYrTZ2.
5. Conclusion
Stripe rust resistance geneYrTZ2, originated from wild emmer, was mapped in a 0.8-cM genetic interval between SNP markerIWB19368and SSR markerXgwm413, and co-segregated with 28 SNP markers on chromosome 1BS.
Acknowledgements
This work was financially supported by the Science and Technology Service Network Initiative of Chinese Academy of Sciences (KFJ-STS-ZDTP-024).
Appendixassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G,Mohler V, Lehmensiek A, Kuchel H, Hayden M J, Howes N,Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A. 2006.Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome.Theoretical and Applied Genetics,113, 1409–1420.
Akhunov E, Nicolet C, Dvorak J. 2009. Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay.Theoretical and Applied Genetics,119, 507–517.
Avni R, Nave M, Eilam T, Sela H, Alekperov C, Peleg Z, Dvorak J, Korol A, Distelfeld A. 2014. Ultra-dense genetic map of durum wheat×wild emmer wheat developed using the 90K iSelect SNP genotyping assay.Molecular Breeding,34,1549–1562.
Blanco A, Bellomo M P, Cenci A, De Giovanni C, D’ovidio R, Iacono E, Laddomada B, Pagnotta M A, Porceddu E, Sciancalepore A, Simeone R, Tanzarella O A. 1998.A genetic linkage map of durum wheat.Theoretical and Applied Genetics,97, 721–728.
Brenchley R, Spannagl M, Pfeifer M, Barker G L, D’Amore R,Allen A M, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo N, Luo M C, Sehgal S, Gill B, Kianian S,et al. 2012. Analysis of the bread wheat genome using whole-genome shotgun sequencing.Nature,491, 705–710.
Cavanagh C R, Chao S, Wang S, Huang B E, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira G L, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, da Silva M L, Bockelman H, Talbert L,et al. 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars.Proceedings of the National Academy of Sciences of the United States of America,110,8057–8062.
Chagué V, Fahima T, Dahan A, Sun G L, Korol A B, Ronin Y I, Grama A, R?der M S, Nevo E. 1999. Isolation of microsatellite and RAPD markers flanking theYr15gene of wheat using NILs bulked segregant analysis.Genome,42, 1050–1056.
Cheng P, Xu L S, Wang M N, See D R, Chen X M. 2014.Molecular mapping of genesYr64andYr65for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016.Theoretical and Applied Genetics,127, 2267–2277.
Dvorak J, Terlizzi P, Zhang H B, Resta P. 1993. The evolution of polyploid wheats: Identification of the a genome donor species.Genome,36, 21–31.
Dvorak J, Zhang H B. 1990. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes.Proceedings of the National Academy of Sciences of the United States of America,87, 9640–9644.
Feldman M. 2001. The origin of cultivated wheat. In: Benjean A P, Angus J, eds.,The Wheat Book:A History of Wheat Breeding. Lavoisier Publishing, Paris. pp. 3–56.
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. 2009. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust.Science,323, 1357–1360.
Ganal M W, Durstewitz G, Polley A, Bérard A, Buckler E S,Charcosset A, Clarke J D, Graner E M, Hansen M, Joets J, Le Paslier M C, McMullen M D, Montalent P, Rose M, Sch?n C C, Sun Q, Walter H, Martin O C, Falque M.2011. A large maize (Zea maysL.) SNP genotyping array:Development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome.PLoS ONE,6, e28334.
Griffiths S, Sharp R, Foote T N, Bertin I, Wanous M, Reader S,Colas I, Moore G. 2006. Molecular characterization ofPh1as a major chromosome pairing locus in polyploid wheat.Nature,439, 749–752.
Han D J, Wang N, Jiang Z, Wang Q L, Wang X J, Kang Z S.2012. Characterization and inheritance of resistance to stripe rust in the wheat line Guinong 775.Hereditas,34,1607–1613.
IWGSC (The International Wheat Genome Sequencing Consortium). 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome.Science,345, 1251788.
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer K F, Li D, Pan S, Zheng F,et al.2013.Aegilops tauschiidraft genome sequence reveals a gene repertoire for wheat adaptation.Nature,496, 91–95.
Krattinger S G, Lagudah E S, Spielmeyer W, Singh R P,Huerta-Espino J, McFadden H, Bossolini E, Selter L L,Keller B. 2009. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat.Science,323, 1360–1363.
Li G Q, Li Z F, Yang W Y, Zhang Y, He Z H, Xu S C, Singh R P, Qu Y Y, Xia X C. 2006. Molecular mapping of stripe rust resistance geneYrCH42in Chinese wheat cultivar Chuanmai 42 and its allelism withYr24andYr26.Theoretical and Applied Genetics,112, 1434–1440.
Liang Y, Zhang D Y, Ouyang S, Xie J, Wu Q, Wang Z, Cui Y,Lu P, Zhang D, Liu Z J, Zhu J, Chen Y X, Zhang Y, Luo M C, Dvorak J, Huo N, Sun Q, Gu Y Q, Liu Z. 2015. Dynamic evolution of resistance gene analogs in the orthologous genomic regions of powdery mildew resistance geneMlIW170inTriticum dicoccoidesandAegilops tauschii.Theoretical and Applied Genetics,128, 1617–1629.
Lin F, Chen X M. 2007. Genetics and molecular mapping of genes for race-specific all-stage resistance and non-racespecific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa.Theoretical and Applied Genetics,114, 1277–1287.
Ling H Q, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D,Dong L, Tao Y, Gao C, Wu H, Li Y, Cui Y, Guo X, Zheng S,Wang B, Yu K, Liang Q, Yang W,et al. 2013. Draft genome of the wheat A-genome progenitorTriticum urartu.Nature,496, 87–90.
Liu R H, Meng J L. 2003. MapDraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data.Hereditas(Beijing),25, 317–321.
Liu Z, Zhu J, Cui Y, Liang Y, Wu H, Song W, Liu Q, Yang T, Sun Q, Liu Z. 2012. Identification and comparative mapping of a powdery mildew resistance gene derived from wild emmer(Triticum turgidumvar.dicoccoides) on chromosome 2BS.Theoretical and Applied Genetics,124, 1041–1049.
Lu P, Qin J, Wang G, Wang L, Wang Z, Wu Q, Xie J, Liang Y, Wang Y, Zhang D, Sun Q, Liu Z. 2015. Comparative fine mapping of the Wax 1 (W1) locus in hexaploid wheat.Theoretical and Applied Genetics,128, 1595–1603.
Luo M C, Deal K R, Akhunov E D, Akhunova A R, Anderson O D, Anderson J A, Blake N, Clegg M T, Coleman-Derr D,Conley E J, Crossman C C, Dubcovsky J, Gill B S, Gu Y Q, Hadam J, Heo H Y, Huo N, Lazo G, Ma Y, Matthews D E,et al. 2009. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae.Proceedings of the National Academy of Sciences of the United States of America,106, 15780–15785.
Luo M C, Gu Y Q, You F M, Deal K R, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen C M, Zhang Y, McGuire P E, Pasternak S, Stein J C, Ware D, Kramer M, McCombie W R, Kianian S F, Martis M M,et al. 2013. A 4-gigabase physical map unlocks the structure and evolution of the complex genome ofAegilops tauschii, the wheat D-genome progenitor.Proceedings of the National Academy of Sciences of the United States of America,110, 7940–7945.
Ma J X, Zhou R H, Dong Y S, Wang L F, Wang X M, Jia J Z.2001. Molecular mapping and detection of the yellow rust resistance geneYr26in wheat transferred fromTriticum turgidumL. using microsatellite markers.Euphytica,120,219–226.
McIntosh R A, Dubcovsky J, Rogers W J, Morris C, Appels R, Xia X C. 2014. Catalogue of gene symbols for wheat:2013–2014 supplement. [2016-06-07]. http://shigennigacjp/wheat/komugi/genes/symbolClassListjsp
McIntosh R A, Dubcovksy J, Rogers W J, Morris C, Appels R, Xia X C. 2016. Catalogue of gene symbols for wheat:2015–2016 supplement. [2017-06-07]. http://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
McIntosh R A, Lagudah E S. 2000. Cytogenetical studies in wheat. XVIII. GeneYr24for resistance to stripe rust.Plant Breeding,119, 81–83.
McIntosh R A, Silk J, The T T. 1996. Cytogenetic studies in wheat. XVII. Monosomic analysis and linkage relationships of geneYr15for resistance to stripe rust.Euphytica,89,395–399.
McIntosh R A, Yamazaki Y, Dubcovsky J, Rogers J, Morris C,Appels R, Xia X C. 2013. Catalogue of gene symbols for wheat. In:12th International Wheat Genetics Symposium Yokohama. [2017-06-07]. http://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
Michelmore R W, Paran I, Kesseli R V. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations.Proceedings of the National Academy of Sciences of the United States of America,88, 9828–9832.
Nachit M M, Elouafi I, Pagnotta M A, El Saleh A, Iacono E, Labhilili M, Asbati A, Azrak M, Hazzam H, Benscher D, Khairallah M, Ribaut J M, Tanzarella O A, Porceddu E, Sorrells M E. 2001. Molecular linkage map for an intraspecific recombinant inbred population of durum wheat(Triticum turgidumL. var. durum).Theoretical and Applied Genetics,102, 177–186.
Ouyang S, Zhang D, Han J, Zhao X, Cui Y, Song W, Huo N,Liang Y, Xie J, Wang Z, Wu Q, Chen Y X, Lu P, Zhang D Y, Wang L, Sun H, Yang T, Keeble-Gagnere G, Appels R,Dole?el J,et al. 2014. Fine physical and genetic mapping of powdery mildew resistance geneMlIW172originating from wild emmer (Triticum dicoccoides).PLoS ONE,9, e100160.
Peleg Z, Saranga Y, Suprunova T, Ronin Y, R?der M S, Kilian A, Korol A B, Fahima T. 2008. High-density genetic map of durum wheat×wild emmer wheat based on SSR and DArT markers.Theoretical and Applied Genetics,117, 103–115.
Peng J H, Fahima T, R?der M S, Huang Q Y, Dahan A, Li Y C, Grama A, Nevo E. 2000. High-density molecular map of chromosome region harboring stripe-rust resistance genesYrH52andYr15derived from wild emmer wheat,Triticum dicoccoides.Genetica,109, 199–210.
Peng J H, Fahima T, R?der M S, Li Y C, Dahan A, Grama A,Ronin Y I, Korol A B, Nevo E. 1999. Microsatellite tagging of stripe-rust resistance geneYrH52derived from wild emmer wheat,Triticum dicoccoides, and suggestive negative crossover interference on chromosome 1B.Theoretical and Applied Genetics,98, 862–872.
Qin B, Cao A, Wang H, Chen T, You F M, Liu Y, Ji J, Liu D, Chen P, Wang X E. 2011. Collinearity-based marker mining for the fine mapping ofPm6, a powdery mildew resistance gene in wheat.Theoretical and Applied Genetics,123, 207–218.
Ramirez-Gonzalez R H, Segovia V, Bird N, Fenwick P, Holdgate S, Berry S, Jack P, Caccamo M, Uauy C. 2015. RNA-Seq bulked segregant analysis enables the identification of highresolution genetic markers for breeding in hexaploid wheat.Plant Biotechnology Journal,13, 613–624.
Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger A E. 2012. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize.Proceedings of the National Academy of Sciences of the United States of America,109, 8872–8877.
Schneider A, Molnár I, Molnár-Láng M. 2008. Utilisation ofAegilops(goatgrass) species to widen the genetic diversity of cultivated wheat.Euphytica,163, 1–19.
Somers D J, Isaac P, Edwards K. 2004. A high-density microsatellite consensus map for bread wheat (Triticum aestivumL.).Theoretical and Applied Genetics,109,1105–1114.
Song Q J, Shi J R, Singh S, Fickus E W, Costa J M, Lewis J, Gill B S, Ward R, Cregan P B. 2005. Development and mapping of microsatellite (SSR) markers in wheat.Theoretical and Applied Genetics,110, 550–560.
Sun G L, Fahima T, Korol A B, Turpeinen T, Grama A, Ronin Y I, Nevo E. 1997. Identification of molecular markers linked to theYr15stripe rust resistance gene of wheat originated in wild emmer wheatTriticum dicoccoides.Theoretical and Applied Genetics,95, 622–628.
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. 2006.A NAC gene regulating senescence improves grain protein,zinc, and iron content in wheat.Science,314, 1298–1301.
de Vallavieille-Pope C, Ali S, Leconte M, Enjalbert J, Delos M,Rouzet J. 2012. Virulence dynamics and regional structuring ofPuccinia striiformisf. sp.triticiin France between 1984 and 2009.Plant Disease,96, 131–140.
Wan A M, Chen X M, He Z H. 2007. Wheat stripe rust in China.Australian Journal of Agricultural Research,58, 605–619.
Wan A M, Zhao Z H, Chen X M, He Z H, Jin S L, Jia Q Z, Yao G,Yang J X, Wang B T, Li G B, Bi Y Q, Yuan Z Y. 2004. Wheat stripe rust epidemic and virulence ofPuccinia striiformisf. sp.triticiin China in 2002.Plant Disease,88, 896–904.
Wang L F, Ma J X, Zhou R H, Wang X M, Jia J Z. 2002.Molecular tagging of the yellow rust resistance geneYr10in common wheat, P.I.178383 (Triticum aestivumL.).Euphytica,124, 71–73.
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang B E,Maccaferri M, Salvi S, Milner S G, Cattivelli L, Mastrangelo A M, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, IWGSC (International Wheat Genome Sequencing Consortium), Lillemo M, Mather D, Appels R,et al. 2014.Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array.Plant Biotechnology Journal,12, 787–796.
Wang Z, Cui Y, Chen Y, Zhang D, Liang Y, Zhang D, Wu Q, Xie J, Ouyang S, Li D, Huang Y, Lu P, Wang G, Yu M, Zhou S, Sun Q, Liu Z. 2014. Comparative genetic mapping and genomic region collinearity analysis of the powdery mildew resistance genePm41.Theoretical and Applied Genetics,127, 1741–1751.
Wu Q H, Chen Y X, Zhou S H, Fu L, Chen J J, Xiao Y, Zhang D,Ouyang S H, Zhao X J, Cui Y, Zhang D Y, Liang Y, Wang Z Z, Xie J Z, Qin J X, Wang G X, Li D L, Huang Y L, Yu M H, Lu P,et al. 2015. High-density genetic linkage map construction and QTL mapping of grain shape and size in the wheat population Yanda 1817×Beinong 6.PLoS ONE,10, e0118144.
Xie W L, Nevo E. 2008. Wild emmer: Genetic resources, gene mapping and potential for wheat improvement.Euphytica,164, 603–614.
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. 2006. The wheat and barley vernalization geneVRN3is an orthologue ofFT.Proceedings of the National Academy of Sciences of the United States of America,103, 19581–19586.
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W,SanMiguel P, Bennetzen J L, Echenique V, Dubcovsky J. 2004. The wheatVRN2gene is a flowering repressor down-regulated by vernalization.Science,303, 1640–1644.
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. 2003. Positional cloning of the wheat vernalization geneVRN1.Proceedings of the National Academy of Sciences of the United States of America,100, 6263–6268.
Zhang H, Guan H, Li J, Zhu J, Xie C, Zhou Y, Duan X, Yang T, Sun Q, Liu Z. 2010. Genetic and comparative genomics mapping reveals that a powdery mildew resistance geneMl3D232originating from wild emmer co-segregates with an NBS-LRR analog in common wheat (Triticum aestivumL.).Theoretical and Applied Genetics,121, 1613–1621.
Zhang J Y, Xu S C, Zhang S S, Zhao W S, Zhang J X. 2001.Monosomic analysis of resistance to stripe rust for source wheat line Jinghe 8811.Acta Agronomica Sinica,27,273–277. (in Chinese)
Zhao K, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R. 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits inOryza sativa.Nature Communications,2, 467.
 Journal of Integrative Agriculture2018年6期
Journal of Integrative Agriculture2018年6期
- Journal of Integrative Agriculture的其它文章
- Improve access to the EU market by identifying French consumer preference for fresh fruit from China
- Factors influencing hybrid maize farmers’ risk attitudes and their perceptions in Punjab Province, Pakistan
- Long-term grazing exclusion influences arbuscular mycorrhizal fungi and their association with vegetation in typical steppe of lnner Mongolia, China
- Soil microbial characteristics and yield response to partial substitution of chemical fertilizer with organic amendments in greenhouse vegetable production
- Reducing nitrogen fertilization of intensive kiwifruit orchards decreases nitrate accumulation in soil without compromising crop production
- Ultrastructure of the sensilla on antennae and mouthparts of larval and adult Plutella xylostella (Lepidoptera: Plutellidae)
