Analysis of SSR loci and development of SSR primers in Eucalyptus
Guo Liu?Yaojian Xie?Dangquan Zhang?Hongpeng Chen
Introduction
Simple sequence repeats(SSRs),or microsatellites,are arrays of short motifs 1–6 base pairs long.They are abundantly and uniformly distributed throughoutthe eukaryotic genome and inherited codominantly.They can provide multiple allelic,highly informative content,with good reproducibility and are easy to manipulate(Tautz 1989;Powell et al.1996;Gupta and Varshney 2000).The number of repetitions in SSRs varies greatly,and they are rich in polymorphisms.Because they can be assayed relatively quickly with low technical dif fi culty and cost,SSR markers have found wide application in genetic analyses ofEucalyptusspecies and genotypes,including for fi ngerprinting,genetic mapping,genetic diversity assessment,genetic structure analyses and molecular marker assisted selection(MAS)(Bradbury et al.2013;Zhang et al.2013;Liu and Xie 2012;Maria et al.2011;Zhou 2011;Kengavanar et al.2011;Shanmugapriya et al.2011;He 2010;Wang 2009).
SSR markers are generally classi fi ed into two categories:genomic SSRs(gSSR)and expressed sequence tag SSRs,(EST-SSRs or eSSRs).Genomic SSRs are based on the genome sequences,and EST-SSRs exist in the expressed gene sequences(Xu et al.2014;Zhang et al.2011).EST-SSRs have a higher transferability among closely related species(Varshney et al.2005),and the methods for their development are relatively simple and of lower cost in comparison to genomic SSRs.The latter come from the noncoding sequences in the genome,and they have higher degrees of polymorphism.With the dis coveriesofcontrolfunctions ofsome ‘noncoding sequences’,gSSRs have signi fi cant potential to be linked to genomic sequences in fl uencing phenotypic traits of importance.
At present,the public database GenBank(http://www.ncbi.nlm.nih.gov/genbank/)has published a large number ofEucalyptusgenomes,EST sequences ofEucalyptus,and the complete genome sequence ofEucalyptus grandis(http://www.phytozome.net/cgi-bin/gbrowse/eucalyptus/)(Myburg et al.2014;DOE 2008).This database provides direct access to genome sequences and EST sequences that can be used to developEucalyptusSSR markers.Over the past 20 years,many articles about development of SSR markers ofEucalyptushave been published worldwide.For example,Steane et al.(2001)developed 12E.globulusmicrosatellite loci for fi ngerprinting and future studies in genome mapping,gene fl ow and genetic diversity;Wang(2009)developed 185 pairs of EST-SSR primers to construct genetic maps inEucalyptus;He(2010)developed 206 EST-SSR markers and used 90 genomic-SSR markers to evaluate genetic variation among 20 different genotypes ofEucalyptus;Zhou(2011)developed 295 EST-SSR markers based on pool-cloning sequencing of PCR products;and Kengavanar et al.(2011)developed 179 orthologous genic SSR markers inE.camaldulensis.
In contrast,comprehensive studies about developing genomic SSRs and EST SSRs have rarely been published.Yang et al.(2011)developed 37 pairs of SSR primers based on an EST library and 95 pairs based on a genomic library;Zhang et al.(2011)explored genetic differences between genomic SSRs and EST SSRs in 15 species ofPoplarwith 48 pairs of genomic SSR primers and 48 pairs of EST SSR primers;Ding et al.(2015)explored genetic differences between genomic SSR and EST SSR in 12 species ofStylosanthesusing 20 pairs of genomic SSR primers and 20 pairs of EST SSR primers;and Wen et al.(2010)developed 20 genomic SSRs and 36 EST SSRs to analyze the genetic diversity among 45Jatropha curcasaccessions.
The research reported here was aimed at developing genomic SSRs and EST SSRs ofEucalyptusby detecting and analyzing SSR and EST sequences in the genomes ofEucalyptusspecies and screening the effectiveness of primers.PhyML3.0 software was then used to construct a maximum likelihood phylogentic tree for six accessions of eucalypts,represented by fi ve species of the genusEucalyptusand one of the genusCorymbia.The results will provide resources and theoretical foundations for evaluating the genetic diversity and phylogenetics of eucalypts by SSR markers.
Materials and methods
Plant material and DNA isolation
DNA samples were obtained from fi ve species ofEucalyptussubgenusSymphyomyrtusand one species ofCorymbia(Table 1).The taxonomy of samples was based on the classi fi cations of Pryor and Johnson(1971)and Hill and Johnson(1995).
TotalgenomicDNAwasextractedfromfreshleaveswith DNeasy Plant Mini Kit(QIAGEN,Germany).The quantity of DNA was checked on 1.5%agrose gels and determined using a Nanodrop nucleic acid-protein analyzer.The DNA templates constituted a genomic DNA pool,obtained by combining the DNA from the fi ve accessions ofEucalyptusspecies and oneCorymbiaspecies.
SSR mining and primer design
A total of 45,557 sequences ofEucalyptuswere obtained from GenBank(http://www.ncbi.nlm.nih.gov/genbank/),including 28,691 genome sequences and 16,566 EST sequences ofEucalyptus.These sequences were checked to remove redundancies,and then assembled and clustered using DNAStar 7.1 software(http://www.dnastar.com/).Next,the sequences were searched for the presence of SSR repeats using SSRHunter 1.03 software(http://en.bio-soft.net/dna/SSRHunter.html).For SSR identi fi cation,a criterion of a minimum length of 18 bases was adopted.
PCR primer pairs fl anking the SSR repeats were designed using softwarePrimer5.0 (http://www.pre mierbiosoft.com/).For designing PCR primers,the length ranged from 18-to 25-mer,and the optimum annealing temperature ranged from 50 to 60°C,the optimum GC content was 40–60%,and the rest of the parameters were set at default values.And these primers were checked with Oligo 6.0 software(http://oligo.net/).The primers were then synthesized by Invitrogen Trading(Shanghai).
SSR analysis
The SSRs were classi fi ed based on the length of the SSR motifs in their sequences.The different repeats of the SSR motifs and the frequencies of SSR motifs were used to analyse the characteristics of the SSRs.The types of SSR motifs were analysed to evaluate the speci fi city of SSRs inEucalyptus.
SSR screening
To obtain SSR-PCR products with higher speci fi cities and more stable rates,the SSR-PCR conditions for primer screening were optimized by adjusting Mg2+concentration and the annealing temperature(Tm).The PCR system and PCR program used followed methodologies described by Li et al.(2010):Mg2+concentration was set to 1.5,2.0,2.5 mmol L-1,and annealing temperature(Tm)were set to 50,56 and 60°C.After the optimum Mg2+concentration and annealing temperature were determined,effective primers that yielded good PCR results were reserved at each round,and ineffective primers that did not yield ideal products were discarded.
PCR was performed in a volume of 20 μL,containing genomic DNA(about 100 ng),0.1 μM of each primer,TaqDNA polymerase(2 U),0.2 mM of each dNTP,MgCl2(1.5,2.0 or 2.5 mM).The PCR pro fi le consisted of denaturing the template DNA at 94°C for 4 min.,followed by 35 cycles,each at 94 °C for 30 s,50,56 or 60 °C for 30 s,and 72 °C for 1 min,followed by 72 °C for 10 min.The PCR products were separated by electrophoresis in 1.5%agarose.
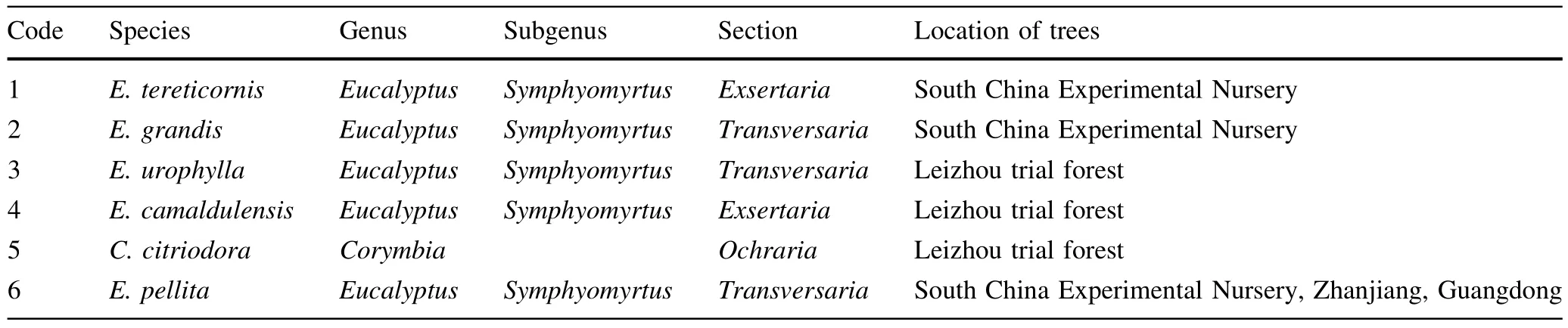
Table 1 Information on Eucalyptus and Corymbia species used in this study
Statistical analyses
Data for length of SSRs,the numbers of repeats,the frequencies of SSRs,and the SSRs types,were collated and statistics analyzed using Excel 2010 software(Microsoft,WA,USA).
Phylogenetic analyses
Phylogenetic analyses to examine the genetic relationships of the fi veEucalyptusand oneCorymbiaspecies were conducted using maximum likelihood and 1000 times bootstrapping was used to statistically support the groups using PhyML 3.0 software.The SSR-PCR sequences of six species were presented using the tree- fi gure drawing tool FigTree version 3.0 from the output fi le of PhyML 3.0.Information on the morphological features ofEucalyptusin China is from Qi(2002)and was compared with the results of phylogenetic analyses.
Results
Characterization of SSRs in Eucalyptus genome sequences and EST sequences of Eucalyptus
Statistics on the 45,257 DNA sequences fromEucalyptusdownloaded from GenBank are presented in Table 2.The genomic sequences included 13,373 fromE.camaldulensis,1206 fromE.globulus,5751 fromE.grandis,7803 fromE.gunnii,265 fromE.nitensand 293 fromE.urophylla.The EST sequences included 588 fromE.globulusand 16,008 fromE.grandis.After removing redundancies,14,121 contigs were obtained and 1785 effective sequences detected that contained SSRs,accounting for 12.6% of the total contigs.The effectiveEucalyptussequences obtained consisted of820 EST sequencesand 965 genome sequences.
The frequencies of different nucleotide repeat motifs in the SSRs contained within theEucalyptusDNA sequences obtained from the GenBank database are shown in Fig.1.Frequencies of motifs varied greatly;dinucleotide SSRs were the most common(44.5% of total),followed by trinucleotide SSRs(32.9%).The proportion of tetranucleotide and pentanucleotide SSRs were relatively low(3.2 and 2.8% of total,respectively).
In the analysis conducted with SSRHunter software,435 SSR repeat motifs were found among the 2292 SSRs obtained.Of these SSR repeat motifs,12 were dinucleotides,50 were trinucleotides,51 were tetranucleotides,54 were pentanucleotides and 268 were hexanucleotides.
Among the repeat motifs,CT/GA,TC/AG repeat motifs were the most common of the di-nucleotide types,occurring in 21.3 and 20.7% of SSRs,respectively.The same repeat motifs also accounted for 94.3% of all the dinucleotide repeat motifs obtained.In contrast,CG/GC repeat motifs accounted for only 0.4% of the dinucleotide repeat motifs.The CGG/GCC,CCG/CGG repeat motifs were the highest frequency trinucleotide repeat motifs,and occurred in 4.3%,3.9% of the SSRs,respectively.
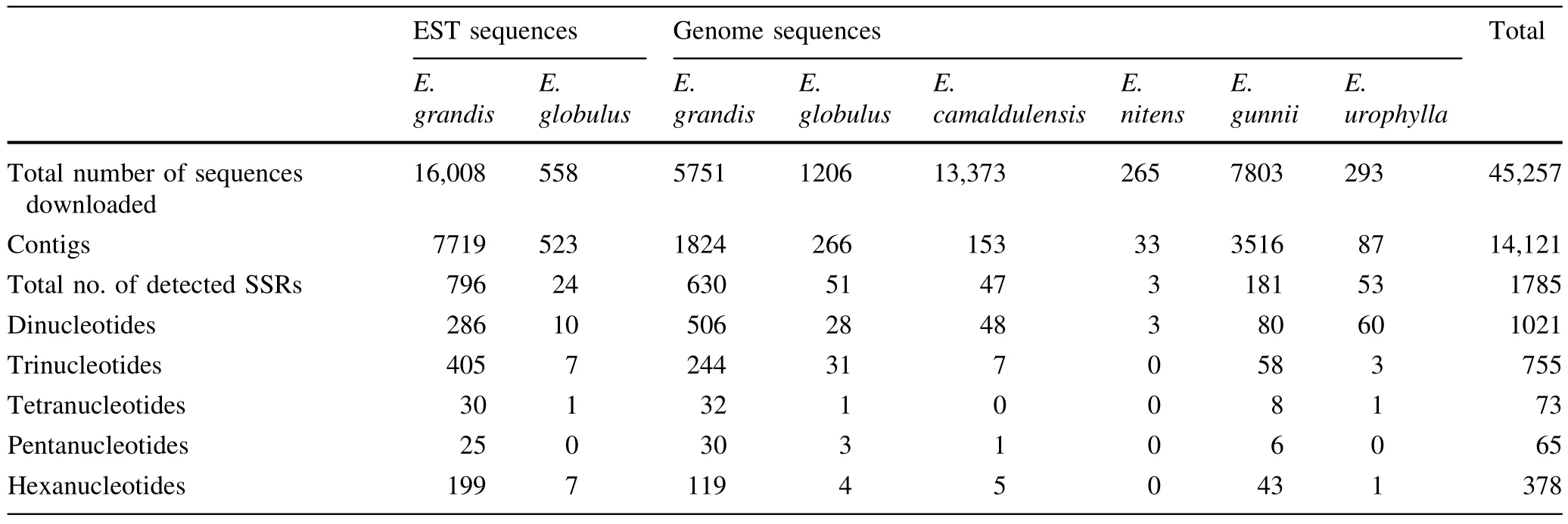
Table 2 Statistics for SSRs generated from Eucalyptus species
Among the genome sequences and EST sequences ofEucalyptus,the repeat numbers varied from 3 to 50.Repeat numbers of 3–9 were predominant in the SSRs,totalling 1382 or 60.3% of the total SSRs.Repeat numbers of 10–20 accounted for 768 or 33.5% of the total number of SSRs,repeat numbers of 21–30 accounted for 130 or 5.7% of SSRs,whilst repeat number of 31–40 and more than 40 were accounted for just 10(0.4%)and 2(0.1%)of the SSRs,respectively(Fig.2).From the SSR repeat motifs analyzed,532 or 52.1% of the di-nucleotide repeat motifs occurred between repeat numbers of 10–15.Of the other repeat motifs,most were concentrated in among the 3–9 motif repeats,including 691 or 91.5% of trinucleotide repeat motifs,72 or 98.6%tetranucleotide repeat motifs,65 or 100% of pentanucleotide repeat motifs and 376 or 99.5% of hexanucleotide repeat motifs.
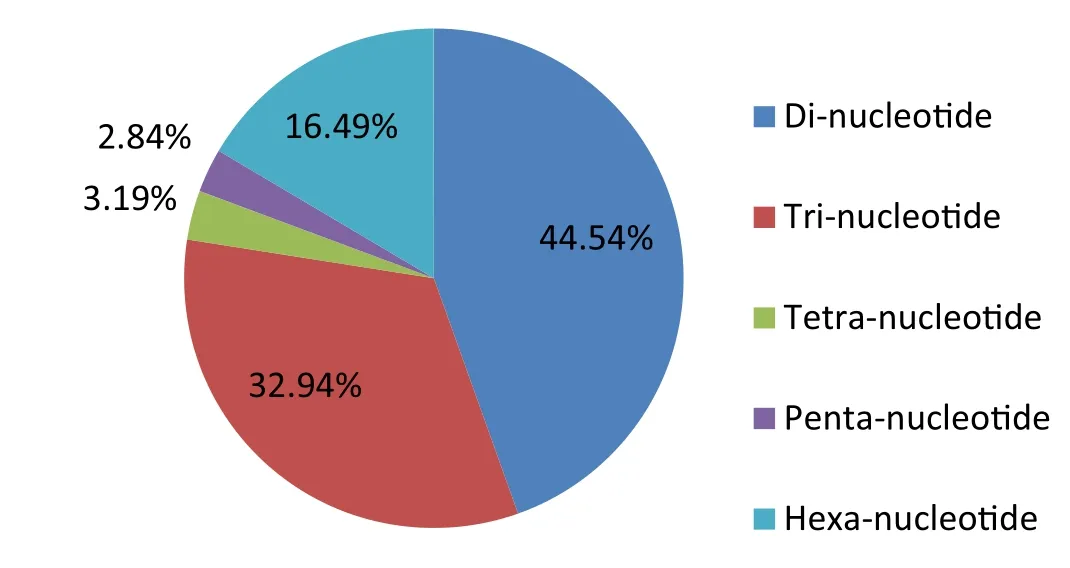
Fig.1 Frequencies of different nucleotide repeat motifs in the SSRs
The characterization of SSRs in genome sequences and EST sequences ofEucalyptusare presented in Table 3.Dinucleotide repeat motifs occurred at higher frequency in the genome sequences(48 repeat motifs),than in EST sequences(20 repeat motifs)ofEucalyptus.In contrast,the frequency of trinucleotide repeat motifs was higher in theEucalyptusgenome sequences(29 repeat motifs)than in the EST sequences(13 repeat motifs),as were the frequencies of both tetranucleotide and hexanucleotide repeat motifs.
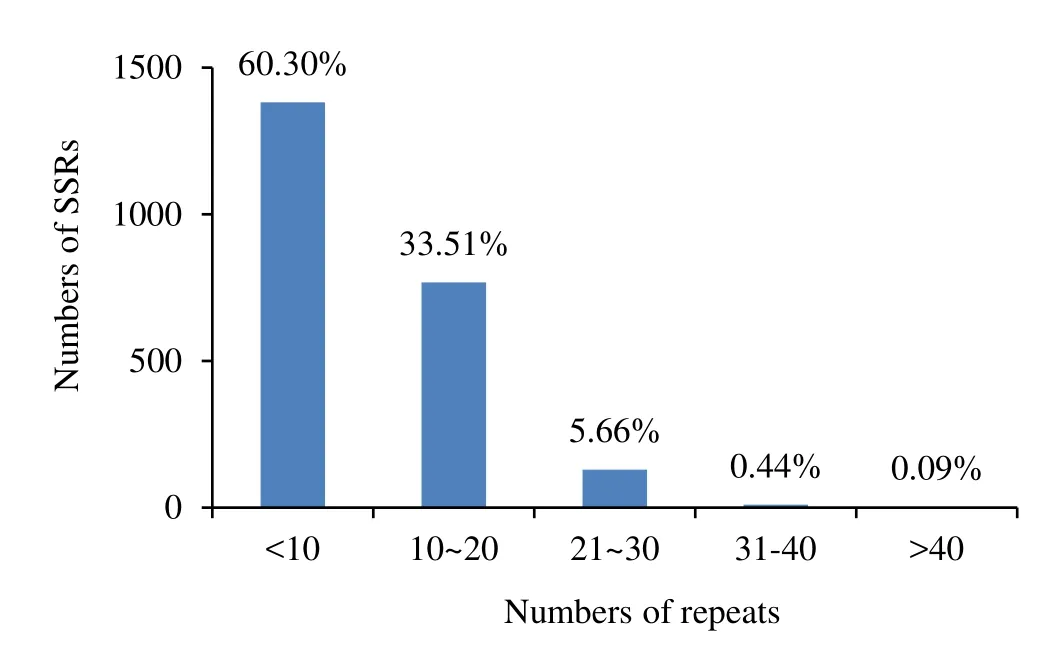
Fig.2 Frequency distributions of SSR repeats
The number of SSRs in repeat motifs of different nucleotide lengths seemed to be negatively correlated with the length of the repeat motifs;the SSRs with longer lengths of nucleotide repeat motifs had lower frequencies ofnucleotide repeatmotifs inEucalyptusgenome sequences.However,the frequencies of tri-nucleotide repeat motifs were higher than other nucleotide repeat motifs in EST sequences ofEucalyptus.
In Table 4,the 970 SSRs found in theEucalyptusEST sequences accounted ig for 42.3% of the total SSRs,and 1322SSRswerefoundintheEucalyptusgenome sequences,accounting for 57.7% of the total SSRs.The number of dinucleotide,tetranucleotide and pentanucleotide repeat motifs found in theEucalyptusgenome sequencesexceededthenumberfoundintheEST sequences ofEucalyptus.In contrast,more tri-and hexanucleotide repeat motifs were found inEucalyptusEST sequences than inEucalyptusgenome sequences.Dinucleotide repeat motifs were predominant inEucalyptusgenome sequences,accounting for 54.8% of total SSRs,and the frequency of trinucleotide repeat motifs was highestamong the EST sequences ofEucalyptus,accounting for 42.5% of the total SSRs.
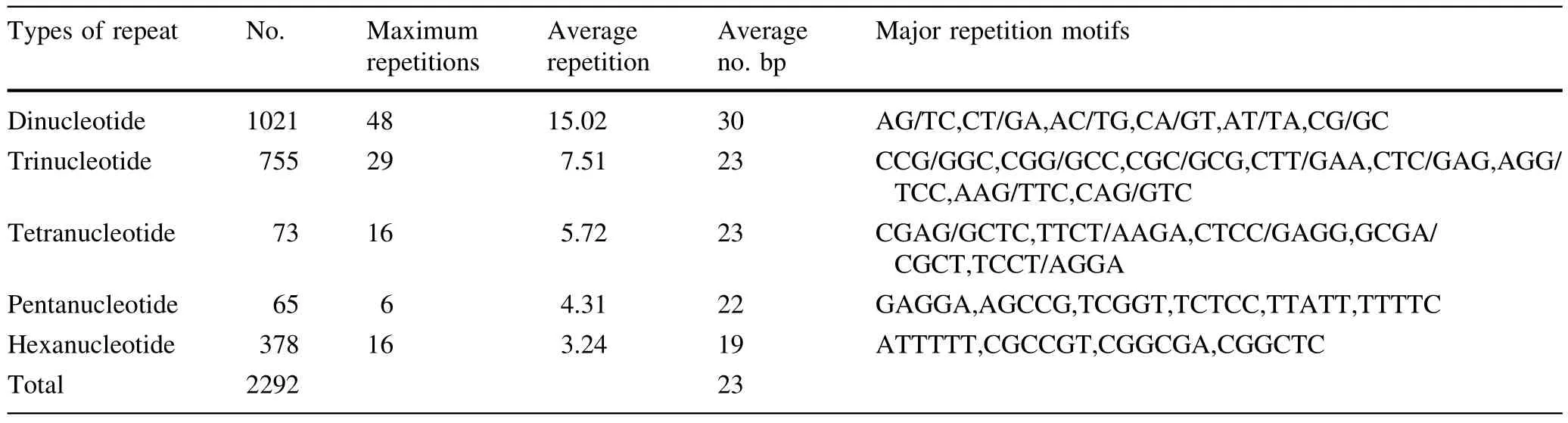
Table 3 Information on SSR motifs in Eucalyptus
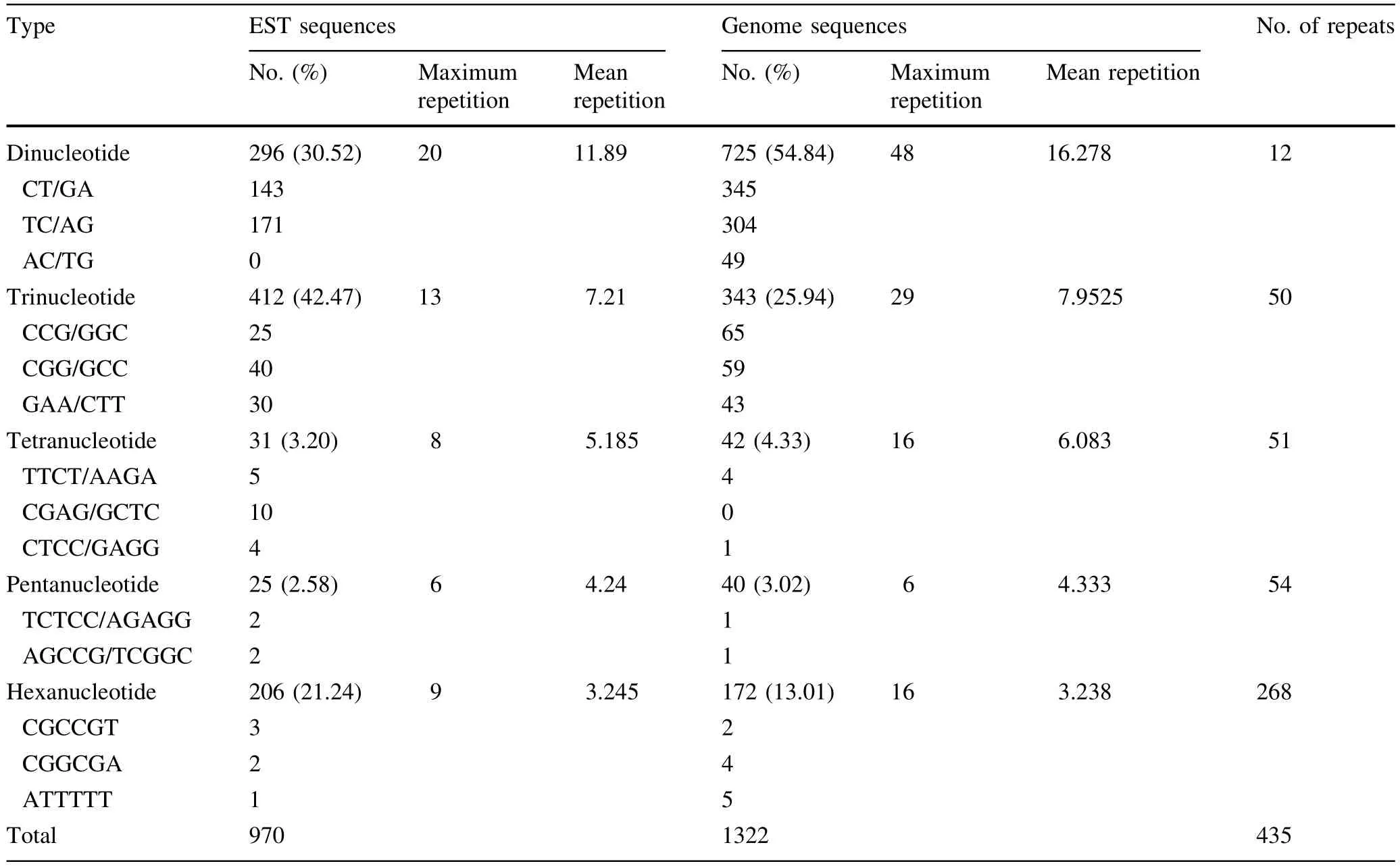
Table 4 Characterization of SSRs in Eucalyptus genome sequences and EST sequences of Eucalyptus
SSR primer design
After primer design and veri fi cation,a total of 395 SSR primers were synthesized for use in subsequent analyses.These included 150 pairs of EST-SSR(eSSR)primers,of which nine pairs were designed using EST sequences ofE.globulus,and 141 pairs were designed using EST sequences ofE.grandis.The 245 pairs of genomic-SSR(gSSR)designed included 168 pairs,which were designed using genome sequences ofE.grandis,13 pairs designed using genome sequences ofE.globulus,17 pairs designed using genome sequences ofE.camaldulensis,36 pairs designed using genome sequences ofE.gunnii,and 11 pairs designed using genome sequences ofE.urophylla.The length of primers varied from 18 to 22 bp,and the length of target fragments varied from 200 to 500 bp.Selection details for these SSR primers are presented in Table 5.
Screening for effective SSR primers of Eucalyptus
The DNA templates were from a mixed genomic DNA pool obtained by combining the DNA from the fi veEucalyptusand oneCorymbiaspecies sampled(see Table 1).After nine rounds of screening by optimizing the PCR conditions,340 pairs of primers successfully ampli fi ed the target fragments with a success ratio up to 86.1%;136 pairs of effective primers were screened from 150 pairs of eSSR primers,with a success ratio of 90.7%;204 pairs of effective primers were screened from 245 pairs of gSSR primers,with a success ratio of 83.3%.Some of the results from screening partial eSSR and gSSR primers are shown in Figs.3 and 4 respectively.
Results from primer screening are presented in Table 6.From among the nine rounds of primer screening,an annealing temperature of 56°C with 2.0 or 2.5 mmol L-1Mg2+of provided the highest success ratios.The maximum eSSR primer success ratio was 56.0%,achieved with an annealing temperature of 56°C and 2.5 mmol L-1Mg2+,while the maximum gSSR primer success ratio was 51.4%,which was achieved at the same annealing temperature but with 2.0 mmol L-1Mg2+.
Phylogenetic analysis using SSR-PCR sequences
To assess the genetic relationships of the six accessions( fi veEucalyptusand oneCorymbiaspecies),data from fi ve pairs of effective SSR primers that had good stability,strong signals and high polymorphisms were analysed.This analysis generated a maximum likelihood phylogenetic tree based on fi ve combined ampliconic sequences of the six species;see Fig.5.
The results of combined-ML phylogenetic analyses revealed thatC.citriodorahad a greater genetic distance from the other fi ve species,consistent with morphological taxonomy.Based on this analysis,the kinship ofE.camaldulensisandE.pellitais somewhat closer,whileE.tereticornisandE.urophyllahad the closest genetic relationship of all the species examined.These results differed somewhat from what was expected based on the taxonomy of these species described by both Pryor and Johnson(1971)and Hill and Johnson(1995).
Further analyses using the six pairs of SSR primers in the fi veEucalyptusspecies(the primer of gSSR-GU023 had no ampliconic sequence inC.citriodora)examined the genetic relationships among only species of subgenusSymphyomyrtus(Fig.6).The combined ML phylogenetic tree of these species shows thatE.pellita,E.camaldulensisandE.grandishad a relatively close genetic relationship,while the shortest genetic distance with this group of fi ve species was betweenE.tereticornisandE.urophylla.This latter result concurs with the combined ML phylogenetic tree developed from data using the fi ve pairs of SSR primers for the six eucalypt species(Fig.5).
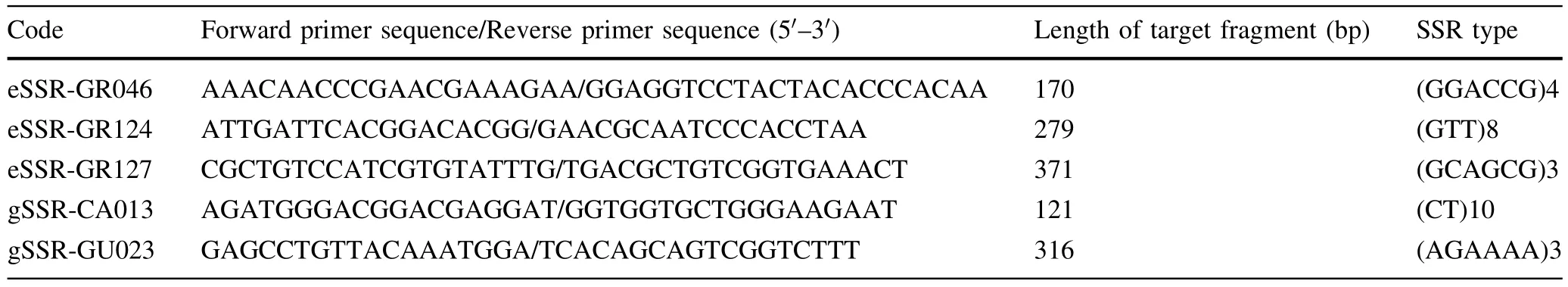
Table 5 Basic information for fi ve pairs of SSR primers
Discussion
Characterization of SSRs in Eucalyptus
Among 14,141 contigs assembled from 45,257 genome and EST sequences ofEucalyptus,1785 SSRs(12.6%)were detected.This frequency is consistent with results obtained by Ellis and Burke(2007)and Yasodha et al.(2008)who obtained frequencies of 12.3 and 12.9%respectively.However,He(2010)reported a somewhat higher frequency of SSRs,25.3%,in EST sequences ofEucalyptus,and Zhou(2011)reported a frequency of SSRs of 21.7%in 36,029 unigenes assembled from EST sequences ofEucalyptus.Similarly,Rabello et al.(2005)and Ceresini et al.(2005)reported frequencies of SSRs inEucalyptusof 25.5 and 25.6%,respectively.
Dinucleotide repeat motifs are generally the most common types of repeat motifs in dicotyledons,and trinucleotide repeat motifs are most common in graminaceous plants(Biet et al.1999).In this current study,dinucleotide and trinucleotide repeat motifs were found to be the most frequent inEucalyptus, accounting for 44.4 and 32.8%,respectively,ofthe totalrepeatmotifs.The tetranucleotide and pentanucleotide repeat motifs
accounted for only 3.2 and 2.8%,respectively.These results agreed closely with those of He(2010),Li(2010)and Zhou(2011),who also examined frequencies of SSRs in EST sequences ofEucalyptus.
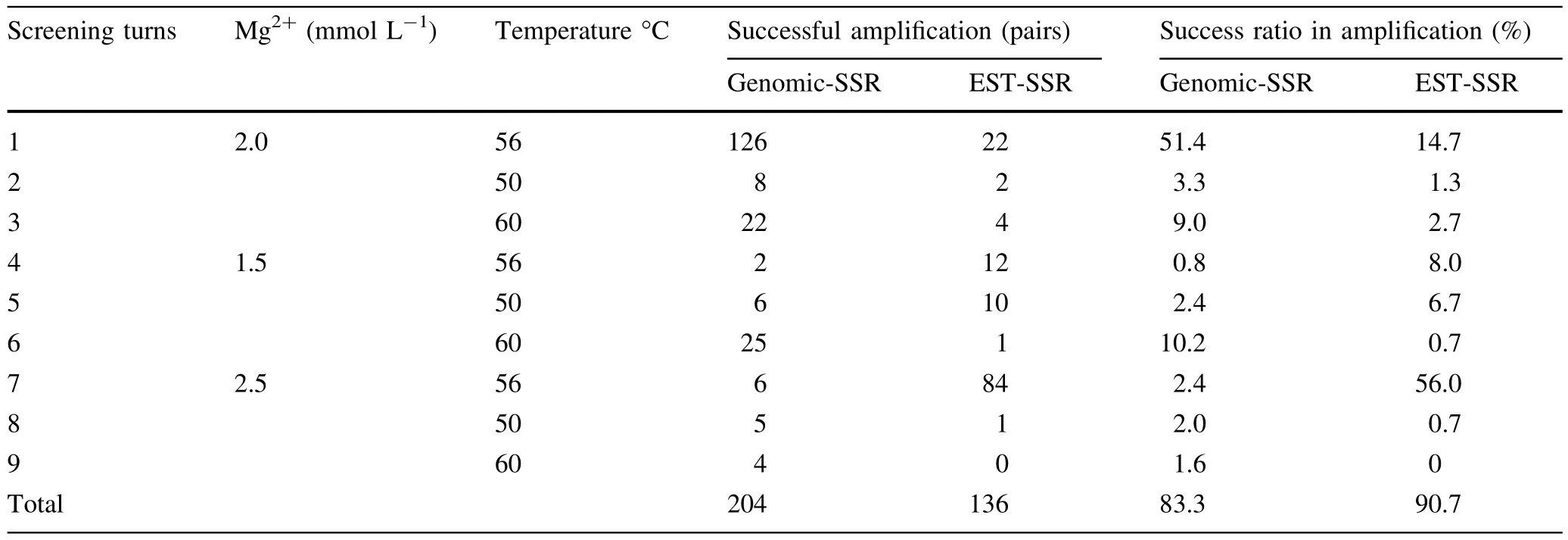
Table 6 Results of primer screening
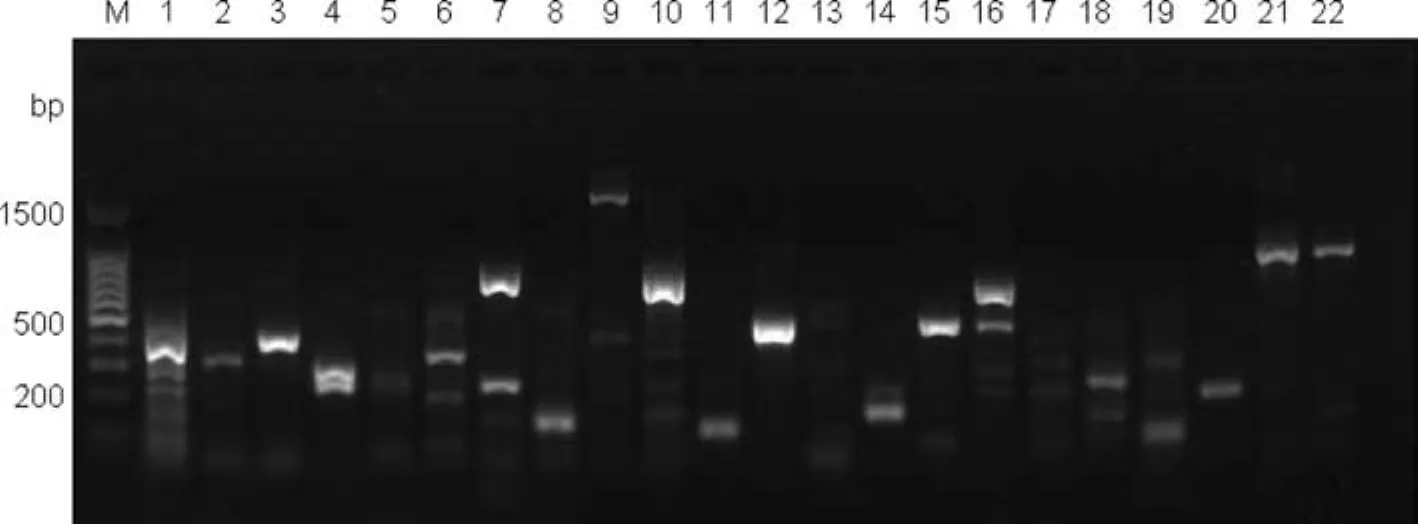
Fig.3 Fast screening result of partial eSSR primers.Lanes 1,2,3,4,7,8,10,11,12,14,15,16,18,20 lane have bands that indicate successfully screens of primers
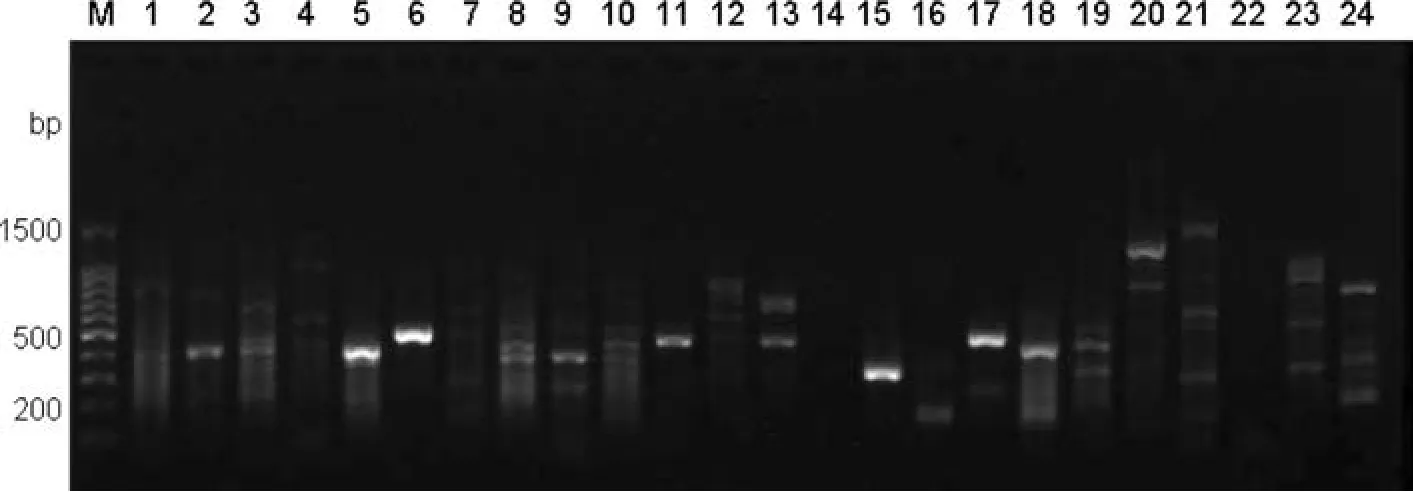
Fig.4 Fast screening result of partial gSSR primers.Lanes 2,5,6,9,11,13,15,17,18,20 have bands that indicate successful screens of primers
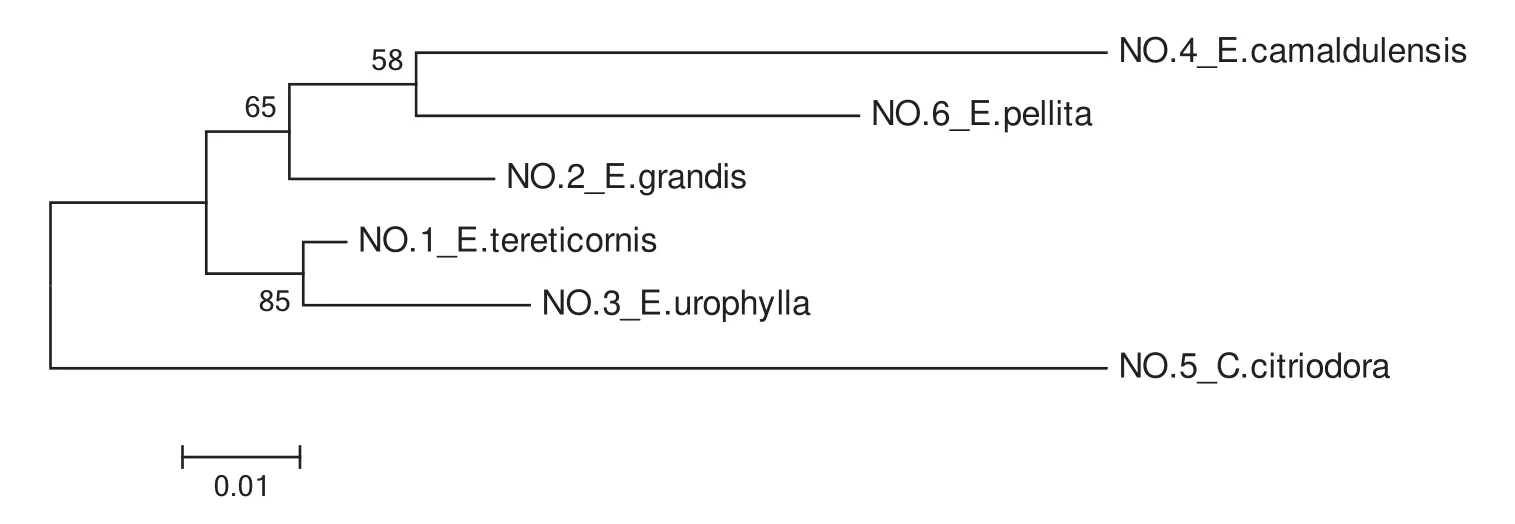
Fig.5 Combined ML phylogenetic analysis of Eucalyptus(including 1 Corymbia species)using 5 combined sequences.Numbers on branch points represent the percentage of 1000 bootstraps by heuristic searching
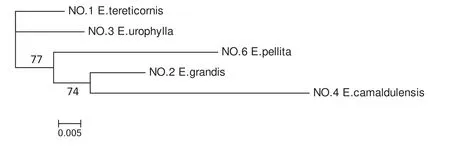
Fig.6 Combined ML phylogenetic analysis of the fi ve Eucalyptus species using the six combined sequences.Numbers on branch points represent the percentage of 1000 bootstraps by heuristic searching
Previous research has shown that the formation of SSR loci might be associated with DNA replication slippage,alternation of nucleic acids and unbalanced recombination(Tóth et al.2000;Ma et al.2015).The combinations of CA,GA,and GT in SSR repeat motifs could affect DNA recombination by impacting the DNA structure(Biet et al.1999).Therefore,the composition of nucleotides in SSRs can affect the activities of life,and hence analyses of the structure of motifs and distributions of SSRs can be of great importance.In this current study,the repeat motifs of AG/TC and GA/CT accounted for the vast majority(94.3%)of dinucleotide repeat motifs,and CCG/CGG repeat motifs were the most common(16.4%)trinucleotide repeat motifs.These results are in perfect agreement with the results of other studies inEucalyptus(Zhou 2011;Yasodha et al.2008).
EST-SSR primers come from the expressed gene conservative region(Ding et al.2015).Numerous studies have shown that polymorphisms in EST-SSR primers are lower than in genomic SSR primers(Qi et al.2009),that EST sequences tend to be more conservative than genome sequences,and that EST-SSR primers are better than genomic SSR primers in transferability across species(Qi et al.2009;Yang et al.2011;Xu et al.2014;Zhang et al.2011).In this current study,trinucleotide repeat motifs(42.5%)were more frequent than the other nucleotide repeat motifs in EST sequences ofEucalyptus,while in genomic sequences dinucleotide repeat motifs were more frequent(54.8%).This result concurs with fi ndings of Li(2010)on the content of microsatellites in EST sequences ofEucalyptus.Trinucleotide repeat motifsis where excessive enrichment might occur because the genetic code only allows triplet repeats to have mutations.
Analyses of the data on the maximum repetition of SSRs in this study revealed that the repetition of di-,tri-,tetraand hexanucleotide repeat motifs was greater in genome sequences than in EST sequences ofEucalyptus.Pentanucleotide repeat motifs were an exception,as the repetitions of these in EST sequences equaled that in genomic sequences.The average repetitions of di-,tri-,tetra-and pentanucleotide repeat motifs in genomic sequences were higher than in EST sequences ofEucalyptus,but the average repetitions of hexanucleotide repeat motifs in genomic sequences was equal to that in EST sequences ofEucalyptus.These results clearly indicate that the length and polymorphism of SSRs in genomic sequences were superior to those in EST sequences ofEucalyptus.
In molecular genetics,development and utilization of molecular markers to diagnose and detect DNA polymorphisms and analyze genetic diversity have valuable applications in understanding genetic relationships,accelerating breeding and facilitating genetic improvement(Zhou 2011).SSR molecular marker technology is based on PCR,and for this,the operability of the assembly sequences must be considered in the design of primers(Yang et al.2011).The research of Temnykh et al.(2001)showed that when the SSRs of lengths greater than or equal to 20 bp tend to have a higher degree polymorphism,SSRs of lengths between 12 and 20 bp tend to have a medium degree of polymorphism,and SSRs of lengths less than 12 bp tend to have extremely low degree of polymorphism.To ensure a high degree of polymorphism in SSR primers in this current study,selection criteria for SSRs included a minimum length of 18 bp.The lengths of the 395 SSR primers used in this current study were 18–22 bp,and the lengths of their target sequences were 200–500 bp.Those lengths can guarantee the quantity of target sequences and the fi delity of SSR primers,both sides of SSR loci must have a certain length of sequences,and it would be more convenient to select the parameters of SSR primers,as an appropriate primer length,GC content,annealing temperature,and target sequence length(Temnykh et al.2001;Picoult-Newberg et al.1999).
The screening of SSR primers
To evaluate the effectiveness of SSR primers ofEucalyptus,we screened 150 pairs of eSSR primers and 245 pairs of gSSR primers.After nine rounds of screening to optimize PCR conditions,340 pairs of primers were screened and found to be effective primers.Among the 340 pairs of primers,136 pairs were eSSR primers,and 204 pairs were gSSR primers;the overall success ratio was 86.1%.This result demonstrated that the SSR primers had good transferability and that one SSR marker could probably be shared among species with a close genetic relationship.This conclusion is agrees with results reported by Cupertino et al.(2011)who found that genomic SSR and EST SSR had no signi fi cant differences among 112 hybrid taxa ofEucalyptus.Ellis and Burke(2007)reported that the length of SSRs and their stability varied between different species and between different SSR loci within the one species.Ellis showed that gSSRs had higher polymorphism than eSSRs,and the transferability of gSSRs was worse than eSSRs.In this current study,the success ratio of screening for eSSRs(90.7%)was somewhat higher than thatforgSSRs (83.3%);a resultthatmightbe attributable to the EST sequences accounting for 57.7% of the genome sequences in this study.
Conclusion
By detecting the SSRs in both genome and EST sequences ofEucalyptus,this study obtained 970 SSRs in EST sequences and 1322 SSRs in genome sequences.The software Primer 5.0 designed 150 pairs of eSSR primers and 245 pairs of gSSR primers.PCR reaction used a pool of genomic DNA,obtained from sixEucalyptusspecies,as a DNA template.PCR conditions were optimized and used to obtain 136 pairs of eSSR primers and 204 pairs of gSSR primers from the screening of 395 pairs of SSR primers,providing screening success ratios of 90.7 and 83.3%,respectively.The 340 pairs of SSR primers developed in the study along with insights on SSRs provide important resources for future studies on genetic diversity,phylogenetics and other genetic aspects inEucalyptus.
The classi fi cation ofEucalyptusby Pryor and Johnson(1971)and Hill and Johnson(1995)acknowledgesC.citriodoraas belonging to a separate genus,Corymbia,and thatE.tereticornis,E.grandis,E.urophylla,E.camaldulensisandE.pellitabelonged to the genusEucalyptus.Their classi fi cations also placeE.tereticornisandE.camaldulensisinto the subgeneric taxon known as sectionExsertaria,whilstE.grandis,E.urophyllaandE.pellitaare placed into sectionLatoangulatae.Within these sections,they placedE.grandisinto seriesSalignae,andE.urophyllaandE.pellitainto seriesResiniferinae.In the combined ML phylogenetic analyses in the current study,the results at the genus level were consistent with thothe classi fi cations of Pryor and Johnson(1971)and Hill and Johnson(1995).
However,the genetic relationships among the fi veEucalyptusrevealed by analyses conducted in this current study differed somewhat from those suggested by Pryor and Johnson(1971)and Hill and Johnson(1995).The phylogenetic trees obtained in this study indicate thatE.camaldulensisandE.grandishave a close genetic relationship even though traditional taxonomy placed them in different subgeneric sections,ExsertariaandLatoangulataerespectively.And the same status applied toE.tereticornisandE.urophylla.
In the analysis of the morphological features of six eucalypt species,the features of young leaves and mature leaves ofE.pellitawere similar to those ofE.camaldulensis:3–4 pairs of young leaves opposite,ovoid to lanceolate,and mature leaves alternate,lanceolate.Species in sectionLatoangulataehave mature leaves,upper and lower sides concolorous;juvenile leaves on seedlings are sessile.Zhang et al.(2010)analyzed the genetic diversity of four species ofEucalyptusand illustrated that genetic similarity ofE.camaldulensisandE.grandiswas greater,in agreement with the phylogenetic trees in the present study.Thus,the reliability of classi fi cation schemes based on foliar features or petiole features need further study.
AcknowledgementsThis research was funded by the Special Fund for Forestry Scienti fi c Research in the Public Interest(201504204).The authors are indebted to numerous persons who provided help for this study,in particular Roger Arnold,Luo Jianzhong,Wu Zhihua,and the forestry administration of Leizhou and South China Experimental Nursery who providedEucalyptussamples for this study.
Open AccessThis article is distributed under the terms of the Creative Commons Attribution 4.0 International License(http://crea tivecommons.org/licenses/by/4.0/),which permits unrestricted use,distribution,and reproduction in any medium,provided you give appropriate credit to the original author(s)and the source,provide a link to the Creative Commons license,and indicate if changes were made.
Biet E,Sun J,Dutreix M(1999)Conserved sequence preference in DNA binding among recombination proteins:an effect of ssDNA secondary structure.Nucleic Acids Res 27:596–600
Bradbury D,Smithson A,Krauss SL(2013)Signatures of diversifying selection at EST-SSR loci and association with climate in naturalEucalyptuspopulations.Mol Ecol 22:5112–5129
Ceresini PC,Silva CLSP,Missio RF,Souza EC,Fischer CN,Guillherme IR,Gregorio I,Silva EHT,Cicarelli RMB,Silva MTA,Garcia JF,Avelar GA,Neto LRP,Marcon AR,Junior MB,Marini DC(2005)Satellypus:analysis and database of microsatellites from ESTs ofEucalyptus.Genet Mol Biol 28(3 suppl):589–600
Cupertino FB,Leal JB,Correa RX,Gaiotto FA(2011)Genetic diversity ofEucalyptushybrids estimated by genomic and EST microsatellite markers.Biol Plant 55(2):379–382
Ding XP,Luo XY,Shao CG,Zhang L,Wang WQ,Bai LJ(2015)Comparative analysis of genetic diversity revealed by genomic-SSR and EST-SSR markers in interspecies ofStylosanthes.Guangdong Agric Sci 14:106–113
DOE Joint Genome Institute Announces(2008)Genome Sequencing Targets.Eucalyptus,Foxtail Millet,Red Algae,and Novel Microbial Communities Added to Growing Bioenergy and Carbon Cycling Portfolio
Ellis JR,Burke JM(2007)EST-SSRs as a resource for population genetic analyses.Heredity 99:125–132
Gupta PK,Varshney RK(2000)The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat.Euphytica 113(3):163–185
He XD(2010)Heterosis onEucalyptus hybrids and molecular marker-assisted selection.Dissertation,Nanjing Forestry University,Nanjing
Hill KD,Johnson LAS(1995)Systematic studies in theEucalyptus-7 A revision of the bloodwoods,genusCorymbia(Myratceae).Telopea 6(2/3):185–504
Kengavanar N,Prasad SH,Navin S,Rajkumar R(2011)Novel design and deployment of orthologous genetic SSR markers inEucalyptus camaldulensisDehnh.BMC Proc 5(Suppl 7):51.doi:10.1186/1753-6561-5-S7-P51
Li FG(2010)Linkage map construction and growth QTL detection inEucalyptus urophyllaandE.tereticornisbased on STS markers.Dissertation,Chinese Academy of Forestry,Beijing
Li SX,Zhang XY,Wang YY,Yin TM(2010)Content and characteristics of microsatellites detected in expressed sequence tag sequences inEucalyptus.Chin Bull Bot 45(3):363–371
Liu G,Xie YJ(2012)Application of molecular marking technologies in heredity breeding ofEucalyptus.World For Res 25(3):19–25
Ma QY,Liao ZY,Zhang DF,Dai XG,Chen YN,Li SX(2015)Deep sequenced-based transcriptome analysis of microsatellites inPeach(Prunus persica cv.duplex) fl owers.J Nanjing For Univ(Nat Sci Ed)39(1):1–8
Maria MR,Leopoldo S,Carla R,Fátima C,JoséA,Nuno MGB,Cristina M (2011)A case study ofEucalyptus globulusfi ngerprinting for breeding.Ann For Sci 68:701–714.doi:10.1007/s13595-011-0087-x
Myburg AA,Grattapaglia D,Tuskan GA,Hellsten U,Hayes RD,Grimwood J,Jenkins J,Lindquist E,Tice H,Bauer D,Goodstein DM,Dubchak I,Polikov A(2014)The genome ofEucalyptus grandis.Nature 510(19):356–374.doi:10.1038/nature13308
Picoult-Newberg L,Ideker TE,Pohl MG,Taylor SL,Donaldson MA,Nickerson DA,Boyce-Jacino M(1999)Mining SNPs from EST databases.Genome Res 9:167–174
Powell W,Machray GC,Provan J(1996)Polymorphisms revealed by simple sequence repeats.Trends Plant Sci 7(1):215–222
Pryor LD,Johnson LAS(1971)A classi fi cation of theEucalyptus.The Australian National University,Canberra,p 102
Qi SX(2002)Eucalyptus in China,vol 2.China Forestry Publishing House,Beijing,pp 42–51
Qi J,Wang KJ,Wu CL,Wang WX,Hao YB,Leng P(2009)Development of EST-SSR markers inJuglans regia.J Agric Biotechnol 17(5):872–876
Rabello E,Souza AN,Saito D,Tsai SM(2005)In silico characterization of microsatellites inEucalyptusspp.:abundance,length variation and transpose on associations.Genet Mol Biol 28(3 suppl):582–588.doi:10.1590/S1415-47572005000400013
Shanmugapriya A,Modhumita G,Sivakumar V,Yasodha R(2011)Species discrimination,population structure and linkage disequilibrium inEucalyptus camaldulensisandeucalyptus tereticornisusing SSR markers.PLoS ONE 6(12):1–8
Steane DA,Vaillancourt RE,Russell J,Powell W,Maeshall D,Potts BM(2001)Development and characterisation of microsatellite lociinEucalyptusglobulus(Myrtaceae).Silvae Genet 50(2):89–91
Tautz D(1989)Hyper variability of simple sequences as a general sourceforpolymorphicDNA markers.NuclAcidsRes 17(16):6463–6471
Temnykh S,DeClerck G,Lukashova A,Lipovich L,Cartinhour S,McCouch S(2001)Computational and experimental analysis of microsatellites inRice(OryzasativaL.).Genome Res 11(8):1441–1452
Tóth G,Gáspári Z,Jurka J(2000)Microsatellites in different eukaryotic genomes:survey and analysis.Genome Res 10(7):967–981
Varshney RK,Graner A,Sorrells ME(2005)Genetic microsatellite markers in plants:features and applications.Trends Biotechnol 23(1):48–55
Wang Y(2009)Development of EST-SSR markers and their application to construction of genetic maps inEucalyptus.Dissertation,Nanjing Forestry University,Nanjing
Wen MF,Wang HY,Xia ZQ,Zou ML,Lu C,Wang WQ(2010)Developent of EST-SSR and genomic-SSR markers to assess genetic diversity inJatropha Curcas L.BMC Res Notes 42(3):1–8
Xu Y,Chen JH,Li Y,Hong Z,Wang Y,Zhao YQ,Wang XM,Shi JS(2014)Development of EST-SSR and genomic-SSR in Chinese fi r.J Nanjing For Univ(Nat Sci Ed)38(1):9–14
Yang H,Chen Q,Wei CL,Shi CY,Fang CB,Wan XC(2011)Analysis on SSR information inCamellia sinensistranscriptome.J Anhui Agric Univ 38(6):882–886
Yasodha R,Sumathi R,Chezhian P,Kavitha S,Ghosh M(2008)Eucalyptusmicrosatellites mined in silico:survey and evaluation.J Genet 87(1):21–25
Zhang DQ,Tian H,Xie YJ,Huang QY,Gu ZJ,Cao JG,Tan XF,Zeng YL,Deng SY,Fan SG (2010)Gentic diversity of fourEucalyptusspecies by ISSR.J Central South Univ For Technol 30(1):12–17
Zhang YD,Peng C,Li ZF,Yang YL,Hu XY(2011)Genetic Diversity of Genomic-SSR and EST-SSR Markers in Interspecies ofPoplar.J Northeast For Univ 39(12):8–11
Zhang ZY,Xiang DY,Deng ZY,Tang QL,Chen JB,Lan J(2013)Establishment of Fingerprinting for 24 Clones inEucalyptus.J Northwest For Univ 28(2):74–78
Zhou CP(2011)Development of a new set of EST-SSR markers and their application to integrated linkage map inEucalyptus.Dissertation,Chinese Academy of Forestry,Beijing
Zhou CP,Li FG,Weng QJ,Yu XL,Li M,Gan SM(2010)Comparison between direct sequencing and pool-cloning-based sequencing of PCR products in EST-SSR marker development inEucalyptus.Fenzi Zhiwu Yuzhong(Online)8(1):1–10.doi:10.5376/mpb.cn.2010.08.0001
 Journal of Forestry Research2018年2期
Journal of Forestry Research2018年2期
- Journal of Forestry Research的其它文章
- Measurement of lumber moisture content based on PCA and GSSVM
- Characterization of mean stem density, fi bre length and lignin from two Acacia species and their hybrid
- Theoretical modeling of the effects of temperature and moisture content on the acoustic velocity of Pinus resinosa wood
- The properties of fl ax fi ber reinforced wood fl our/high density polyethylene composites
- Risks involved in fecal DNA-based genotyping of microsatellite loci in the Amur tiger Panthera tigris altaica:a pilot study
- Genecological zones and selection criteria for natural forest populations for conservation:the case of Boswellia papyrifera in Ethiopia
