Isolation and characterization of a novel wall-associated kinase gene TaWAK5 in wheat(Triticum aestivum)
Kun Yang,Lin Qi,Zengyan Zhang*
The National Key Facility for Crop Gene Resources and Genetic Improvement,Key Laboratory of Biology and Genetic Improvement of Triticeae Crops,Ministry of Agriculture,Institute of Crop Science,Chinese Academy of Agricultural Sciences,Beijing 100081,China
1.Introduction
Plants are continuously threatened by a broad range of pathogens,including fungi,oomycetes,viruses,and bacteria.To defend themselves against pathogen attack,plants have evolved an array of response systems,in which external cues are deciphered and translated into effective defense responses [1].Receptor-like kinases (RLKs) play fundamental roles in the perception of external stimuli and activate defense-associated signaling pathways,thereby regulating cellular responses to pathogen infection[1].For example,FLAGELLIN SENSTIVE2 (FLS2) and bacterial translation elongation factor EF-Tu receptor(EFR)act as pattern-recognition receptors (PRRs) that recognize pathogen-associated molecular patterns(PAMPs)and play key roles in PAMP-triggered immunity in Arabidopsis thaliana[2,3].The cell surface receptor chitin elicitor receptor kinase 1 of Arabidopsis (AtCERK1) directly binds chitin through its lysine motif (LysM)-containing ectodomain(AtCERK1-ECD)to activate defense responses[4].
Wall-associated kinases (WAKs) and WAK-like kinases(WAKLs) are a unique RLK subfamily that contains excellent candidates which may directly link and enable communication between the extracellular matrix (ECM) and the cytoplasm[5,6].WAK proteins possess a typical cytoplasmic Ser/Thr kinase signature,and have an extracellular domain(ectodomain) with similarity to vertebrate epidermal growth factor (EGF)-like domains [7].WAKs have been shown to perceive damage-associated molecular patterns (DAMPs),which are comprised of the pectin and oligogalacturonide(OG)molecules that are released from the plant cell wall following damage caused by pathogen attack.WAKs then function to communicate these damage signals,thereby modulating both plant defense and development[5,8].
In Arabidopsis,26 WAK/WAKL genes have been identified.Five of these WAK genes(AtWAK1–5)were shown to be clustered on chromosome 1.Certain WAK homologues have been identified in rice(Oryza sativa),tobacco(Nicotiana tabacum),maize(Zea mays),barley (Hordeum vulgare),and wheat(Triticum aestivum)[9].AtWAK1 in Arabidopsis is the most studied WAK receptor kinase.The transcription of AtWAK1 is induced by OG molecules and salicylic acid(SA)[10].AtWAK1 was shown to bind OG molecules and to mediate the perception of OG molecules [5].Transgenic plants overexpressing AtWAK1 showed elevated resistance to the necrotrophic pathogen Botrytis cinerea[5].Both AtWAK1 and AtWAK2 were shown to bind pectin in vitro.AtWAK2 was shown to be required for the pectin-induced activation of numerous genes,many of which were involved in defense responses [8].OsWAK1 transcript was significantly induced after infection with the rice blast fungus (Magnaporthe oryzae) and also induced following treatment with exogenous SA or methyl jasmonate(MeJA).Transgenic rice lines overexpressing OsWAK1 showed enhanced resistance to M.oryzae strain P007[11].Although four WAKs (TaWAK1–4) and two WAKLs (TaWAKL1 and TaWAKL2)have been isolated from wheat[12],their functional roles remain poorly understood.
Phyto-hormones,including SA,JA,ethylene,and abscisic acid(ABA),are known to play important roles in plant responses to biotic and abiotic stresses[13–19].Upon microbial attack,plants modify the relative abundance of these hormones as a defense mechanism that can then activate efficient defense responses at the molecular genetic level,enabling plant survival [20].SA is involved in recognition of pathogen-derived components and the subsequent establishment of local and systemic acquired resistance [21,22].JA and ethylene signaling act synergistically and regulate induced systemic resistance.ABA also plays an important role in plant defense response,and the ABA signaling pathway interacts with other phyto-hormone signaling pathways in plant defense responses[23–25].
Wheat is one of the most important staple crops in the world and plays a fundamental role in food security.Sharp eyespot disease,mainly caused by the necrotrophic fungal pathogen R.cerealis,is one of the most devastating diseases in wheat production [26,27].In infected wheat plants,R.cerealis may destroy the stems and sheaths of host plants and can cause lodging and dead spikes[27].Few wheat genes involved in wheat defense responses to R.cerealis have been identified or characterized to date.Moreover,it is not known whether protein kinases participate in wheat responses to the pathogen infection during the developing process of sharp eyespot disease.
The goal of this research was to understand the roles of WAKs in wheat defense responses to R.cerealis infection.By using the Agilent wheat microarrays,we studied the transcriptomic profiles of WAK/WAKL genes in resistant and susceptible wheat lines following inoculation with R.cerealis.A WAK gene named TaWAK5 was identified to be significantly up-regulated at 21 days post inoculation (dpi) in the resistant wheat line CI12633 as compared with susceptible wheat cultivar Wenmai 6.This paper reports the identification,molecular characterization,and functional analysis of TaWAK5.The transcript abundance of TaWAK5 was markedly induced after R.cerealis infection.Its expression was also induced following exogenous application of SA,ABA,and MeJA.The protein was localized at the sub-cellular level to the plasma membrane in onion cells.We further analyzed the function of TaWAK5 in wheat defense responses to R.cerealis using virus-induced gene silencing(VIGS)technique.
2.Materials and methods
2.1.Plant and fungal materials and treatments
Six wheat (T.aestivum L.) lines/cultivars exhibiting different levels of resistance and susceptibility to R.cerealis were used in this study.They included CI12633 and Shanhongmai(resistant to R.cerealis); Navit 14,and Shannong 0431 (moderately resistant to R.cerealis);Wenmai 6(susceptible to R.cerealis);and Yangmai 158(moderately susceptible to R.cerealis)[28].
A major Jiangsu virulent isolate strain of pathogen fungus R.cerealis causing the sharp eyespot disease,R0301,was provided by Profs.Huaigu Chen and Shibin Cai from Jiangsu Academy of Agricultural Sciences,China.
Wheat plants were grown in a 14 h light/10 h dark(22 °C/10 °C) regime.At the tillering stage,the 2nd base sheath of each wheat plant was inoculated with small toothpick fragments harboring well-developed mycelia of the pathogen R.cerealis following Chen [27].Mock treatment (control) plants were inoculated with small toothpick fragments soaked in liquid potato dextrose agar(PDA).Inoculated plants were grown at 90%relative humidity for 4 days.The inoculated stems were sampled at 0,4,7,10,14,and 21 days post inoculation,quickly frozen in liquid nitrogen,and stored at-80 °C prior to total RNA extraction.At 4 dpi,the roots,sheaths,stems,and leaves of the inoculated CI12633 plants were collected,respectively.At 45 dpi,the roots,stems,leaves,and young spikes of the inoculated CI12633 plants were separately sampled and used for RNA extraction and the tissue expression profiles of TaWAK5.
In additional experiments,the seedlings at the three-leaf stage of the resistant line CI12633 were treated with phyto-hormones,including 1.0 mmol L-1SA,0.1 mmol L-1MeJA (JA analog),ethylene released from 0.2 mmol L-1ethephon,and 0.2 mmol L-1ABA,following Zhang et al.[29].Leaves were collected for RNA extraction at 0,1,3,6,12,and 24 h after treatment with these hormones.
2.2.RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent(Qiagen,China)according to the manufacturer's instructions.DNase I treatment was used to remove genomic DNA.First-strand cDNA was synthesized using 2 μg purified RNA,AMV reverse transcriptase,and oligo (dT15) primers (TaKaRa Inc.,Tokyo,Japan) according to the manufacturer's instructions for the cDNA synthesis kit.
2.3.Cloning of full-length sequence of TaWAK5 cDNA
Based on microarray analysis results,a partial cDNA fragment(GenBank accession number CA642360),which was differentially expressed between the resistant wheat genotype CI12633 and the susceptible wheat Wenmai 6,was identified.Based on the sequence of CA642360 and using a 3′-Full RACE Core Set kit v.2.0 from TaKaRa Inc.,the sequence of the 3′ untranslated region (UTR) was amplified from cDNA of CI12633 stems that had been challenged with the pathogen R.cerealis for 21 days.Using CA642360 as a query sequence,the DNA sequence was retrieved from the Cereals Data Base (Cereals DB,http://www.cerealsdb.uk.net/CerealsDB/Documents/ DOC_CerealsDB.php),and the gene structure was analyzed using SoftBerry FGENESH software program in LINUX system (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind).Gene-specific primers TaWAK5-ORF-F/TaWAK5-ORF-R were then designed and used to amplify the full-length open reading frame(ORF)sequence of TaWAK5 from the cDNA of the CI12633.The purified PCR products were cloned to the pMD-18T vector from TaKaRa Inc.and selected to identify the positive clones.Five positive clones were then sequenced with an ABI PRISM 3130XL Genetic analyzer (Applied Biosystems,Foster City,CA).The full-length cDNA sequence of the resulting TaWAK5 gene with 2282 bp length was obtained by analyzing the aligned sequences.
The TaWAK5 gene was analyzed using several bioinformatics tools.First,the cDNA sequence data was analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and ORF Finder(http://www.ncbi.nlm.nih.gov/gorf/).The deduced protein sequence was then analyzed with the Compute pI/Mw tool(http://web.expasy.org/compute_pi/)which is used for computation of the theoretical iso-electric point and protein molecular weight,InterPro-Scan(http://www.ebi.ac.uk/interpro/)for domain identification and Smart software(http://smart.embl-heidelberg.de/smart/set_mode.cgi? GENOMIC = 1) for prediction of the conserved motifs of domains.DNAMAN software was then used for sequence alignment and MEGA 5.0 software for constructing a phylogenetic tree.The region upstream (1000 bp) of the start codon was analyzed using the plant cis-acting regulatory DNA element(PLACE)database(http://www.dna.affrc.go.jp/PLACE/).
2.4.Subcellular localization of TaWAK5
The coding region of TaWAK5 lacking the stop codon was amplified using gene-specific primers TaWAK5-GFP-F/TaWAK5-GFP-R.The amplified fragment was digested with restriction enzymes Pst I and Xba I,then subcloned in-frame into the 5′-terminus of the GFP(green fluorescent protein)coding region in the pCaMV35S:GFP vector (kindly provided by Dr.Daowen Wang,Chinese Academy of Sciences),resulting in the TaWAK5-GFP fusion construct pCaMV35S:TaWAK5-GFP.
The p35S:TaWAK5-GFP fusion construct or p35S:GFP control construct was separately bombarded into epidermal cells of a white onion according to the protocol described by Zhang et al.[30].To induce the expression of the introduced GFP proteins,the transformed onion cells were incubated at 25 °C for 16 h.The GFP signals were then observed and photographed using a Confocal Laser Scanning Microscope (Zeiss LSM 700,Germany) with a Fluar 10X/0.50M27 objective lens and an SP640 filter.The plasmolysis of the onion cells was undertaken by addition of 0.8 mol L-1sucrose solution for 5 min,as described by Lang-Pauluzzi and Gunning[31].
2.5.Functional analysis of TaWAK5 through virus-induced gene silencing
A virus-induced gene silencing (VIGS) technique was previously developed with barley stripe mosaic virus (BSMV) and found to be an effective reverse genetics tool for investigating rapidly the functions of some genes in barley and wheat[32,33].To generate a BSMV:TaWAK5 construct,a 298 bp sequence of TaWAK5 (from nucleotide position 1913 to 2211 in the TaWAK5 cDNA sequence)was amplified from the cDNA sequence for TaWAK5 from the genotype CI12633 with the primers TaWAK5-VIGS-F/TaWAK5-VIGS-R.PCR-amplified cDNA fragments were digested with Pac I and Not I,then ligated into the BSMV:RNAγ vector digested with Pac I-Not I,resulting in the recombinant construct RNAγ:TaWAK5-as.
Following a previously described protocol [32],the tripartite cDNA chains of BMSV:TaWAK5,or the control virus BMSV:GFP genome,were separately transcribed into the RNAs,then mixed and used to infect CI12633 plants at the 2-leaf stage.At the same time,CI12633 plants were inoculated with only the buffer without virus.Hereafter,these plants treated only with buffer are referred to as mock treatments.The 4th leaves of the inoculated seedlings were collected and analyzed for the virus infection based on the RNA transcript presence of the BSMV coat protein gene using primers BSMV-CP-F/BSMV-CP-R.These tissues were also evaluated for changes in TaWAK5 expression with primers TaWAK5-Q-F/TaWAK5-Q-R at 10 days after BSMV infection.
For R.cerealis inoculation,the fungus was cultured on potato dextrose agar at 25 °C for 10 days,then 1 cm2plugs from the edge of R.cerealis colonies were placed into liquid PDA medium and cultured at 25 °C for 2 weeks,to develop the mycelia.The 4th base sheath of wheat plants was inoculated with 15 μL of the R.cerealis liquid culture at 20 days after BSMV virus inoculation.
Inoculated plants were grown at 90%relative humidity for 4 days.Sharp eyespot symptoms were observed respectively at 14 days and 40 days after fungal inoculation.These are the times when sharp eyespot symptoms are normally present at the infected sheaths and stems,respectively,of the susceptible cultivar Wenmai 6.
2.6.RT-PCR and real-time quantitative RT-PCR (qRT-PCR)analysis
RT-PCR was performed with 20 μL reaction volumes from the TaKaRa Inc.kit containing 1× PCR buffer,2.0 μL 10× first strand cDNA,150 μmol L-1of each dNTP,and 1 U Taq polymerase,plus 0.25 μmol L-1of each primer.The program used was as follows: initial denaturation at 94 °C for 5 min;followed by 30 cycles of 30 s at 94 °C,30 s at 60 °C,and 30 s at 72 °C;and final extension at 72 °C for 5 min.The PCR products were detected on 2%agarose gels.In all the semi-quantitative RT-PCR experiments,wheat elongation factor 1 alpha-subunit(TaEF-1a) was used to normalize the cDNA contents among various samples.
qRT-PCR was performed using SYBR Green I Master Mix from TaKaRa Inc.in a volume of 25 μL on an ABI 7300 RT-PCR system (Applied Biosystems Corp.).Reactions were set up with the following thermocycling profile: 95 °C for 5 min,followed by 41 cycles of 95 °C for 15 s and 60 °C for 31 s.The products were continuously examined with a melting curve analysis program.All qRT-PCR reactions were repeated three times.The relative expression of the gene TaWAK5 was calculated with the 2-ΔΔCTmethod [34],where the wheat TaActin gene was used as the internal reference.
The sequences of primers used are listed in Table S1.
3.Results
3.1.The transcription of TaWAK5 is induced by R.cerealis infection
Microarray analysis is a frequently used molecular genetic technique for the identification of target genes that are expressed differentially between different plant tissue samples or the same samples under different treatments.In this study,we used Agilent wheat microarrays to identify WAK genes that were differentially expressed between the resistant wheat genotype CI12633 and susceptible wheat cultivar Wenmai 6 following infection with R.cerealis.Based on differentially-expressed gene analysis,a wheat cDNA fragment CA642360 had a 30-fold increase in transcript level in the resistant CI12633 as compared with the susceptible Wenmai 6 at 21 dpi.BLAST searching against the GenBank database showed that this gene was homologous to the genes encoding WAKs in plants.As four WAK genes,TaWAK1,TaWAK2,TaWAK3,and TaWAK4,were isolated from wheat in a previous study [12],hereafter,this novel wheat WAK gene induced by R.cerealisis designated as TaWAK5.
To further investigate the involvement of TaWAK5 in wheat responses against R.cerealis,qRT-PCR was used to analyze the transcript profile of TaWAK5 in wheat infected with the fungal pathogen R.cerealis.The analysis over a 21-day pathogen inoculation time-series showed that TaWAK5 was induced by R.cerealis infection in both the resistant CI12633 and in the susceptible Wenmai 6,whereas the induction degree was higher in CI12633 as compared to Wenmai 6 (Fig.1-A).The expression level of TaWAK5 in CI12633 was about 15 times higher than the level in Wenmai 6 at 21 dpi,consistent with the result of the microarray analysis and with the level of resistance displayed by the genotypes.Following R.cerealis infection,TaWAK5 transcripts in the resistant CI12633 were induced at 4 dpi,reached a first peak at 10 dpi (about 24-fold increase over 0 dpi),decreased at 14 dpi,and reached a second peak at 21 dpi (about 33-fold increase over 0 dpi).
Meanwhile,the expression of TaWAK5 in different tissues of the R.cerealis-inoculated CI12633 was assessed using qRT-PCR (Fig.1-B).At 4 dpi,the TaWAK5 gene was expressed most highly in the roots(10-fold over in the stems)than in the sheaths and leaves.The lowest expression was found in the stems.The expression level of TaWAK5 in the sheaths was 7 times higher than that in the stems.At 45 dpi,the transcriptional level of TaWAK5 was the highest in the root samples and lowest in the young spike tissue,with 107 times higher expression level in the former root tissue.The expression level of TaWAK5 was elevated 2-fold in stems and 1.99-fold in leaves compared with the young spike.
A more detailed analysis of the expression patterns of TaWAK5 was carried out in R.cerealis-resistant lines (CI12633,Shanhongmai,Navit14,and Shannong 0431) and susceptible lines (Wenmai 6 and Yangmai 158).As shown in Fig.1-C,transcript abundances of TaWAK5 were higher in the resistant lines than in the susceptible lines at 7 dpi,with highest level found in the highly resistant wheat CI12633,and lowest level found in the moderately-susceptible wheat Yangmai 158.The above results suggested that TaWAK5 may be involved in wheat defense response to R.cerealis infection.
3.2.TaWAK5 encodes a wall-associated kinase
The full-length cDNA sequence (2282 bp) of TaWAK5 was obtained from the resistant wheat genotype CI12633 and deposited in the GenBank database (accession number KF710462).The cDNA contained an ORF of 2112 nucleotides(from 21 to 2132 bp),encoding a protein of 703 amino acids with an estimated molecular mass of 77.0 kDa and a predicted pI of 6.7.BLAST searching against the GenBank database indicated that the TaWAK5 gene was homologous to WAK genes from Aegilops tauschii (GenBank entry,EMT17650)with 67% identity,from Triticum urartu (GenBank entry,EMS57881) with 60% identity,from Setaria italic (GenBank entry,XP_004959009) with 55% identity,and from O.sativa(GenBank entry,AAX95007)with 42%identity.
The deduced amino acid sequence of TaWAK5 was found to contain various signals and protein domains(Fig.2).In the N-terminal region,there was a predicted signal peptide at amino acids 1–37,which may cause membrane targeting.Two EGF-like repeats at amino acids 268–319 and 323–363 were identified in the putative extracellular domain of the sequence.Additionally,the TaWAK5 protein had a putative protein kinase catalytic domain (residues 429–694) that included an ATP binding site and a Ser/Thr kinase active site(ILHGDVKPANILL,residues 549–561).TaWAK5 is non-arginine aspartate (RD)-type protein,as it carries a glycine (G) rather than an arginine (R) residue immediately preceding the conserved aspartate (D)in the catalytically-active subdomain VIb.
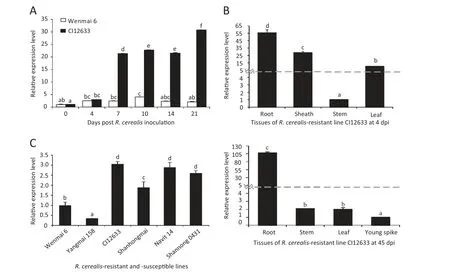
Fig.1-Expression patterns of TaWAK5 in wheat responding to R.cerealis inoculation.A:Transcript levels of the TaWAK5 gene in the R.cerealis-resistant line CI12633 and the susceptible cultivar Wenmai 6 over a 21 day pathogen inoculation time-series.Relative expression of TaWAK5 indicated the changing fold expression level of the gene transcript in CI12633 over Wenmai 6 at 0 dpi.B:Expression patterns of TaWAK5 in different tissues of R.cerealis-inoculated CI12633 plant at 4 dpi and 45 dpi.C:Expression patterns of TaWAK5 in R.cerealis-resistant lines and susceptible lines after R.cerealis inoculation for 7 days.Transcript levels of TaWAK5 in wheat lines were measured by qRT-PCR relative to highly susceptible cultivar Wenmai 6.The mean and standard error(SE) of three replicates are presented.The transcript abundances with different letters are significantly different from each other,based on inferential statistical comparison using SPSS19.
Phylogenetic analysis was performed to decipher the relationship between TaWAK5 and any related RLKs.Twenty-one available RLK sequences from different plant species were used to construct a rooted phylogenetic tree.These RLK sequences formed four different subgroups of RLKs including WAK,leucine-rich repeat (LRR)-RLK,LysM-RLK,and lectin-RLK.In the first group,the proteins for TaWAK5,TaWAK1,TaWAK2,TaWAK3,TaWAK4,OsWAK,HvWAK,AtWAK1,AtWAK2,AtWAK3,AtWAK4,and AtWAK5 were clustered into a single WAK clade(Fig.3-A).
We performed a comparison of amino acid sequences of WAK proteins to determine their similarity.TaWAK5 was found to be closely related to HvWAK from H.vulgare (56.6%identity),followed by OsWAK from O.sativa (47.0% identity),suggesting that these are orthologs of each other from different cereals in the Gramineae family.Meanwhile,TaWAK5 shared 31.5–38.6% protein sequence identities with the four reported wheat WAK paralogs,TaWAK1,TaWAK2,TaWAK3,and TaWAK4.The sequence identities between TaWAK5 and Arabidopsis WAKs were only 30.6–32.8%,showing the distance between monocot and dicot homologues of WAK genes.We then carried out a multiple alignment of EGF-like repeats of TaWAK5 and WAKs from wheat,barley,rice,and Arabidopsis,in which each EGF-like repeat contained six conserved cysteine residues (Fig.3-B).The positions of the six cysteine residues are conserved in TaWAK5 and the other tested WAKs,although the amino acid sequences between the cysteine residues varied.
3.3.TaWAK5 was localized to the plasma membrane in planta
To study the subcellular localization of TaWAK5,the p35S:TaWAK5-GFP and p35S:GFP constructs were separately introduced into onion epidermal cells.As presented in Fig.4-A,the TaWAK5-GFP fusion proteins were localized on the cell periphery,whereas the fluorescence of GFP alone as a control was distributed throughout the cell.To verify the nature of the subcellular localization of TaWAK5,a plasmolysis experiment was performed.When onion cells expressing TaWAK5-GFP were plasmolyzed in a 0.8 mol L-1sucrose solution,TaWAK5-directed GFP fluorescence signal was observed on the plasma membrane(Fig.4-B).Thus,TaWAK5 may be a plasma membrane-localized protein.
3.4.TaWAK5 was induced by exogenous SA,ABA,or MeJA application
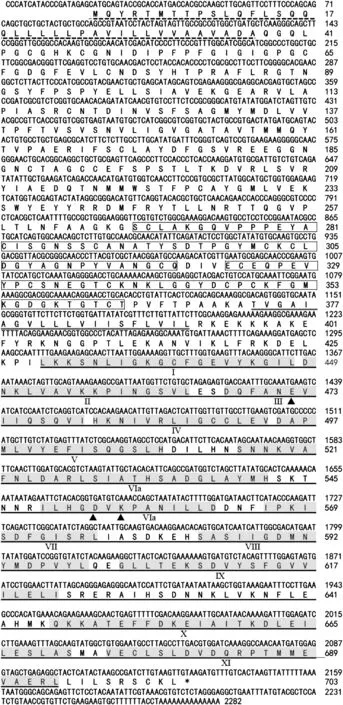
Fig.2-Sequenced nucleotide and deduced amino acid sequences of wheat wall-associated kinase TaWAK5.The conserved EGF-like motif is marked by the boxed residues and located between the signal peptide(indicated by dotted line)and the transmembrane domain (underlined by a double line).The kinase domain(underlined by a single line)follows the transmembrane domain.The roman numerals mark the eleven subdomains(shaded residues)conserved in the plant serine/threonine protein kinase family.Arrowheads indicate the three kinase catalytic sites.
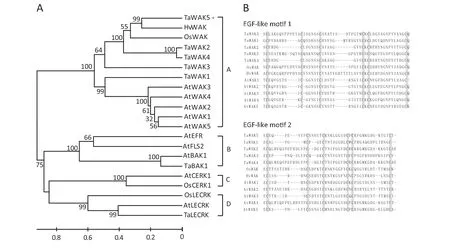
Fig.3-Phylogenetic analysis of the deduced amino acid sequences and comparison of EGF-like repeat sequences.A:Phylogenetic tree constructed by neighbor-joining algorithms with multiple RLK protein sequences.Bootstrapping was performed 1000 times to obtain support values for each branch.Four groups of RLK proteins,WAK,LRR-RLK,LysM-RLK,and Lectin-RLK were found and are represented,respectively,by the letters A,B,C,and D.The GenBank accession numbers of RLK protein sequences are shown in Table S2.The scale bar indicates sequence divergence.B:Amino acid sequence alignment of EGF-like motifs between TaWAK5 and WAK.Boxes in gray represent the six conserved cysteine residues.
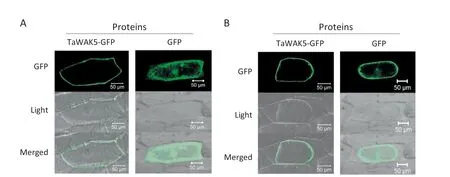
Fig.4-Plasma membrane localization of TaWAK5-GFP fusion protein.A:Subcellular localization of the fused 35S:TaWAK5-GFP in onion epidermal cells.The 35S:TaWAK5-GFP and 35S:GFP constructs,respectively,were introduced into onion epidermal cells by bombardment and expressed under control of the CaMV35S promoter.Bars = 50 μm.B:TaWAK-GFP fusion protein or GFP alone expressed under the control of the CaMV35S promoter in onion cells treated with 0.8 mol L-1 sucrose solution.Bars = 50 μm.
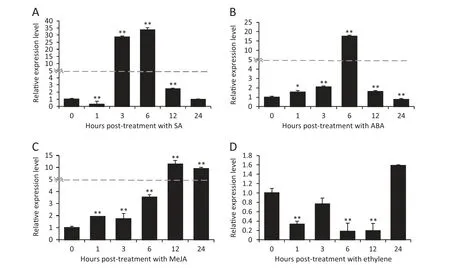
Fig.5-Expression patterns of TaWAK5 in wheat before(0 h)and after treatment with exogenous phyto-hormones SA(A),ABA(B),MeJA(C),and ET(D) for 0,1,3,6,12,and 24 h.Relative expression of TaWAK5 as fold change of the transcript over the control(0 hpt).Three biological replicates for each time point were averaged,with standard error of mean indicated.Asterisks indicate statistically significant variation calculated using Student's t-test(*P <0.05,**P <0.01).
To determine if TaWAK5 was responsive to various phytohormone (SA,ABA,ethylene,or MeJA) treatments,we used qRT-PCR to monitor the transcriptional patterns of TaWAK5 in wheat following treatment for 0,1,3,6,12,and 24 h with exogenous SA,ABA,ethylene or MeJA.As shown in Fig.5,the expression of TaWAK5 was significantly induced by SA,ABA,or MeJA treatment.The greatest induction effect was observed with the SA treatment.Upon SA treatment,the expression of TaWAK5 was induced at 1–12 h post-treatment (hpt),reached a peak at 6 hpt(about 33-fold over that of 0 hpt),and then decreased to a normal level by 24 hpt (Fig.5-A).The expression pattern of TaWAK5 after treatment with ABA was similar to that induced by SA;the induction reached a peak(about 17-fold over that of 0 hpt)at 6 hpt (Fig.5-B).Upon MeJA treatment,the transcript of TaWAK5 was induced from 1 to 24 hpt,and peaked at 12 hpt(more than 11-fold over that of 0 hpt) (Fig.5-C).Upon ethylene treatment,the transcriptional level of TaWAK5 decreased from 1 to 24 hpt(Fig.5-D).These results suggested that TaWAK5 may be responsive to the SA,ABA,and MeJA signals.
3.5.Promoter characterization for TaWAK5
Transcriptional regulation is important in mediating the responses of plants to external stimuli.To study which stimuli TaWAK5 may respond to,we analyzed cis-acting elements in the TaWAK5 promoter region using the PLACE database.Many important transcriptional motifs were identified in the promoter of TaWAK5,including a TATA box (at position 956),basal transcription,transcription factor binding site,hormone (ABA,SA,gibberellins,cytokinins,and auxin)responsiveness sites,and sites for responsiveness to elicitors and other processes(Table S3).
3.6.TaWAK5-silencing did not obviously impair wheat resistance to R.cerealis
To investigate whether TaWAK5 plays a critical role in wheat resistance response to R.cerealis,we used a BSMV-based VIGS technique to down-regulate TaWAK5 transcript levels in resistant wheat CI12633.At 10 days after the virus inoculation,the BSMV CP transcript was detected in plants inoculated with BSMV virus,but not in the mock plants,revealing successful virus infection.As expected,the TaWAK5 transcript levels were considerably reduced in CI12633 plants infected by BSMV:TaWAK5 (Fig.6-A),suggesting that the TaWAK5 transcripts were silenced in these CI12633 plants infected by BSMV:TaWAK5.In disease screening tests of non-infected plants,the 4th sheaths of mock-treated CI12633 and those infected with the BSMV:TaWAK5 virus were inoculated with mycelia of R.cerealis.The 4th sheaths of the susceptible cultivar Wenmai 6 were used as a positive control to show successful R.cerealis inoculation.At 2 weeks post R.cerealis inoculation,lesions with dark-brown margins (an early symptom of sharp eyespot disease) were observed on the 4th sheaths of the susceptible Wenmai 6,but not on the BSMV:TaWAK5-inoculated,BSMV:GFP-inoculated,or mock-treated CI12633 plants (Fig.6-B).The resistance continued to be present through more mature stages.No sharp eyespot symptoms were observed at 4th sheaths and stems of BSMV:TaWAK5-inoculated,BSMV:GFP-inoculated,and mock CI12633 plants,but obvious symptoms were present on the 4th sheaths and stems of Wenmai 6 plants.These results suggested that the silencing of TaWAK5 did not directly compromise wheat resistance to R.cerealis in CI12633.
4.Discussion
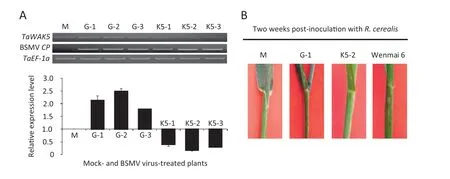
Fig.6-Effect of silencing TaWAK5 on the resistance response of CI12633 to the necrotrophic fungal pathogen R.cerealis,causal agent for sharp eyespot disease.A:Relative transcript levels of TaWAK5 and BSMV CP genes in the 4th leaves of the mock-treated plants,or those infected by BSMV:GFP and BSMV:TaWAK5 as evaluated by semi-quantitative RT-PCR using gene-specific primers.Amplification of the wheat TaEF-1a gene served as the internal control.B:Response of the 4th sheaths of the mock-treated,BSMV virus-inoculated CI12633 genotype,and positive control Wenmai 6 reactions to R.cerealis.The photographs were taken 2 weeks after R.cerealis inoculation.M:Mock plant;G-1:BSMV:GFP-1 plant;G-2:BSMV:-GFP-2 plant;G-3:BSMV:GFP-3 plant;K5-1:BSMV:TaWAK5-1 plant;K5-2:BSMV:TaWAK5-2 plant;K5-3:BSMV:TaWAK5-3 plant.
In this study,we isolated a novel wheat WAK gene,TaWAK5,from R.cerealis-resistant wheat CI12633,based on a cDNA transcript that was differentially expressed between resistant wheat genotype CI12633 and susceptible wheat cultivar Wenmai 6.qRT-PCR analysis revealed that the transcript abundance of TaWAK5 in wheat was rapidly increased by R.cerealis infection.Additionally,TaWAK5 in the R.cerealis-resistant lines was induced to higher levels than in R.cerealis-susceptible lines at 7 dpi with R.cerealis.These results suggested that TaWAK5 may be involved in wheat defense responses to R.cerealis infection.
Sequence analysis and phylogenetic analysis revealed that TaWAK5 was a member of the WAK sub-group of the RLK family in wheat.Several WAK genes have been shown to play important roles in regulating plant defense responses.WAK1 from Arabidopsis and OsWAK1 from rice were shown to enhance resistance to the pathogens B.cinerea and M.oryzae,respectively[5,11].TaWAK5 is a non-RD-type protein,as it has an HGD motif in its subdomain VIb.Out of 38 receptor kinases tested in plants,the six which fall into the non-RD class all function in disease resistance and act as PRRs,while the remaining 32 kinases are RD or alternative catalytic function(ACF) kinases and are involved in developmental processes[35],suggesting that all the non-RD RLKs seemed to participate in innate immunity.The TaWAK5 protein was localized to the plasma membrane in onion epidermal cells,consistent with the signal sequence and receptor responses.
Many WAKs have been shown to be involved in hormonal signals.Arabidopsis WAK1 is induced by both SA and the SA analog 2,2-dichloroisonicotinic acid(INA),and ectopic expression of the entire WAK1 or the kinase domain alone was shown to provide resistance to lethal SA levels[36].According to cDNA microarray analysis in Arabidopsis,AtWAK1 is induced by MeJA and ethylene [37].In this study,qRT-PCR analyses revealed that TaWAK5 could be induced by application of exogenous SA,ABA,and MeJA.Although an antagonistic interaction between SA-and JA-dependent signaling has been suggested[38–40],in some cases,SA does not inhibit JA biosynthesis and may even contribute to JA-mediated signaling pathway function [41].In Arabidopsis,concentrations of both SA and JA and the timing of initiation of SA and JA signaling are important for the outcome of the complex SA-JA signal interaction [42,43].ABA has been shown to interact with the SA-JA network.ABA has been suggested to affect JA biosynthesis and resistance against the JA-inducing,necrotrophic pathogen Pythium irregular[23,24],and to suppress SA-dependent disease resistance [44].Related to the role of phyto-hormones in WAK expression,the region upstream of the start codon(1000 bp)of TaWAK5 was analyzed in this study.The promoter region contained one ABRE-like motif (ACGTG),but no SA-,or JA-responsive elements (shown in Table S3).Several studies have suggested that modulation of gene expression is accomplished through the interaction of induced regulatory proteins and specific DNA regions [45–47].For instance,the induction of a dehydration-responsive gene,rd22,is mediated by ABA.MYC and MYB recognition sites in the rd22 promoter region function as cis-acting elements that interact specifically with AtMYC2 and AtMYB2; transgenic plants overexpressing AtMYC2 and/or AtMYB2 cDNAs have higher sensitivity to ABA[47].In this study,TaWAK5 promoter had five binding sites of an ABA-regulated protein,two of a SA-regulated protein,and one of a JA-regulated protein(Fig.S1),suggesting that TaWAK5 was also regulated possibly through SA-,ABA-,and MeJA-hormones.
In this study,VIGS,which has been an efficient tool for rapidly analyzing the functions of plant genes [48–51],was also used to evaluate the disease resistance role of TaWAK5.In wheat,infection with barley stripe mosaic virus (BSMV)constructs carrying a fragment of the resistance gene Lr21 caused conversion of incompatible interactions of wheat and leaf rust pathogen to compatible reactions after the gene silencing,whereas infection with a control construct or one that silences phytoene desaturase gene had no effect on resistance or susceptibility[33].Knocking down the transcript levels of three wheat RLK genes TaRLK-R1,TaRLK-R2,or TaRLKR3 individually or all together by VIGS and the suppression of TaHsp90.2 or TaHsp90.3 genes via VIGS compromised the wheat hypersensitive reaction to stripe rust fungus,suggesting that TaRLK-R1,TaRLK-R2,TaRLK-R3,and TaHsp90.2 and TaHsp90.3 were positive contributors in the wheat hypersensitive reaction to stripe rust fungus[50,51].These studies suggested that VIGS is an effective reverse genetic tool for investigating the functions of genes in wheat by knocking down the transcripts of target genes during the development of disease resistance.Conventional methods for gene functional analysis of plant genes,including transformation are not easily accomplished given wheat's large genome [52].Transformation is also time-consuming because the function of a target gene should be tested over multiple generations [53].In contrast to the conventional methods,the main advantage of VIGS is the generation of a rapid phenotype without the need for plant transformation[54].Moreover,the VIGS method provides a large-scale screening of genes for functional analysis; only a single plant is enough to follow phenotype with targeted silencing [55].In this study,the VIGS approach was utilized to investigate the function of TaWAK5 in wheat defense response to R.cerealis.Although the TaWAK5 transcript level was reduced in CI12633 plants infected by BSMV:TaWAK5,down-regulation of TaWAK5 in resistant CI12633 did not result in an obvious impairment of wheat resistance to R.cerealis.Plant defense is a complicated network in which some components and network sectors interact with each other in complex ways.The function of an individual component of a network can be compensated for by some other component.Therefore,functional characterization of disease resistance components by knockout of an individual component is difficult and multi-gene knock outs or gene × gene interactions need to be considered [56].In Arabidopsis,it has been suggested that there is functional redundancy between the WAKs,as induced silencing of individual AtWAK1or AtWAK2 using gene-specific antisense transcripts did not cause any phenotypic alteration[57].In this study,knocking down TaWAK5 expression did not cause the compromised resistance phenotype of the host CI12633 to R.cerealis.The reason might be that TaWAK5 is not the major gene controlling wheat defense response to R.cerealis,or that TaWAK5 is functionally redundant with other wheat WAK genes that help replace its functionalities when it is knocked out by VIGS experiments.
5.Conclusions
A wheat WAK gene,TaWAK5,was identified by microarray analysis of differentially expressed genes between R.cerealisresistant line CI12633 and susceptible cv.Wenmai 6 and characterized.TaWAK5 was rapidly induced by R.cerealis infection,and by exogenous SA,MeJA,or ABA application.The deduced TaWAK5 protein shares the structural characteristic of a wall-associated kinase,possessing two EGF-like repeats and a kinase catalytic domain.The TaWAK5 protein was localized to the plasma membranes in onion epidermal cells.Our results provide new insights into the WAK sub-group of the RLK family and will provide the foundation for further research into functions of WAKs in wheat.
This work was funded by the National Natural Science Foundation of China (31271799),and the National “Key Sci-Tech”program,China (2013ZX08002-001-004),and the China–Czech Government Science and Technology Cooperation Project (40–3 and LH12196).Editing assistance from Chinese Academy of Agricultural Sciences (CAAS) and from M.Blair is gratefully acknowledged.
Supplementary material
Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.cj.2014.04.010.
[1] M.Antolín-Llovera,M.K.Ried,A.Binder,M.Parniske,Receptor kinase signaling pathways in plant-microbe interactions,Annu.Rev.Phytopathol.50(2012) 451–473.
[2] D.Chinchilla,C.Zipfel,S.Robatzek,B.Kemmerling,T.Nurnberger,J.D.Jones,G.Felix,T.Boller,A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence,Nature 448 (2007) 497–500.
[3] L.Gómez-Gómez,T.Boller,FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis,Mol.Cell 5(2000) 1003–1011.
[4] T.Liu,Z.Liu,C.Song,Y.Hu,Z.Han,J.She,F.Fan,J.Wang,C.Jin,J.Chang,J.M.Zhou,J.Chai,Chitin-induced dimerization activates a plant immune receptor,Science 336 (2012)1160–1164.
[5] A.Brutus,F.Sicilia,A.Macone,F.Cervone,G.De Lorenzo,A domain swap approach reveals a role of the plant wall-associated kinase 1(WAK1)as a receptor of oligogalacturonides,Proc.Natl.Acad.Sci.U.S.A.107 (2010)9452–9457.
[6] B.D.Kohorn,Plasma membrane-cell wall contacts,Plant Physiol.124 (2000) 31–38.
[7] Z.H.He,M.Fujiki,B.D.Kohorn,A cell wall-associated,receptor-like protein kinase,J.Biol.Chem.271 (1996)19789–19793.
[8] B.D.Kohorn,S.Johansen,A.Shishido,T.Todorova,R.Martinez,E.Defeo,P.Obregon,Pectin activation of MAP kinase and gene expression is WAK2 dependent,Plant J.60(2009)97–982.
[9] V.Kanneganti,A.K.Gupta,Wall associated kinases from plants–an overview,Physiol.Mol.Biol.Plant 14(2008)109–118.
[10] C.Denoux,R.Galletti,N.Mammarella,S.Gopalan,D.Werck,G.De Lorenzo,S.Ferrari,F.M.Ausubel,J.Dewdney,Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings,Mol.Plant 1(2008) 423–445.
[11] H.Li,S.Y.Zhou,W.S.Zhao,S.C.Su,Y.L.Peng,A novel wall-associated receptor-like protein kinase gene,OsWAK1,plays important roles in rice blast disease resistance,Plant Mol.Biol.69 (2009) 337–346.
[12] Y.Liu,D.Liu,H.Zhang,H.Gao,X.Guo,X.Fu,A.Zhang,Isolation and characterisation of six putative wheat cell wall-associated kinases,Funct.Plant Biol.33(2006) 811–821.
[13] B.Mauch-Mani,F.Mauch,The role of abscisic acid in plant–pathogen interactions,Curr.Opin.Plant Biol.8(2005)409–414.
[14] L.C.Van Loon,B.P.Geraats,H.J.Linthorst,Ethylene as a modulator of disease resistance in plants,Trends Plant Sci.11(2006) 184–191.
[15] M.Pozo,L.C.Loon,C.J.Pieterse,Jasmonates–signals in plant-microbe interactions,J.Plant Growth Regul.23(2004)211–222.
[16] G.Loake,M.Grant,Salicylic acid in plant defence–the players and protagonists,Curr.Opin.Plant Biol.10(2007)466–472.
[17] B.Vernooij,S.Uknes,E.Ward,J.Ryals,Salicylic acid as a signal molecule in plant–pathogen interactions,Curr.Opin.Cell Biol.6(1994) 275–279.
[18] R.A.Creelman,J.E.Mullet,Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress,Proc.Natl.Acad.Sci.U.S.A.92 (1995) 4114–4119.
[19] J.R.Ecker,The ethylene signal transduction pathway in plants,Science 268 (1995) 667–675.
[20] M.A.Lopez,G.Bannenberg,C.Castresana,Controlling hormone signaling is a plant and pathogen challenge for growth and survival,Curr.Opin.Plant Biol.11 (2008) 420–427.
[21] J.A.Ryals,U.H.Neuenschwander,M.G.Willits,A.Molina,H.Y.Steiner,M.D.Hunt,Systemic acquired resistance,Plant Cell 8(1996) 1809–1819.
[22] K.E.Hammond-Kosack,J.D.Jones,Resistance genedependent plant defense responses,Plant Cell 8 (1996)1773–1791.
[23] V.Flors,J.Ton,R.van Doorn,G.Jakab,P.Garcia-Agustin,B.Mauch-Mani,Interplay between JA,SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola,Plant J.54(2008) 81–92.
[24] B.A.Adie,J.Pérez-Pérez,M.M.Pérez-Pérez,M.Godoy,J.J.Sánchez-Serrano,E.A.Schmelz,R.Solano,ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis,Plant Cell 19 (2007) 1665–1681.
[25] J.P.Anderson,E.Badruzsaufari,P.M.Schenk,J.M.Manners,O.J.Desmond,C.Ehlert,D.J.Maclean,P.R.Ebert,K.Kazan,Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis,Plant Cell 16(2004) 3460–3479.
[26] M.S.Hamada,Y.Yin,H.Chen,Z.Ma,The escalating threat of Rhizoctonia cerealis,the causal agent of sharp eyespot in wheat,Pest Manag.Sci.67(2011) 1411–1419.
[27] L.Chen,Z.Zhang,H.Liang,H.Liu,L.Du,H.Xu,Z.Xin,Overexpression of TiERF1 enhances resistance to sharp eyespot in transgenic wheat,J.Exp.Bot.59(2008) 4195–4204.
[28] K.Yang,W.Rong,L.Qi,J.Li,X.Wei,Z.Zhang,Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis,Sci.Rep.3(2013)3021,http://dx.doi.org/10.1038/srep03021.
[29] Z.Zhang,X.Liu,X.Wang,M.Zhou,X.Zhou,X.Ye,X.Wei,An R2R3 MYB transcription factor in wheat,TaPIMP1,mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense-and stress-related genes,New Phytol.196 (2012) 1155–1170.
[30] Z.Zhang,W.Yao,N.Dong,H.Liang,H.Liu,R.Huang,A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments,J.Exp.Bot.58 (2007) 2993–3003.
[31] I.Lang-Pauluzzi,B.Gunning,A plasmolytic cycle:the fate of cytoskeletal elements,Protoplasma 212 (2000) 174–185.
[32] S.Holzberg,P.Brosio,C.Gross,G.P.Pogue,Barley stripe mosaic virus-induced gene silencing in a monocot plant,Plant J.30(2002) 315–327.
[33] S.R.Scofield,L.Huang,A.S.Brandt,B.S.Gill,Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway,Plant Physiol.138 (2005) 2165–2173.
[34] K.J.Livak,T.D.Schmittgen,Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod,Methods 25(2001) 402–408.
[35] C.Dardick,P.Ronald,Plant and animal pathogen recognition receptors signal through non-RD kinases,PLoS Pathog.2(2006) e2.
[36] Z.H.He,D.He,B.D.Kohorn,Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response,Plant J.14(1998)55–63.
[37] P.M.Schenk,K.Kazan,I.Wilson,J.P.Anderson,T.Richmond,S.C.Somerville,J.M.Manners,Coordinated plant defense responses in Arabidopsis revealed by microarray analysis,Proc.Natl.Acad.Sci.U.S.A.97(2000) 11655–11660.
[38] C.A.Preston,C.Lewandowski,A.J.Enyedi,I.T.Baldwin,Tobacco mosaic virus inoculation inhibits wound-induced jasmonic acid-mediated responses within but not between plants,Planta 209 (1999) 87–95.
[39] J.Cui,A.K.Bahrami,E.G.Pringle,G.Hernandez-Guzman,C.L.Bender,N.E.Pierce,F.M.Ausubel,Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores,Proc.Natl.Acad.Sci.U.S.A.102(2005)1791–1796.
[40] A.P.Kloek,M.L.Verbsky,S.B.Sharma,J.E.Schoelz,J.Vogel,D.F.Klessig,B.N.Kunkel,Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive(coi1) mutation occurs through two distinct mechanisms,Plant J.26 (2001) 509–522.
[41] J.?a?niewska,V.K.Macioszek,C.B.Lawrence,A.K.Kononowicz,Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens,Acta Physiol.Plant.32(2010) 1–10.
[42] L.A.Mur,P.Kenton,R.Atzorn,O.Miersch,C.Wasternack,The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy,antagonism,and oxidative stress leading to cell death,Plant Physiol.140 (2006) 249–262.
[43] A.Koornneef,A.Leon-Reyes,T.Ritsema,A.Verhage,F.C.Den Otter,L.C.Van Loon,C.M.Pieterse,Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation,Plant Physiol.147 (2008)1358–1368.
[44] M.Yasuda,A.Ishikawa,Y.Jikumaru,M.Seki,T.Umezawa,T.Asami,A.Maruyama-Nakashita,T.Kudo,K.Shinozaki,S.Yoshida,H.Nakashita,Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis,Plant Cell 20(2008)1678–1692.
[45] S.H.Spoel,A.Koornneef,S.M.Claessens,J.P.Korzelius,J.A.Van Pelt,M.J.Mueller,A.J.Buchala,J.P.Metraux,R.Brown,K.Kazan,L.C.Van Loon,X.Dong,C.M.Pieterse,NPR1 modulates cross-talk between salicylate-and jasmonate-dependent defense pathways through a novel function in the cytosol,Plant Cell 15(2003) 760–770.
[46] S.Nakamura,T.J.Lynch,R.R.Finkelstein,Physical interactions between ABA response loci of Arabidopsis,Plant J.26(2001) 627–635.
[47] H.Abe,T.Urao,T.Ito,M.Seki,K.Shinozaki,K.Yamaguchi-Shinozaki,Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling,Plant Cell 15 (2003) 63–78.
[48] S.H.Gabriels,J.H.Vossen,S.K.Ekengren,G.van Ooijen,A.M.Abd-El-Haliem,G.C.van den Berg,D.Y.Rainey,G.B.Martin,F.L.Takken,P.J.de Wit,M.H.Joosten,An NB-LRR protein required for HR signalling mediated by both extra-and intracellular resistance proteins,Plant J.50(2007) 14–28.
[49] I.Hein,M.Barciszewska-Pacak,K.Hrubikova,S.Williamson,M.Dinesen,I.E.Soenderby,S.Sundar,A.Jarmolowski,K.Shirasu,C.Lacomme,Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley,Plant Physiol.138(2005)2155–2164.
[50] G.F.Wang,X.Wei,R.Fan,H.Zhou,X.Wang,C.Yu,L.Dong,Z.Dong,Z.Kang,H.Ling,Q.H.Shen,D.Wang,X.Zhang,Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90(Hsp90):functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance,New Phytol.191(2011)418–431.
[51] H.Zhou,S.Li,Z.Deng,X.Wang,T.Chen,J.Zhang,S.Chen,H.Ling,A.Zhang,D.Wang,X.Zhang,Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection,Plant J.52(2007) 420–434.
[52] H.D.Jones,Wheat transformation: current technology and applications to grain development and composition,J.Cereal Sci.41(2005) 137–147.
[53] C.A.Newell,Plant transformation technology,Mol.Biotechnol.16(2000) 53–65.
[54] T.M.Burch-Smith,J.C.Anderson,G.B.Martin,S.P.Dinesh-Kumar,Applications and advantages of virus-induced gene silencing for gene function studies in plants,Plant J.39(2004) 734–746.
[55] A.Becker,M.Lange,VIGS–genomics goes functional,Trends Plant Sci.15(2010) 1–4.
[56] M.Sato,K.Tsuda,L.Wang,J.Coller,Y.Watanabe,J.Glazebrook,F.Katagiri,Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling,PLoS Pathog.6(2010)e1001011.
[57] T.A.Wagner,B.D.Kohorn,Wall-associated kinases are expressed throughout plant development and are required for cell expansion,Plant Cell 13(2001) 303–318.
- The Crop Journal的其它文章
- Productivity,quality and soil health as influenced by lime in ricebean cultivars in foothills of northeastern India
- Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava
- Identification of QTL for adult-plant resistance to powdery mildew in Chinese wheat landrace Pingyuan 50
- Effect of subsoil tillage depth on nutrient accumulation,root distribution,and grain yield in spring maize
- Exp2 polymorphisms associated with variation for fiber quality properties in cotton(Gossypium spp.)
- The impacts of conservation agriculture on crop yield in China depend on specific practices,crops and cropping regions

