First Insights into the Viral Communities of the Deep-sea Anoxic Brines of the Red Sea
Andre′Antunes*Intikhab AlambMarta Filipa Simo?escCamille Daniels Ari J.S.FerreiraRania SiamHamza El-DorryVladimir B.Bajic*h
1Computational Bioscience Research Center(CBRC),Computer,Electrical and Mathematical Sciences and Engineering Division(CEMSE),King Abdullah University of Science and Technology(KAUST),Thuwal 23955-6900,Saudi Arabia
2Red Sea Research Center(RSRC),Biological and Environmental Sciences and Engineering Division(BESE), King Abdullah University of Science and Technology(KAUST),Thuwal 23955-6900,Saudi Arabia
3Biology Department,American University in Cairo,New Cairo 11835,Egypt
ORIGINAL RESEARCH
First Insights into the Viral Communities of the Deep-sea Anoxic Brines of the Red Sea
Andre′Antunes1,*,a,Intikhab Alam1,b,Marta Filipa Simo?es1,c,Camille Daniels2,d, Ari J.S.Ferreira3,§,e,Rania Siam3,f,Hamza El-Dorry3,g,Vladimir B.Bajic1,*,h
1Computational Bioscience Research Center(CBRC),Computer,Electrical and Mathematical Sciences and Engineering Division(CEMSE),King Abdullah University of Science and Technology(KAUST),Thuwal 23955-6900,Saudi Arabia
2Red Sea Research Center(RSRC),Biological and Environmental Sciences and Engineering Division(BESE), King Abdullah University of Science and Technology(KAUST),Thuwal 23955-6900,Saudi Arabia
3Biology Department,American University in Cairo,New Cairo 11835,Egypt
Available online 31 October 2015
Handled by Fangqing Zhao
Viraldiversity; Metagenomics; Brine–seawater interface; Caudovirales; Red Sea
The deep-sea brines of the Red Sea include some of the most extreme and unique environments on Earth.They combine high salinities with increases in temperature,heavy metals, hydrostatic pressure,and anoxic conditions,creating unique settings for thriving populations of novel extremophiles.Despite a recent increase of studies focusing on these unusualbiotopes,their viral communities remain unexplored.The current survey explores four metagenomic datasets obtained from different brine–seawater interface samples,focusing specifically on the diversity of their viral communities.Data analysis confirmed that the particle-attached viral communities present in the brine–seawater interfaces were diverse and generally dominated by Caudovirales, yet appearing distinct from sample to sample.With a level of caution,we report the unexpected finding of Phycodnaviridae,which infects algae and plants,and trace amounts of insect-infecting Iridoviridae.Results from Kebrit Deep revealed stratification in the viral communities present inthe interface:the upper-interface was enriched with viruses associated with typicalmarine bacteria, while the lower-interface was enriched with haloviruses and halophages.These results provide first insights into the unexplored viralcommunities present in deep-sea brines of the Red Sea,representing one of the first steps for ongoing and future sampling efforts and studies.
Introduction
The development and widespread use of molecular-based methods in environmental microbiology revealed that microbes dominate our planet.Ocean-dwelling bacteria are estimated to outnumber stars in the universe by several orders of magnitude(total numbers are 1029and 1021,respectively) [1],with even higher values for viruses.Viruses are the most abundant biological entities on Earth(1030for total number of prokaryotic viruses or phages)[2],and harbor the second greatest biomass,after prokaryotes[3,4].Furthermore,they are crucial ecological factors,which affect microbial diversity, population dynamics,and the genomes of their hosts[5].Their impact extends from influencing microbial evolution,to playing an indirect but significant role in the Earth’s biogeochemical cycles[5–7].
Despite a historically stronger focus on medically-relevant viruses,recent years novel technologies brought forth increasing activities in the field of environmental virology, with multiple studies centered in marine and aquatic environments,as well as several extreme environments.Such ongoing efforts have led to the discovery and description of multiple new viruses and increased our understanding of viral communities(e.g.,[2,5,8–10]).Since viruses lack a shared universal phylogenetic marker such as universal ribosomal DNA(rDNA),genetic diversity of environmental viral communities is increasingly assessed through metagenomic sequencing,which provides more and more information about viral diversity and evolution[11,12]. Nonetheless,metagenomic data have shown that we have yet to discover the majority of viruses present in the environment:over 70%of the genes in the oceanic viral fraction cannot be associated with known viruses[7].Furthermore, studies on extreme environments,which include a few metagenomic-based surveys(e.g.,[13,14]),have uncovered that hypersaline environments host the highest viral densities reported for aquatic systems[15],yet still very little is known about them.
The deep-sea brines of the Red Sea include some of the most extreme and inaccessible environments on Earth, combining high salinities with increase in temperature, heavy metals,hydrostatic pressure,and anoxic conditions [16].The microbiology of these brines received considerable attention in the last few years,with studies using an array of culture-dependent[17–21]and molecular-based approaches,including metagenomic studies[22–33]. Nonetheless,the viral communities of these extreme biotopes remain unexplored.
This study makes use of four metagenomic samples, obtained from different brine–seawater interfaces from the Red Sea,providing the first,though partial,insights into the viral diversity and community structure present in these environments.
Results and discussion
In this study,we used the DMAP’s comparison module, associated taxonomic browsing,and filtering capabilities to explore the viral subset of annotations of four metagenomic samples(AT,DD,KU,and KL)obtained from the brine–seawater interfaces of different deeps in the Red Sea.The resulting taxonomic comparison of these samples showed the relative proportions of bacteria,archaea,and viruses (Table 1).While the number of genes associated with viruses might seem relatively low,it should be noted that our source data refers only to reads recovered from the 0.1-μm fraction(i.e.,particle-attached or from infected cells).The ensuing analysis is therefore restricted to only part of the total viral diversity present in these environments.
General viral diversity
Analysis of the metagenomic datasets confirmed that the particle-attached viral communities present in the brine–seawater interfaces were diverse and,despite some similarities, distinct from sample to sample(Figure 1).This is likely a reflection of differences in microbial community profiles, which are specific to each location,and imparted from changes in physicochemical conditions[16,26,30–33].
As a general trend,we observed a clear dominance of dsDNA viruses,which accounted for 45%–85%of the viruses detected(Figure 1).Further scrutinization indicated that dsDNA viruses are mostly Caudovirales(Figure 2).Caudovirales can be further classified as Syphoviridae,Myoviridae, and Podoviridae,while various proportions of them remained unclassified(Figure S1).Caudovirales are tailed bacteriophages,which are known to dominate in marine and other aquatic environments[9,34,35].
Other viral families were detected at much lower abundances,including the unexpected Phycodnaviridae(4%–6%),which infect algae and plants,and trace amounts of insect-infecting Iridoviridae(<1%).These viral taxa are likely derived from the particles originated away from the brines,including host lysis products that act as virus scavengers[5].We hypothesize that these particles sink through the water column,and are eventually trapped and accumulated in the density gradient of the brine–seawater interface.Viruses(and DNA,in general)have been previously reported as having longer-term stability,and can be preserved in such deep-sea brines[36–38].Similar observations have been reported for other marine locations,particularly when transitioning from oxic to anoxic conditions[9].Detection of these unexpected viral taxa would thus be the result of a‘‘pickling”effect,rather than reflecting the presence of specific hosts in close proximity to the brines.
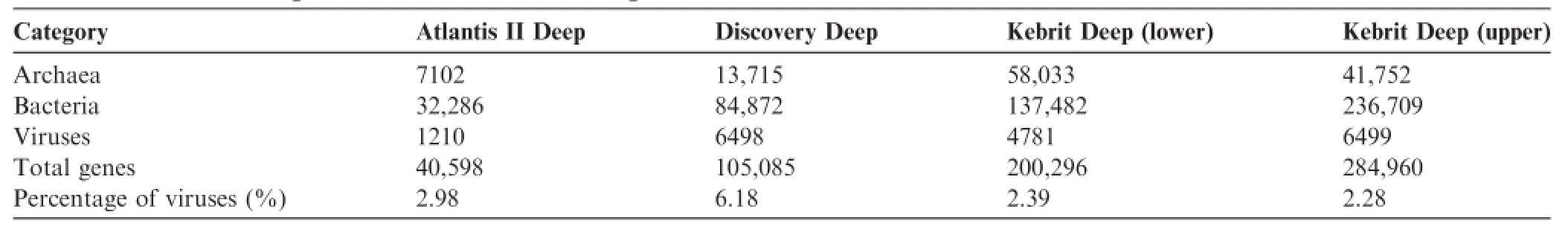
Table 1 Breakdown of genes based on taxonomic assignment
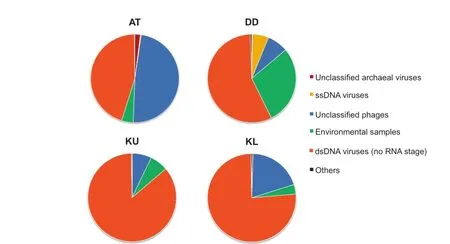
Figure 1 Relative abundances of members of different viral taxaSamples were collected on 0.1-μm filters from the brine–seawater interfaces of the Red Sea at different locations.All taxonomical categories mentioned in this study are based on the NCBI Taxonomy database.‘‘Others”include Adenoviridae,Ascoviridae, Baculoviridae,Bicaudaviridae,Fuselloviridae,Herpesvirales,Iridoviridae,Marseilleviridae,Poxviridae,Polydnaviridae,and Salterproviridae.AT,Atlantis II Deep;DD,Discovery Deep;KU,Kebrit Deep upper brine–seawater interface;KL,Kebrit Deep lower brine–seawater interface;dsDNA,double-stranded DNA;ssDNA,single-stranded DNA.
Samples from Atlantis II Deep had a more divergent profile,with a large,but not dominant proportion of dsDNA viruses(45%)associated with a slightly higher number of unclassified phages(47%;Figure 1).The vast majority of these phages were environmental halophages (data viewable with the taxonomy browser option of DMAP;www.cbrc.kaust.edu.sa/DMAP),which were related to the ones described in a previous study[39].Furthermore,a significant percentage of dsDNA viruses were also unclassified(40%;Figure 2)and belonged mostly to haloviruses(data viewable with the taxonomy browser option of DMAP;www.cbrc.kaust.edu.sa/DMAP).The unusual combination of very high temperatures and salinities at Atlantis II Deep,which creates one of the harshest environments on Earth and provides a unique prokaryotic host community,might be the main reason behind such high numbers of unclassified viruses.Therefore,one expects that the brine–seawater interface of Atlantis II Deep is a particularly interesting environment for future exploration,with a high potential for discovery of novel viruses infecting polyextremophiles.
Stratification in viral communities
An additional highlight of our analysis is the stratification of the viral communities within the brine–seawater interface observed for Kebrit Deep.Differences between upper,and lower interface samples from Kebrit Deep were in accordance with the relative position of these layers.Indeed,the upper interface,which is in closer proximity with seawater,was enriched with viruses associated with more typicalmarine bacteria(e.g.,Pelagibacter and Synechococcus),whereas the lower interface,which is closer to the brine,was enriched in haloviruses and halophages(Table 2).Furthermore,such stratification of viralcommunities is in agreement with previous reports obtained for microbial communities inhabiting brine–seawater interfaces both in the Red Sea and the Mediterranean(e.g., [26,30,40]).
Although there is some agreement between our results and such previous microbial studies,special care must be taken to avoid inferring direct correlations between microbial and viral communities,due to severalwell-known limitations and biases. The main caveat of viral metagenomics is that most sequencesare unique and thus have no matches in databases[13].Indeed, while much of the global microbial metagenome has now been sampled,the same cannot be said for the global viral metagenome[11].
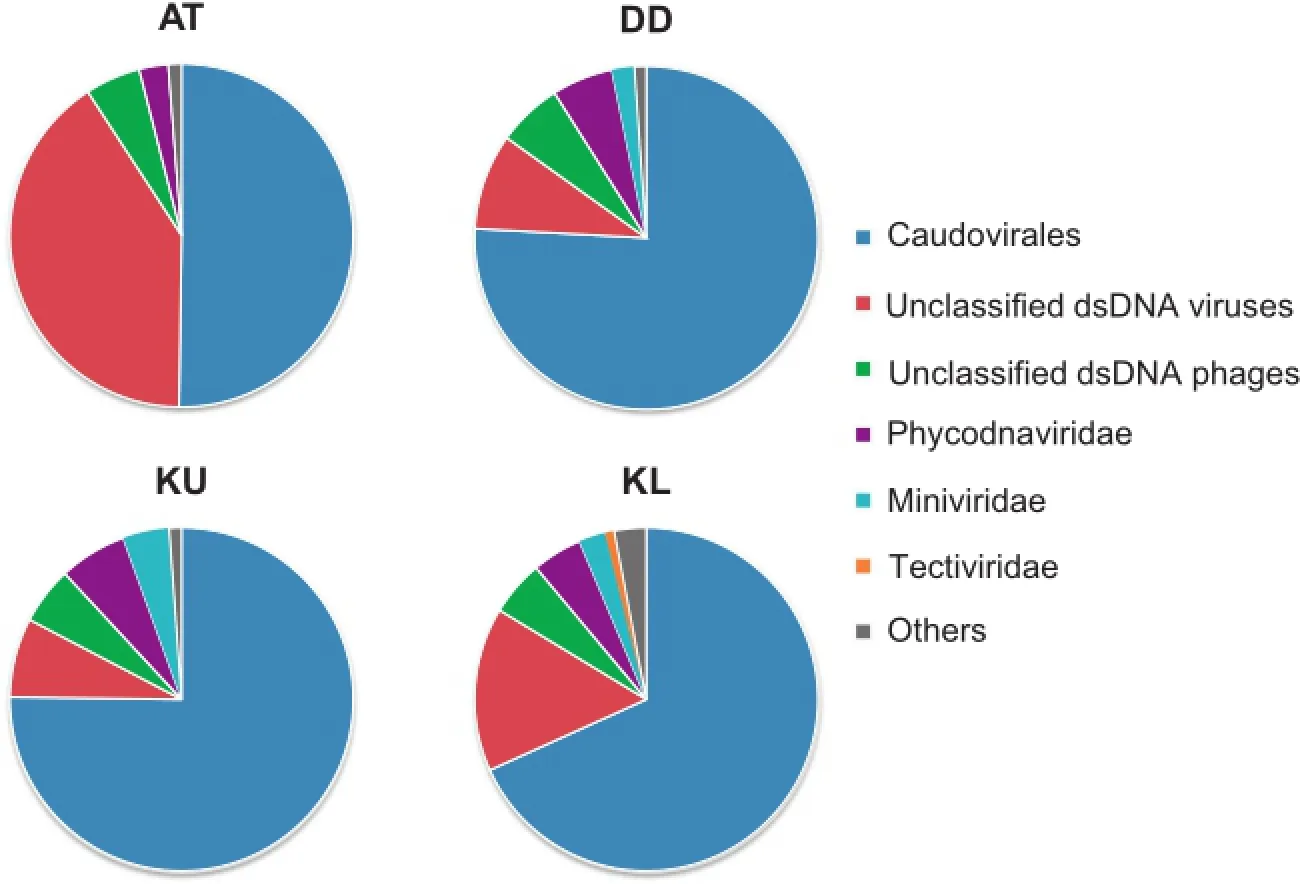
Figure 2 Relative abundances of double-stranded DNA(dsDNA)virusesSamples were collected on 0.1-μm filters from the brine–seawater interfaces of the Red Sea at different locations.All taxonomical categories mentioned in this study are based on the NCBI Taxonomy database.‘‘Others”include retro-transcribing viruses,satellites, ssRNA viruses,unassigned viruses,unclassified virophages,and unclassified viruses.AT,Atlantis II Deep;DD,Discovery Deep;KU, Kebrit Deep upper brine–seawater interface;KL,Kebrit Deep lower brine–seawater interface;dsDNA,double-stranded DNA;ssDNA, single-stranded DNA.
Most of our knowledge on viruses still relies heavily on in vitro cultured phage–host systems.Surveys of viral diversity are therefore bottlenecked by the lack of environmental isolates with ecological relevance,which frequently evade standard cultivation techniques,resulting in the dominance of culture-independent‘‘unknowns”[41].Most viral research focuses on strains amenable to laboratorial manipulation,rather than the most relevant or abundant ones[8].Accordingly,most phage genomes in GenBank are isolated using bacteria from only 3 of the 45 known bacteria phyla(Actinobacteria,Firmicutes,and Gammaproteobacteria),so many others that infect environmental microbes are largely unstudied and unknown[41]. Furthermore,and despite generally being perceived as host-specific predators,information on true host ranges for many viruses is lacking and might be wider than anticipated[5].
Overview and future work
Despite the aforementioned limitations,results from this study provide important first insights into the unexplored viralcommunities present in deep-sea brines of the Red Sea and thus represent the first step for ongoing,and future sampling efforts and studies.Future work should circumvent the constraints of this study by including targeted sampling of the viral community(i.e.,<0.1μm fraction)for metagenomic assessment,as well as isolation/characterization,along with studies to determine viral–host dynamics.
Materials and methods
Metagenomic samples and DNA sequencing
Metagenomic reads from the four brine–seawater interface libraries were obtained from a previous study(see[22]for further details).Briefly,samples were collected on 0.1-μm filters from the brine–seawater interfaces of Atlantis II Deep, Discovery Deep,and Kebrit Deep,Red Sea.Atlantis II Deep and Discovery Deep are both examples of‘‘hot brines”(with temperatures of 68°C and 45°C,respectively),while Kebrit Deep is a colder brine(temperature of 23°C),which is notorious for its very high sulfur concentration(see[16]for detailed information).
DNA extraction and sequencing were carried out at the American University of Cairo using GS FLX Roche Titanium library guide(see[22]for details).
Bioinformatics processing of metagenomic reads
?
Metagenomic data were processed with a focus on viral communities using the Dragon Metagenomic Analysis Platform (DMAP;www.cbrc.kaust.edu.sa/DMAP).Reads were assembled using Newbler software with iterative reference(NCBI RefSeq genomes)and de novo assembly procedure,and annotation was carried out using the DMAP annotation module (www.cbrc.kaust.edu.sa/DMAP).Briefly,the module predicts and annotates RNA and protein-coding genes.During annotation,BLAST best hit genes are considered for assigning taxonomic or function information to predicted genes.For taxonomic assignment to RNA genes,NCBI’s small sub-unit (SSU)RNAs and other non-coding RNAs from the European Bioinformatics Institute(EBI)Rfam database are used.For protein-coding genes,the UniProt Knowledgebase(www. uniprot.org),KEGG(www.kegg.jp),eggNOG(http://eggnogdb. embl.de),Conserved Domain Database(CDD,http://www. ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml),and InterPro (http://www.ebi.ac.uk/interpro)databases are used(see associated help provided at DMAP website;www.cbrc.kaust.edu.sa/ DMAP).From all the databases described above only archaeal,bacterial,and viral sequences were considered.The following parameters were considered when analyzing these datasets.An E value of 1E-3 and BLAST coverage of 50% was considered for BLAST-based analysis,while for other methods,such as InterProscan for domain detection or Infernal for Rfam’s ncRNA predictions,parameters optimized in the source profiles were considered with trusted cutoffs.Annotation results with taxonomic and functional assignments were deposited to the DMAP data warehouse and DMAP comparison module for systematic studies.
Total hit numbers for viral assignments at different taxonomic levels and general statistics for the contigs/singletons are provided as Tables S1 and S2,respectively.All data and analysis tools are openly accessible through the DMAP website at www.cbrc.kaust.edu.sa/DMAP.
Authors’contributions
AA conceived,designed,and performed the experiments;AA and MFS analyzed the data;IA,AJSF,RS and HED contributed reagents/materials/analysis tools;AA,IA,MFS, CD,RS,and VBB wrote the manuscript.All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
The authors acknowledge the support through the KAUST baseline research funds to VBB.The study was partially supported by the KAUST-AUC Global Collaborative Research Program.
Supplementary material
Supplementary material associated with this article can be found,in the online version,at http://dx.doi.org/10.1016/j. gpb.2015.06.004.
References
[1]Smetacek V.The ocean’s veil.Nature 2002;419:565.
[2]Mizuno CM,Rodriguez-Valera F,Kimes NE,GhaiR.Expanding the marine virosphere using metagenomics.PLoS Genet 2013;9: e1003987.
[3]Sabet S.Halophilic viruses.In:Vreeland Russell H,editor. Advances in understanding the biology of halophilic microorganisms.Netherlands:Springer;2012.p.81–116.
[4]Suttle CA.Viruses in the sea.Nature 2005;437:356–61.
[5]Breitbart M.Marine viruses:truth or dare.Ann Rev Mar Sci 2012;4:425–48.
[6]Rodriguez-Valera F,Martin-Cuadrado AB,Rodriguez-Brito B, Pasˇic′L,Thingstad TF,Rohwer F,et al.Explaining microbial population genomics through phage predation.Nat Rev Microbiol 2009;7:828–36.
[7]Sabehi G,Shaulov L,Silver DH,Yanai I,Harel A,Lindell D.A novellineage of myoviruses infecting cyanobacteria is widespread in the oceans.Proc Natl Acad Sci U S A 2012;109:2037–42.
[8]Luk AW,Williams TJ,Erdmann S,Papke RT,Cavicchioli R. Viruses of Haloarchaea.Life(Basel)2014;4:681–715.
[9]Weinbauer MG.Ecology of prokaryotic viruses.FEMS Microbiol Rev 2004;28:127–81.
[10]Brum JR,Sullivan MB.Rising to the challenge:accelerated pace of discovery transforms marine virology.Nat Rev Microbiol 2015;13:147–59.
[11]Edwards RA,Rohwer F.Viralmetagenomics.Nat Rev Microbiol 2005;3:504–10.
[12]Mokili JL,Rohwer F,Dutilh BE.Metagenomics and future perspectives in virus discovery.Curr Opin Virol 2012;2:63–77.
[13]Rodriguez-Brito B,Li L,Wegley L,Furlan M,Angly F,Breitbart M,et al.Viraland microbialcommunity dynamics in four aquatic environments.ISME J 2010;4:739–51.
[14]Narasingarao P,Podell S,Ugalde JA,Brochier-Armanet C, Emerson JB,Brocks JJ,et al.De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbialcommunities.ISME J 2012;6:81–93.
[15]Santos F,Yarza P,Parro V,Meseguer I,Rossello′-Mo′ra R,Anto′n J.Culture-independent approaches for studying viruses from hypersaline environments.Appl Environ Microbiol 2012;78: 1635–43.
[16]Antunes A,Ngugi DK,Stingl U.Microbiology of the Red Sea (and other)deep-sea anoxic brine lakes.Environ Microbiol Rep 2011;3:416–33.
[17]Antunes A,Eder W,Fareleira P,Santos H,Huber R.Salinisphaera shabanensis gen.nov.,sp.nov.,a novel,moderately halophilic bacterium from the brine–seawater interface of the Shaban Deep,Red Sea.Extremophiles 2003;7:29–34.
[18]Antunes A,Franc?a L,Rainey FA,Huber R,Nobre MF,Edwards KJ,et al.Marinobacter salsuginis sp.nov.,a novelspecies from the brine–seawater interface of the Shaban Deep,Red Sea.Int J Syst Evol Microbiol 2007;57:1035–40.
[19]Antunes A,Taborda M,Huber R,Moissl C,Nobre MF,da Costa MS.Halorhabdus tiamatea sp.nov.,a non-pigmented,extremely halophilic archaeon from a deep-sea hypersaline anoxic basin of the Red Sea,and emended description of the genus Halorhabdus. Int J Syst Evol Microbiol 2008;58:215–20.
[20]Antunes A,Rainey F,Wanner G,Taborda M,Pa¨tzold J,Nobre MF,et al.A new lineage of halophilic,wall-less,contractile bacteria from a brine-filled Deep of the Red Sea.J Bacteriol 2008;190:3580–7.
[21]Fiala G,Woese CR,Langworthy TA,Stetter KO.Flexistipes sinusarabici,a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea.Arch Microbiol 1990;154:120–6.
[22]Abdallah RZ,Adel M,Ouf A,Sayed A,Ghazy MA,Alam I,et al. Aerobic methanotrophic communities at the Red Sea brine–seawater interface.Front Microbiol 2014;5:487.
[23]Antunes A,Alam I,Bajic VB,Stingl U.Genome sequence of Halorhabdus tiamatea,the first archaeon isolated from a deep-sea anoxic brine lake.J Bacteriol 2011;193:4553–4.
[24]Antunes A,Alam I,Bajic VB,Stingl U.Genome sequence of Salinisphaera shabanensis,a gammaproteobacterium from the harsh,variable environment of the brine–seawater interface of the Shaban Deep in the Red Sea.J Bacteriol 2011;193:4555–6.
[25]Antunes A,Alam I,El Dorry H,Siam R,Robertson A,Bajic VB, et al.Genome sequence of Haloplasma contractile,an unusual contractile bacterium from a deep-sea anoxic brine lake.J Bacteriol 2011;193:4551–2.
[26]Bougouffa S,Yang JK,Lee OO,Wang Y,Batang Z,Al-Suwailem A,et al.Distinctive microbial community structure in highly stratified deep-sea brine water columns.Appl Environ Microbiol 2013;79:3425–37.
[27]Eder W,Jahnke LL,Schmidt M,Huber R.Microbialdiversity of the brine–seawater interface of the Kebrit Deep,Red Sea,studied via 16S rRNA gene sequences and cultivation methods.Appl Environ Microbiol 2001;67:3077–85.
[28]Eder W,Ludwig W,Huber R.Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep,Red Sea.Arch Microbiol 1999;172:213–8.
[29]Eder W,Schmidt M,Koch M,Garbe-Scho¨nberg D,Huber R. Prokaryotic phylogenetic diversity and corresponding geochemicaldata of the brine–seawater interface of the Shaban Deep,Red Sea.Environ Microbiol2002;4:758–63.
[30]Guan Y,Hikmawan T,Antunes A,Ngugi D,Stingl U.Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea.Res Microbiol 2015;166:688–99.
[31]Siam R,Mustafa GA,Sharaf H,Moustafa A,Ramadan AR, Antunes A,et al.Unique prokaryotic consortia in geochemically distinct sediments from Red Sea Atlantis II and Discovery Deep brine pools.PLoS One 2012;7:e42872.
[32]Wang Y,Yang J,Lee OO,Dash S,Lau SC,Al-Suwailem A,et al. Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea.ISME J 2011;5:1652–9.
[33]Wang Y,Cao H,Zhang G,Bougouffa S,Lee OO,Al-Suwailem A,et al.Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools.Sci Rep 2013;3:1748.
[34]Winter C,Garcia JA,Weinbauer MG,DuBow MS,Herndl GJ. Comparison of deep-water viromes from the Atlantic Ocean and the Mediterranean Sea.PLoS One 2014;9:e100600.
[35]Wommack KE,Colwell RR.Virioplankton:viruses in aquatic ecosystems.Microbiol Mol Biol Rev 2000;64:69–114.
[36]Borin S,Crotti E,Mapelli F,Tamagnini I,Corselli C,Daffonchio D.DNA is preserved and maintains transforming potential after contact with brines of the deep anoxic hypersaline lakes of the Eastern Mediterranean Sea.Saline Systems 2008;4:10.
[37]Corinaldesi C,Tangherlini M,Luna GM,Dell’Anno A.Extracellular DNA can preserve the genetic signatures of present and past viralinfection events in deep hypersaline anoxic basins.Proc Biol Sci 2014;281:20133299.
[38]Danovaro R,Corinaldesi C,Dell’Anno A,Fabiano M,CorselliC. Viruses,prokaryotes and DNA in the sediments of a deephypersaline anoxic basin(DHAB)of the Mediterranean Sea. Environ Microbiol 2005;7:586–92.
[39]Garcia-Heredia I,Martin-Cuadrado AB,Mojica FJ,Santos F, Mira A,Anto′n J,et al.Reconstructing viral genomes from the environment using fosmid clones:the case of haloviruses.PLoS One 2012;7:e33802.
[40]Daffonchio D,Borin S,Brusa T,Brusetti L,van der Wiedjen PWJJ,Bolhuis H,et al.Stratified prokaryote network in the oxic–anoxic transition of a deep-sea halocline.Nature 2006;440: 203–7.
[41]Holmfeldt K,Solonenko N,Shah M,Corrier K,Riemann L, VerBerkmoes NC,etal.Twelve previously unknown phage genera are ubiquitous in global oceans.Proc Natl Acad Sci U S A 2013;110:12798–803.
29 March 2015;revised 9 June 2015;accepted 29 June 2015
s.
E-mail:andre.antunes@kaust.edu.sa(Antunes A),vladimir.bajic@kaust.edu.sa(Bajic VB).§Current address:Engineering and Exact Science Department,FIEO University Center,Osasco,SP 06020-190,Brazil.aORCID:0000-0001-7668-9842.bORCID:0000-0001-5306-847X.cORCID:0000-0002-8767-9487.dORCID:0000-0002-5227-5394.eORCID:0000-0002-7526-5543.fORCID:0000-0002-2879-6368.gORCID:0000-0002-6457-2185.hORCID:0000-0001-5435-4750.
Peer review under responsibility of Beijing Institute of Genomics,Chinese Academy of Sciences and Genetics Society of China.
 Genomics,Proteomics & Bioinformatics2015年5期
Genomics,Proteomics & Bioinformatics2015年5期
- Genomics,Proteomics & Bioinformatics的其它文章
- CVTree3 Web Server for Whole-genome-based and Alignment-free Prokaryotic Phylogeny and Taxonomy
- Soil and Rhizosphere Associated Fungi in Gray Mangroves(Avicennia marina)from theRed Sea—A Metagenomic Approach
- Web Resources for Metagenomics Studies
- Marine Metagenome as A Resource for Novel Enzymes
- PacBio Sequencing and Its Applications
- The Value and Significance of Metagenomics of Marine Environments
