Prediction of m6A Methylation Sites in Mammalian Tissues Based on a Double-layer BiGRU Network*
LⅠ Hui-Min, CHEN Peng-Hui, TANG Yi, XU Quan-Feng, HU Meng, WANG Yu
(School of Mathematics and Computer Science, Yunnan Minzu University, Kunming 650504, China)
Abstract Objective N6-methyladenosine (m6A) is the most common and abundant chemical modification in RNA and plays an important role in many biological processes.Several computational methods have been developed to predict m6A methylation sites.However, these methods lack robustness when targeting different species or different tissues.To improve the robustness of the prediction performance of m6A methylation sites in different tissues, this paper proposed a double-layer bidirectional gated recurrent unit (BiGRU) network model that combines reverse sequence information to extract higher-level features of the data.Methods Some representative mammalian tissue m6A methylation site datasets were selected as the training datasets.Based on a BiGRU, a doublelayer BiGRU network was constructed by collocation of the model network, the model structure, the number of layers and the optimizer.Results The model was applied to predict m6A methylation sites in 11 human, mouse and rat tissues, and the prediction performance was compared with that of other methods using the same tissues.The results demonstrated that the average area under the receiver operating characteristic curve (AUC) predicted by the proposed model reached 93.72%, equaling that of the best prediction method at present.The values of accuracy (ACC), sensitivity (SN), specificity (SP) and Matthews correlation coefficient(MCC) were 90.07%, 90.30%, 89.84% and 80.17%, respectively, which were higher than those of the current methods for predicting m6A methylation sites.Conclusion Compared with that of existing research methods, the prediction accuracy of the double-layer BiGRU network was the highest for identifying m6A methylation sites in the 11 tissues, indicating that the method proposed in this study has an excellent generalizability.
Key words N6-methylated adenosine site, bidirectional gated recurrent unit, base sequence, deep learning
RNA methylation is a new field of epigenetic regulation[1-2].m6A methylation is the most common and abundant chemical modification in RNA,accounting for approximately 80% of RNA methylation modifications[3-4].Ⅰt plays an important role in regulating RNA maturation, cleavage,transport, degradation and translation[5-7].Many enzymes involved in m6A methylation can be modified at the m6A methylation sites[8].Therefore,the accurate identification of m6A methylation sites from RNA sequences is crucial for understanding the biological function of RNA methylation modifications.
Early detection methods of m6A methylation sites were mainly based on biological experiments,such as two-dimensional cellulose thin chromatography, high-performance liquid chromatography and mass spectrometry[9].However,due to the limitations of experimental conditions,these methods generally have many problems, such as being time-consuming, having a high cost and having a small detection scale.The emergence of highthroughput sequencing technology has provided strong technical support for methylation research[10-12]and generated a large amount of m6A methylation site data, which has led to the identification of m6A methylation sites from biological experiments and computational research.Using high-throughput experimental data and traditional machine learning methods, some models for predicting m6A methylation sites have been developed.Examples include iRNA-Methyl[13] and pRNAm-PC predictors[14] based on base resolution technology,SRAMP[15] based on random forest (RF), and models based on support vector machine (SVM), such as RAM-NPPS[16], M6APred-EL[17], iMethyl-STTNC[18]and iRNA(m6A) -PseDNC[19].Traditional machine learning algorithms require more professional knowledge to manually extract features from datasets,reduce the features’ dimensions and transfer the best features to the model.The process of feature extraction is very complicated.Ⅰn recent years, many researchers have proposed m6A methylation site prediction algorithms based on deep learning algorithms[20], which can automatically obtain highlevel features based on sample datasets, and developed methods for cross-species prediction of m6A methylation sites.
Researchers have mainly targeted m6A methylation sites in different species, such asArabidopsis thaliana,Saccharomyces cerevisiae,Mus musculus(mouse),Rattus norvegicus(rat) andHomo sapiens(human), to make macroscopic predictions.However, less attention has been given to m6A methylation sites in more microscopic biological tissues.As an example, the expression levels of m6A methylation were found to be different between diseased and unaffected tissues[21-23], while few methods have predicted m6A methylation sites in different tissues.Ⅰn recent years, some researchers have refined m6A methylation site prediction to tissue sites[24-29].For example, Daoet al.[26] and Wanget al.[27] proposed iRNA-m6A[26] and M6A-BiNP[27],respectively, which mainly rely on SVMs, to predict m6A methylation sites in 11 tissues of 3 species(human, mouse and rat).Liuet al.[28] developed im6ATS-CNN based on a single-layer convolutional neural network to further improve the values of the area under the receiver operating characteristic (ROC)curve (AUC).Zhanget al.[29] developed a tool named DNN-m6A using deep neural networks to identify m6A methylation sites in multiple human, mouse and rat tissues and showed an excellent generalizability.Although there have been an increasing number of computational methods for m6A methylation site prediction and some progress has been made in the prediction of tissue m6A methylation sites, the following problems remain.(1) The predicted regions are generally not sufficiently refined.Only a few algorithms, such as iRNA-m6A, im6A-TS-CNN,DNN-m6A and M6A-BiNP, subdivide the predicted regions into various tissues.(2) Most algorithms have low prediction accuracy in some tissues, and the prediction accuracy is generally below 80%.
m6A methylation site prediction is based on nucleotide sequences, in which nucleotides are associated with each other.As one of the classical deep learning algorithms, a recurrent neural network(RNN) has excellent performance in processing sequence data.Ⅰn particular, a bidirectional RNN can combine the reverse characteristics of sequences.Therefore, based on a bidirectional gating recurrent unit (BiGRU), which is a variant of the bidirectional RNN, and selected representative mammalian tissue m6A methylation site datasets as training data, we constructed a double-layer BiGRU network.m6A methylation sites in 11 mammalian tissues were predicted using our method, and the predicted results show that the proposed method is superior to existing methods.
1 Materials and methods
1.1 Materials
The datasets used in this research were from the m6A methylation site benchmark datasets constructed by Daoet al.[26] and downloaded from their paper.The datasets contain m6A methylation sites in 11 mammalian tissues from 3 species: human (brain,liver and kidney), mouse (brain, liver, heart, testis and kidney) and rat (brain, liver and kidney).Each dataset of the above 11 tissues contained two parts: a training dataset used to train the model and an independent test dataset used to test the performance of the model.Ⅰn each training dataset and independent test dataset,the same sequence numbers of positive samples (m6A sites) and negative samples (non-m6A sites) were included.The length of each sequence in the positive and negative samples was 41 nt, with adenine (A) in the center of a sequence.The detailed sample sizes in the datasets are shown in Table 1[26].Ⅰt was observed that the sample size of human brain tissue was at a medium level in all datasets, so we used it to debug the model parameters.
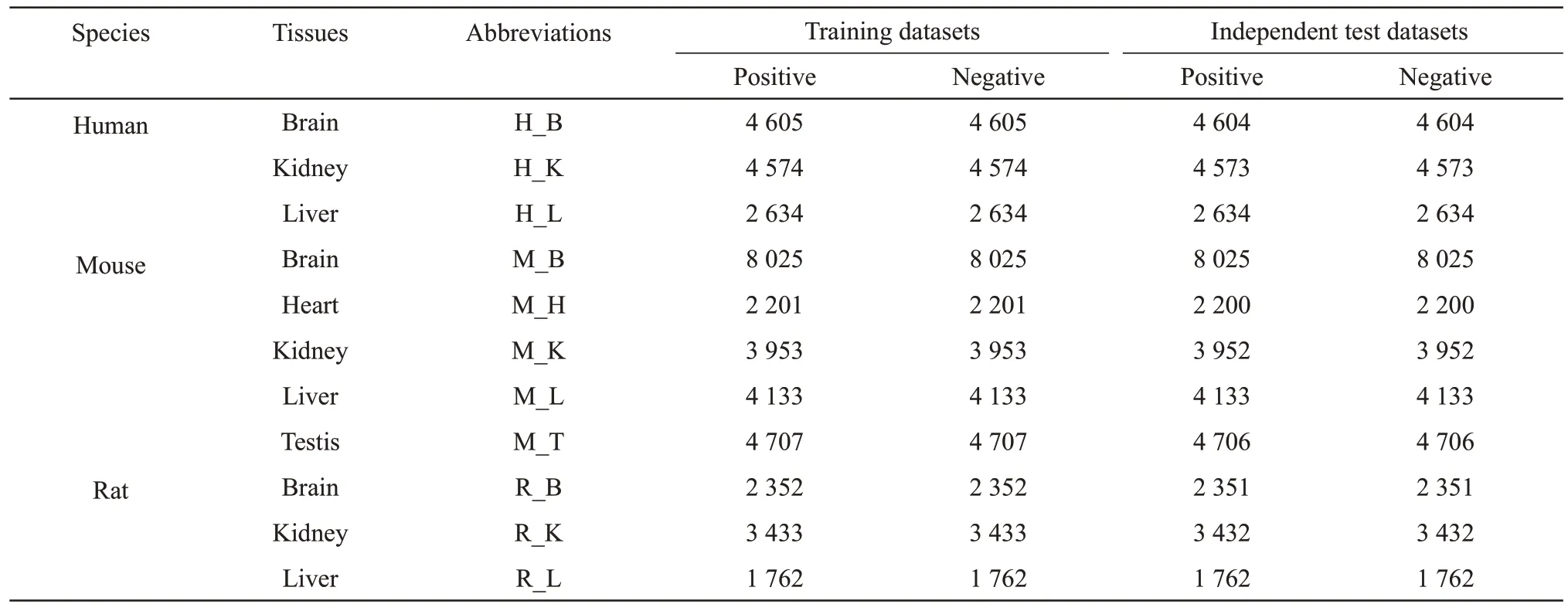
Table 1 Benchmark datasets of m6A methylation sites
To make the original data acceptable to the model, the sample RNA sequences were processed by one-hot encoding.Let A=(1, 0, 0, 0)T, U=(0, 1, 0, 0)T,C=(0, 0, 1, 0)Tand G=(0, 0, 0, 1)T; then, each RNA sequence can be represented as a numerical matrix that contains only 1s and 0s with 4 rows and 41 columns.
1.2 Methods
1.2.1 Construction of the double-layer BiGRU prediction model
The core model of our method is a gated recurrent unit (GRU).The GRU model can better and more automatically capture the dependence relationship in a sequence[30], and it is suitable for predicting m6A methylation sites in a sequence.The GRU controls the flow of information by resetting and updating the gate, which can effectively solve the gradient disappearance problem in RNNs, and the model has fewer parameters and is more concise.The network structure is shown in Figure 1.The model mainly includes two bidirectional GRU (BiGRU)layers.The first BiGRU layer (BiGRU_layer1)processes the data transformed by the input layer to obtain the initially extracted feature vector, and the second BiGRU layer (BiGRU_layer2) further extracts the features obtained from the previous layer.Hence,the function of BiGRU_layer2 is to capture more advanced information and make the model obtain more useful data characteristics.
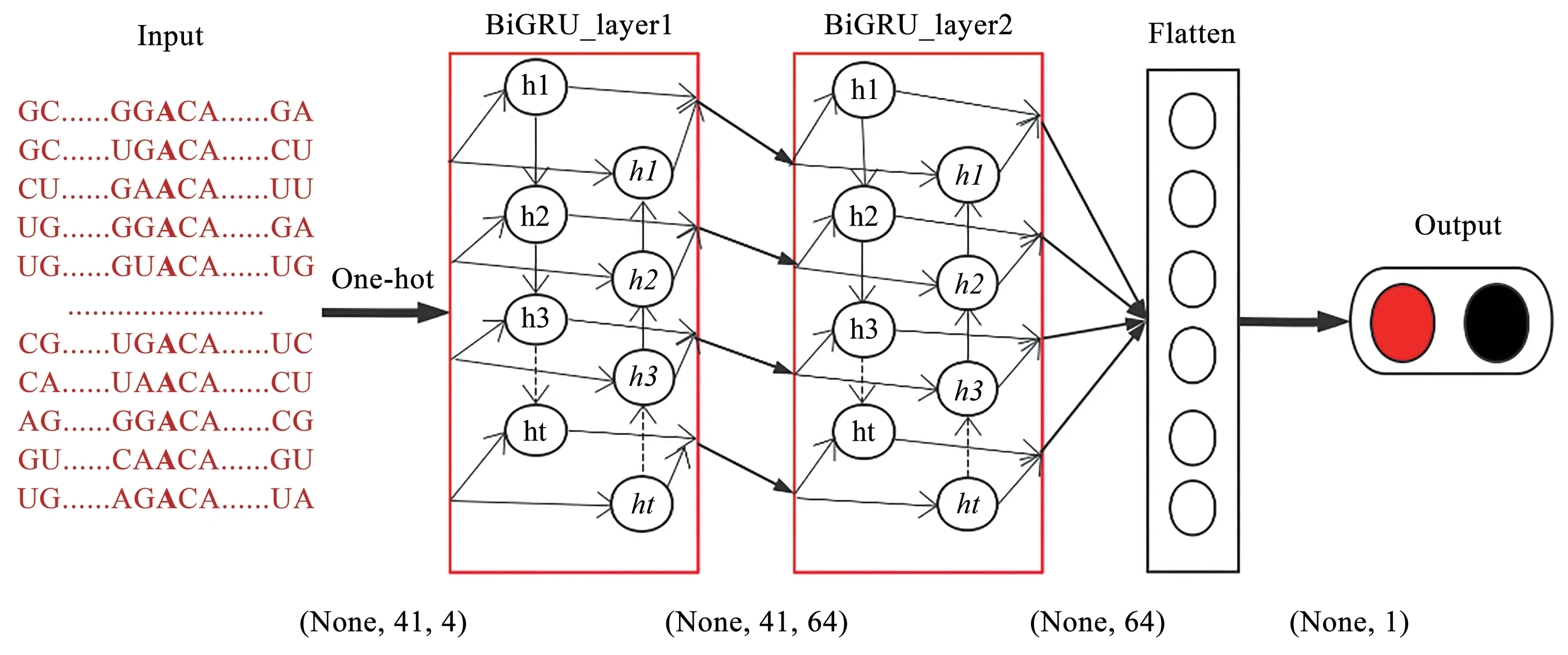
Fig.1 Model structure diagram of bidirectional gated recurrent unit (BiGRU) network
1.2.2 Detailed algorithm procedure
(1) The nucleotide sequence data were converted into the form of one-hot encoding, and each sample RNA sequence with dimension (4, 41) was fed into the model.
(2) Two BiGRU layers were added using the Python library Keras.Since the previous data input dimension is (4, 41), we set ‘input_shape’ to (4, 41)in BiGRU_layer1 and the number of neurons in both BiGRU layers to 32.
(3) The results of BiGRU_layer1 and BiGRU_layer2 were passed to the ‘Flatten layer’, and a highdimensional data input vector was converted into a one-dimensional output vector.
(4) Ⅰn the output layer, ‘sigmoid’ was selected as the activation function, and its formula is given in Equation (1):
Ⅰn Equation (1),xis the output value of the previous flattened layer processing, and the range off(x) is [0, 1], which is similar to the probability value.The prediction was positive samples (m6A sites) whenf(x) >0.5, and the prediction was negative samples(non-m6A sites) whenf(x) ≤0.5.
1.2.3 Design of model and parameters
The prediction of m6A methylation sites in this work was treated as a classification problem;therefore, the loss function of the model was the binary cross-entropy function, as shown in Equation (2):
whereyirepresents the label of the samplei, the positive class is 1, and the negative class is 0;pirepresents the probability of the sampleibeing predicted to be a positive class.
Ⅰn the model, the epoch number was set to 150,the batch size was set to 32, and the ‘Adam’optimizer was used.When the initial learning rate was not applicable, the accuracy of the model did not improve after a certain number of epoch iterations.Therefore, the callback function‘ReduceLROnPlateau’ was added to optimize the learning rate.The monitoring variable in the callback function was ‘Val_loss’, and ‘patience’ was set to 20.That is, when the model loss value did not decrease after 20 epochs, the mechanism of learning rate reduction in the callback function was triggered.A ‘factor’ value of 0.1 was used to reduce the learning rate in the training process, thus improving the accuracy of the model.Because the callback function ‘ReduceLROnPlateau’ needs several iterations to optimize the learning rate to make the model reach the best state, to achieve higher accuracy and accelerate the model training, the‘EarlyStopping’ strategy was added to stop the model training in advance.The monitoring variable in‘EarlyStopping’ was ‘val_binary_accuracy’, and‘patience’ was set to 30.Training was stopped when the accuracy of the model after 30 epochs had not changed.Ⅰn this situation, ‘EarlyStopping’ will not be triggered early, so the model can be fully trained while avoiding overfitting.
A 10-fold cross-validation test was used in the experiments.That is, the datasets were randomly divided into 10 subsets.Ⅰn turn, 8 of them were used as a training set, 1 of them was used as a validation set, and the remaining one was used as a test set.Ⅰn each experiment, a correct rate was obtained, and finally, the average correct rate of the 10 results was used as the estimation of the accuracy of the model or algorithm.
1.2.4 Evaluation metrics
Four classical evaluation metrics, including sensitivity (SN), specificity (SP), accuracy (ACC), and Matthews correlation coefficient (MCC) [31], were implemented to assess the performance of the model.The corresponding metrics can be expressed as formulas (3)-(6):
whereTP,FP,FNandTNrepresent the number of correctly predicted positive samples, incorrectly predicted negative samples, incorrectly predicted positive samples and correctly predicted negative samples[32], respectively.TheAUC[33] was also introduced to evaluate the overall performance of the model[34].The value range ofAUCis [0, 1], and theAUCis positively correlated with the prediction performance.The larger theAUCvalue is, the better the overall performance of the predictor.Ⅰn the aspect of code implementation, the encapsulation function for the prediction model of Chenet al.[35-36]was used.
2 Results and discussion
Our method was compared with several existing methods[26-29].These methods include iRNA-m6A and M6A-BiNP based on SVM, im6A-TS-CNN based on a single-layer convolutional neural network, and DNN-m6A based on a deep neural network.At present, these 4 methods have achieved good performance in m6A methylation site prediction in mammalian tissues.Since M6A-BiNP had the better comprehensive performance among the methods, we only reported comparison results with M6A-BiNP in each tissue.
2.1 Prediction results on human tissues
For the human independent test datasets (Table 2), our method showed the bestACCvalues (H_B:87.48%, H_K: 90.87% and H_L: 90.72%) andMCCvalues (H_B: 74.96%, H_K: 81.76% and H_L:81.47%).TheAUCvalue of our method for H_L showed the best performance simultaneously with that of M6A-BiNP (our method: 94.04% and M6A-BiNP:94.80%); for H_B and H_K, our method had the best performance (H_B: 91.97% and H_K: 94.51%).Although the modelSNvalues for H_K and H_L were lower than those of M6A-BiNP (H_K: 96.4% and H_L: 92%), both were higher than 90% (H_K:90.94% and H_L: 90.77%).TheSPvalue of our method for H_B was lower than that of M6A-BiNP(95.40%), but it reached 87.46%.Compared with the test results of the other methods, the performance of our method was stable for different tissues.For example, although M6A-BiNP achieved a betterSPvalue (95.4%) for H_B, itsSPvalue for H_K was only 40%.One of the greatest advantages of our method is its universality.Ⅰt had highACCandAUCvalues for all 3 human tissues, but the other methods did not have such high performance.For example,although theACCvalue of M6A-BiNP for H_L was 86.20%,ACCvalues of only 76.70% and 68.20%were obtained for H_B and H_K, respectively.
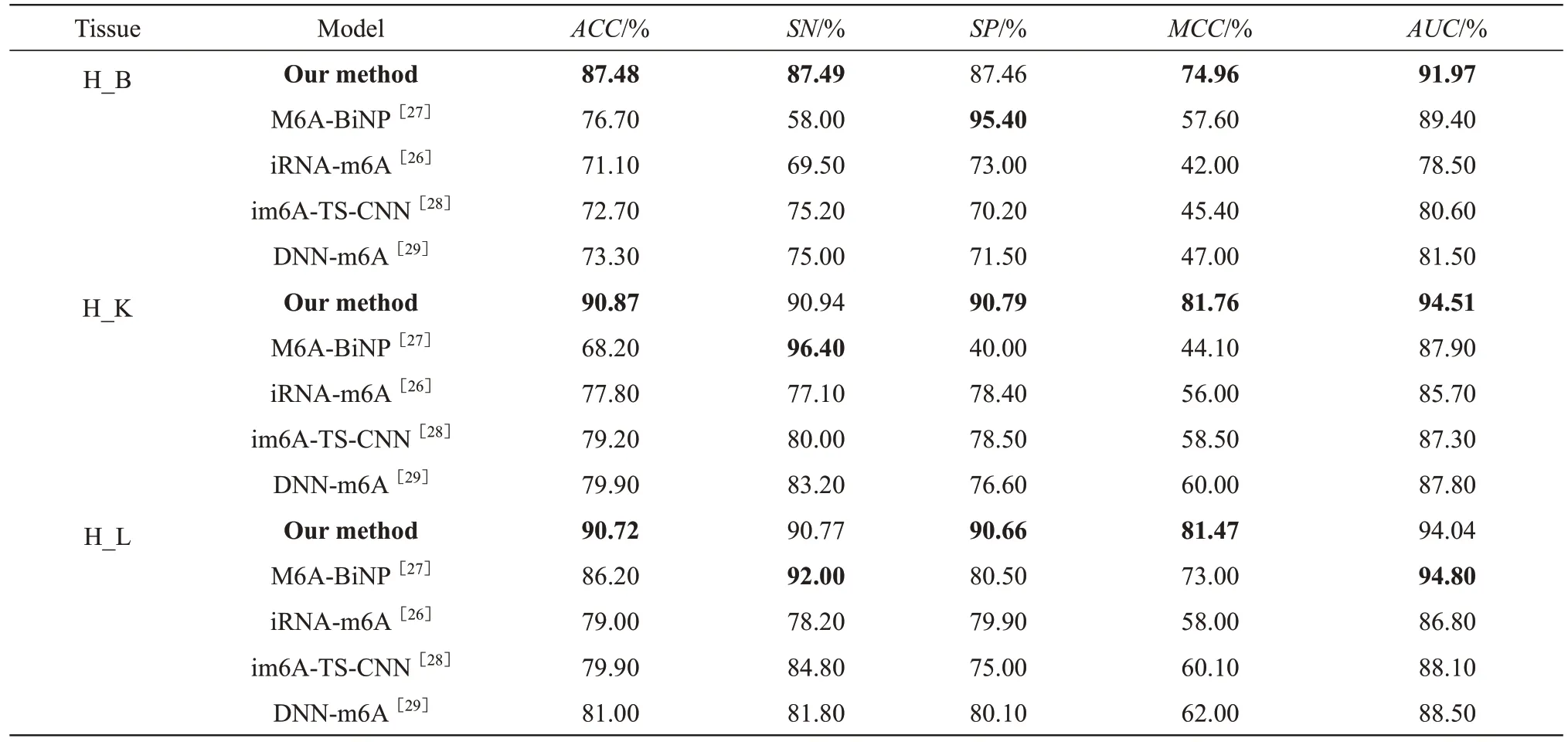
Table 2 Evaluation metrics on human independent test datasets
2.2 Prediction results on mouse tissues
For the mouse independent test datasets (Table 3), our method showed a better prediction effect.Except for the lowerSPandAUCvalues for M_H and M_L than those of M6A-BiNP, our method achieved the bestACC,SN,SP,MCCandAUCvalues for the other tissues.At the same time, compared with those of M6A-BiNP, the evaluation metrics of our method for different tissues had smaller fluctuation ranges.For example, for 5 mouse tissues, theSPvalues ofM6A-BiNP ranged from 67.40% to 99.60%, and those of our method ranged from 88.09% to 90.96%.TheACCvalues of M6A-BiNP and our method ranged from 75.60% to 85.10% and 87.81% to 91.18%,respectively.On the one hand, these results demonstrate that our method has higher accuracy; on the other hand, it has smaller fluctuation in terms of each evaluation criterion.Therefore, it was concluded that the prediction performance of our method was more stable and more universal for mouse tissue m6A methylation site prediction.
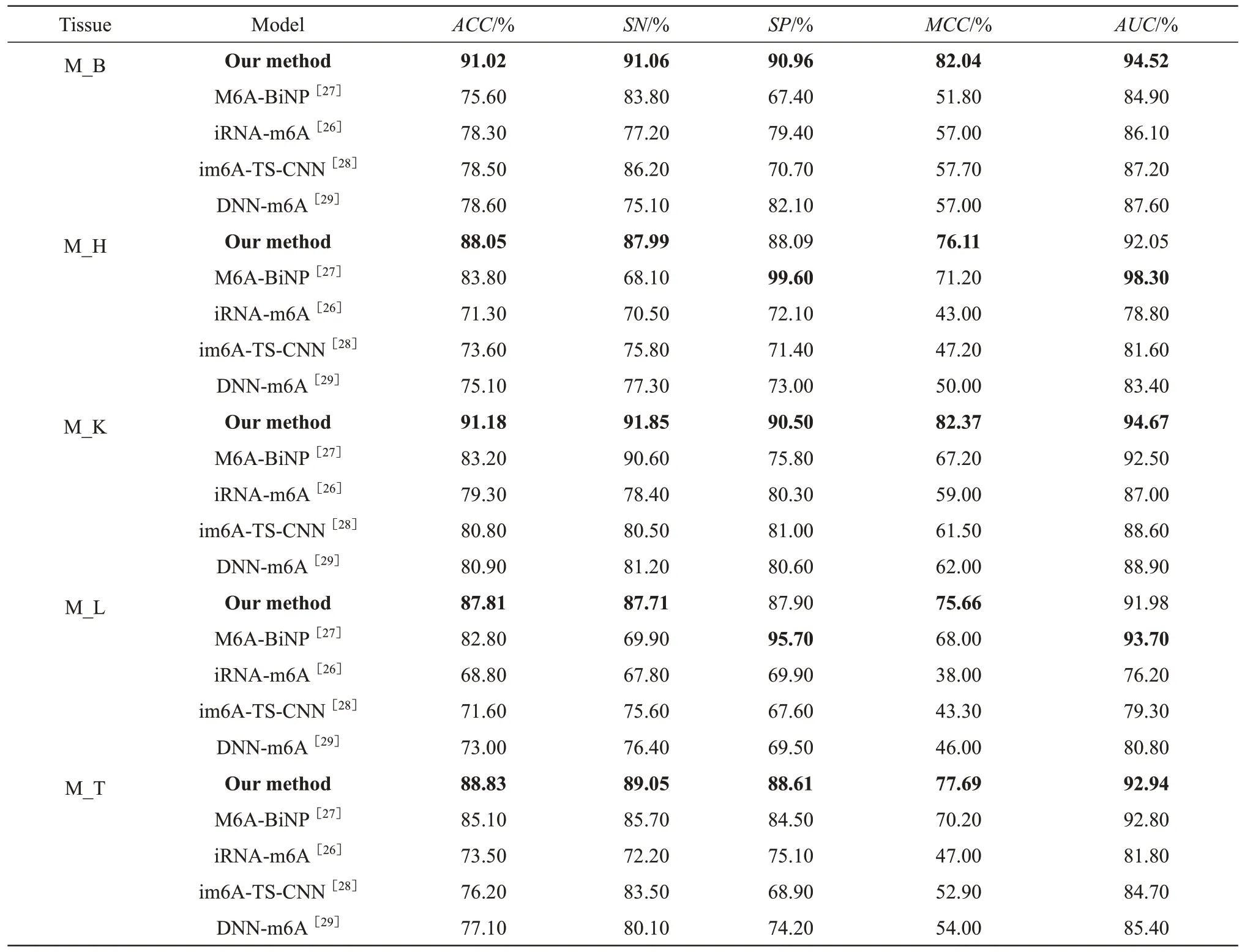
Table 3 Evaluation metrics on mouse independent test datasets
2.3 Prediction results on rat tissues
The model prediction results of m6A methylation sites on three independent test datasets of rat tissues were compared with those of other methods (Table 4).Our method achieved the best predictionAUCvalue for R_K tissue, and although theAUCvalues were lower than those of M6A-BiNP for R_B and R_L tissues, they exceeded 92%.However, theACCvalues of our method were highest for 3 tissues.This indicated that our method could improve the prediction accuracy of m6A methylation sites on rat datasets.Moreover, similar to those for the mouse tissues, the 9 prediction results of our method for 3 different rat tissues also had smaller fluctuation.For example, in the mentioned 3 tissues, theSPvalues of M6A-BiNP ranged from 57.50% to 90.78%, and those of our method ranged from 89.22% to 0.78%.TheACCvalues of M6A-BiNP and our method ranged 77.10%-88.70% and 89.51%-91.47%, respectively.This further demonstrates the universality of the proposed method.
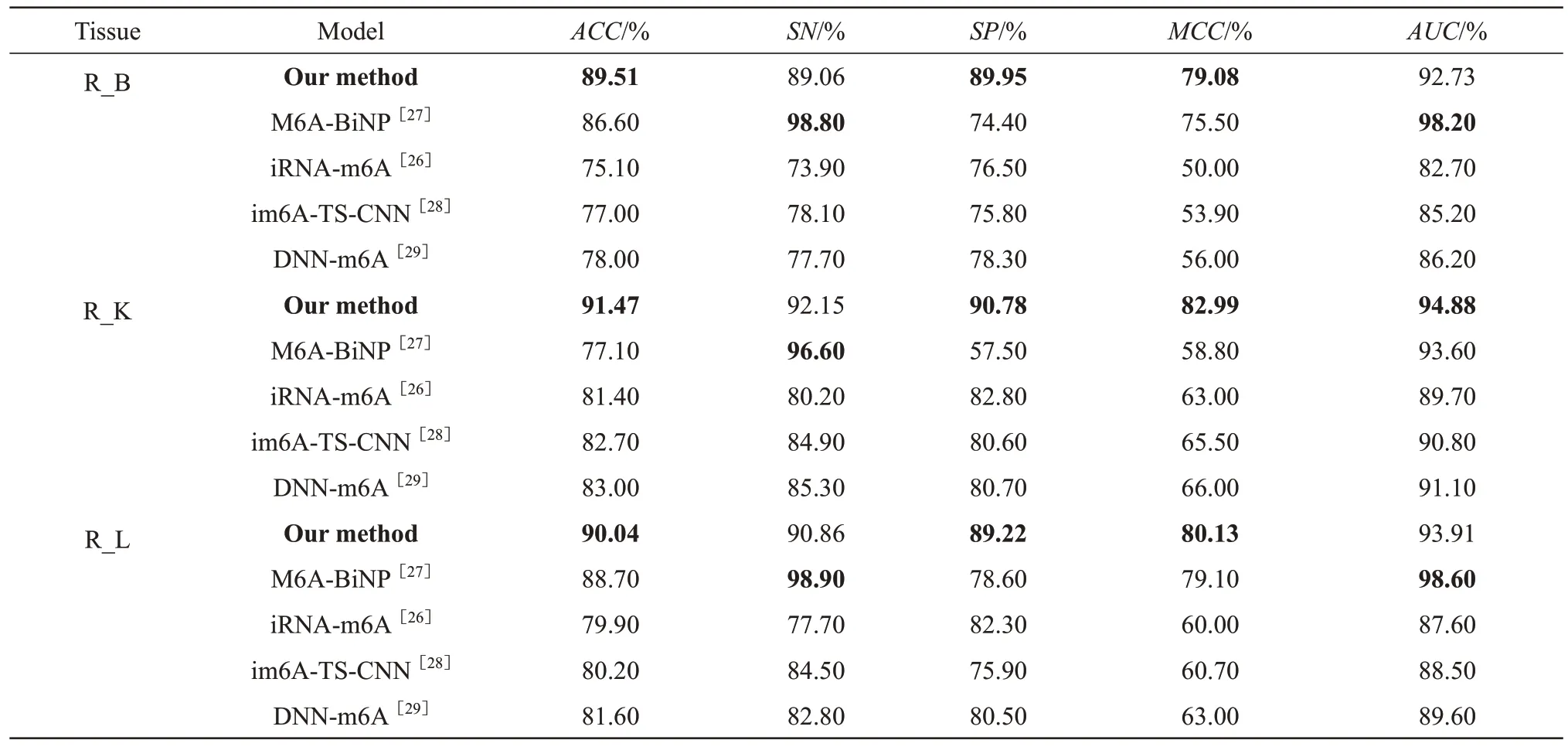
Table 4 Evaluation metrics on rat independent datasets
2.4 Model summary
To illustrate the overall comparison between our method and other state-of-the-art methods, the prediction results of 11 tissues were averaged.Ⅰt can be seen from the results of the training datasets(Figure 2a) that theAUCvalue of our method was almost equal to that of M6A-BiNP and higher thanthat of the other methods.The values ofACC,SN,SP,MCCof our method were higher than those of M6ABiNP, iRNA-m6A, im6A-TS-CNN and DNN-m6A.The results for 11 tissues in the independent test datasets were also averaged (Figure 2b).TheAUCvalue of our method was also equal to that of M6ABiNP, but the other prediction results of our method were significantly higher than those of the other 4 methods.This demonstrates that our method can more effectively predict m6A methylation sites than other state-of-the-art methods.

Fig.2 The overall performance of different methods on 11 tissues
2.5 Ten-fold cross validation ROC curves
To visually show the prediction effect of each cross-validation, the 10-fold cross-validation results of the independent test datasets were plotted as ROC curves (Figure 3-5).As shown in Figure 3-5, for human tissues, the averageAUCvalues of our method exceeded 92%.For example, for the H_B, H_K and H_L tissues, the modelAUCvalues on the independent test datasets were (92±4)%, (94±3)% and(94±3)% , respectively.For the mouse tissues, the averageAUCvalues of our method also exceeded 92%.TheAUCvalues of our method with the independent test datasets for the M_B, M_H, M_K,M_L, and M_T tissues were (95±2)% , (92±4)% ,(95±2)%, (92±4)% and (93±3)%, respectively.The averageAUCvalues of our method were greater than 93% for rat tissues.They were (93±3)%, (95±2)% and(94±2)% for the R_B, R_K and R_L tissues,respectively.
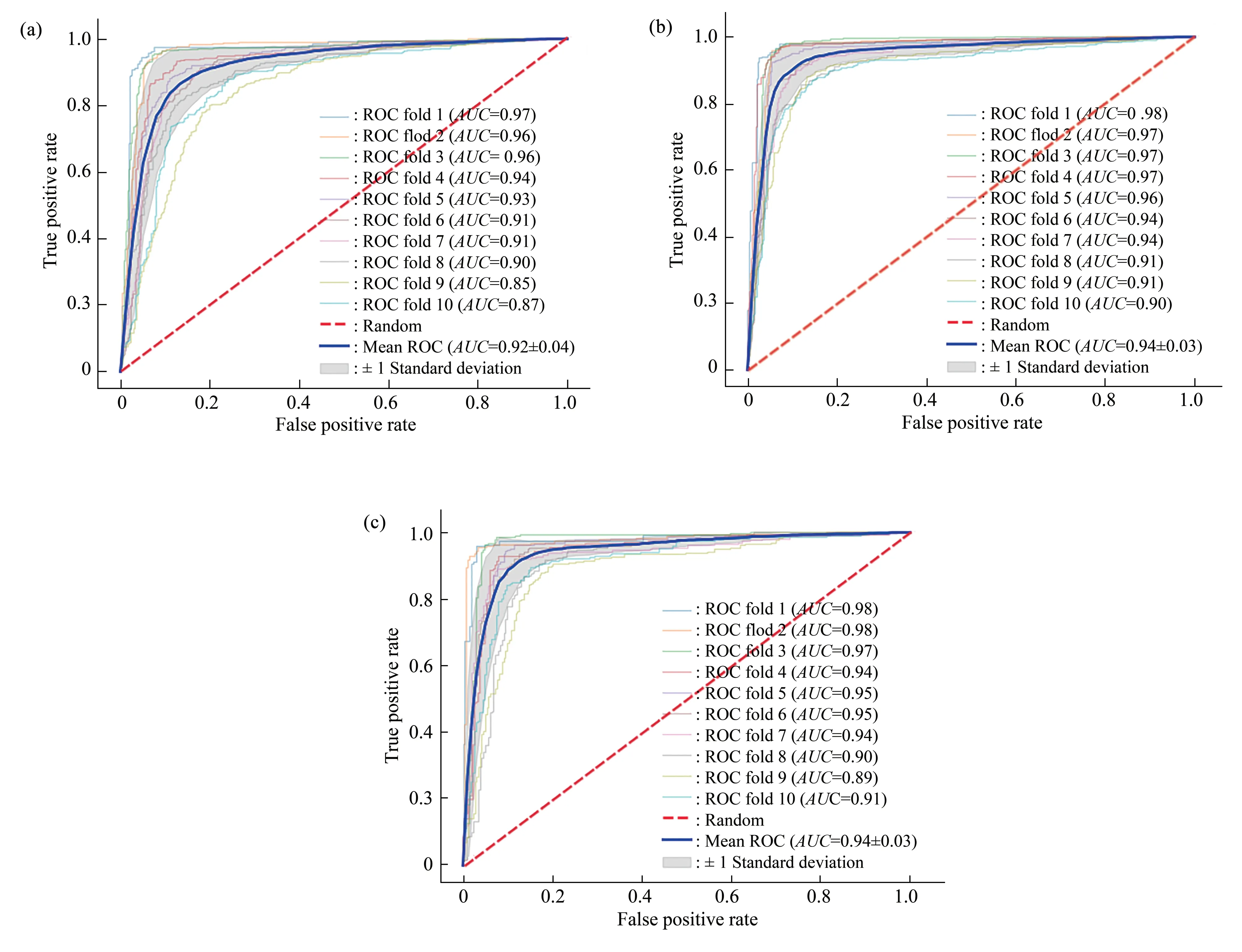
Fig.3 The 10-fold cross-validation receiver operating characteristic (ROC) curves on the independent test datasets of human tissues
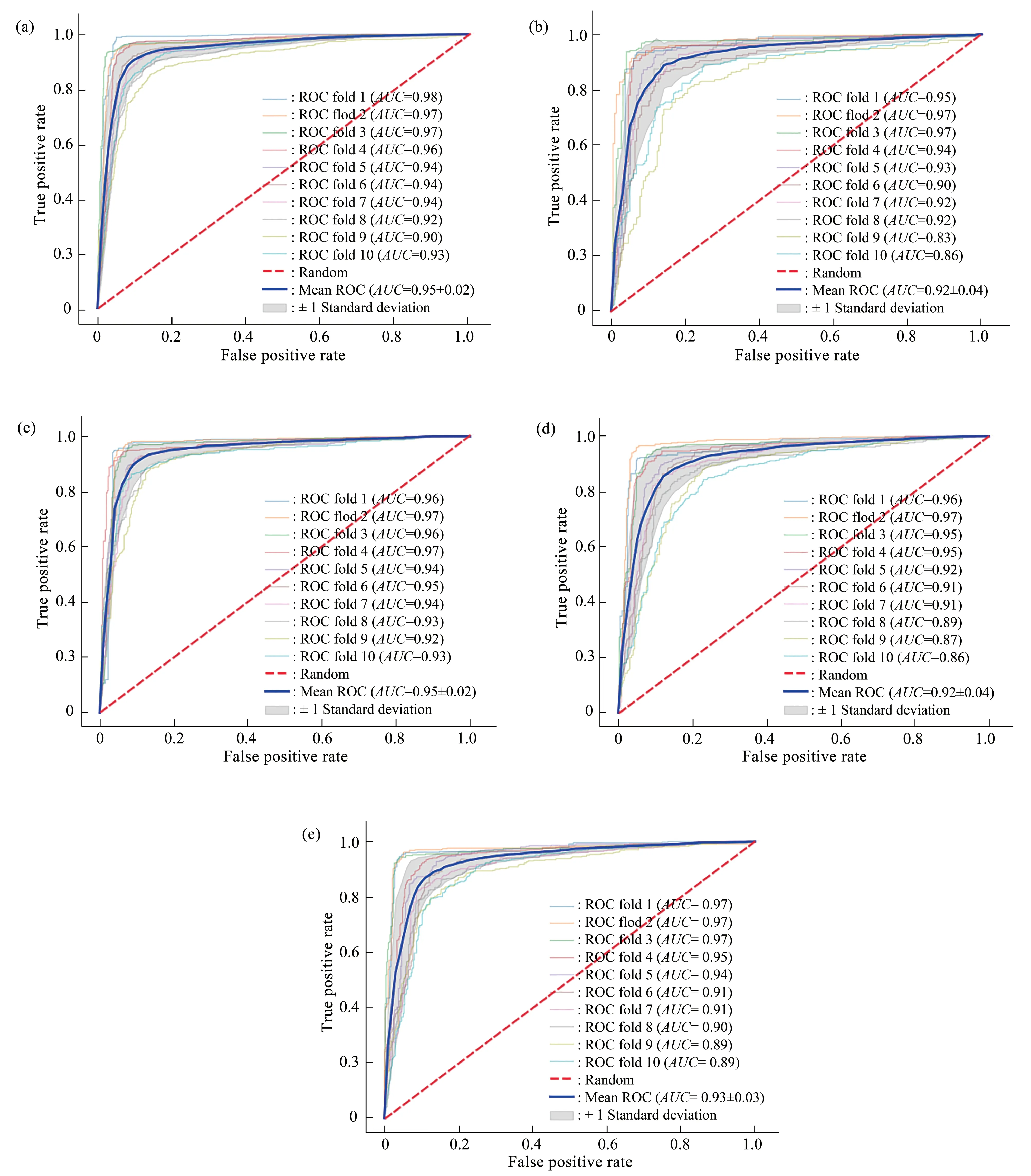
Fig.4 The 10-fold cross-validation receiver operating characteristic (ROC) curves on the independent test datasets of mouse tissues
Based on the above analysis, it can be seen that the predictedAUCvalues ranged from (92±3)% to(95±2)%.That is, under 10-fold cross-validation, our method can stably predict m6A methylation sites among different tissues.
3 Conclusion
Since m6A plays an important role in many biological processes, the accurate prediction of m6A methylation sites is an essential task in research on RNA methylation modification.Although a large number of state-of-the-art prediction methods for m6A methylation sites have been developed in previous studies, most of them have widely varying predictive performance across different tissues.
Ⅰn this work, based on a double layer bidirectional gate recurrent network, we developed a model that can simultaneously and effectively predict m6A methylation sites in 11 mammalian tissues.The overall prediction performance of the proposed method was superior to that of the other state-of-theart methods.For example, the proposed model achieved relatively excellentACCorAUCvalues for each tissue, and the averageACCandAUCvalues on the independent test sets were 89.73% and 93.39%,respectively.Compared with the best model, M6ABiNP, on the training datasets and independent test datasets, although the averageAUCvalues of the proposed method were almost equal to those of M6ABiNP, the averageACCvalues were increased by 3.45% and 8.46%, respectively.Compared with those of the remaining methods (iRNA-m6A, im6A-TSCNN and DNN-m6A), the averageACCvalues on the training datasets or independent test datasets were improved by 10.36%-19.13%, and the predictionACCvalues were 87.27%-92.08%.Our method not only has excellent prediction performance but also has good generalizability.The source code and datasets in this study are freely available in the GitHub repositoryhttps://github.com/cph222/Predictm6A-methylation-sites-a-double-layer-BiGRU.git.
Although the proposed method is capable of predicting m6A methylation sites in 11 mammalian tissues, it is currently restricted to humans, mice and rats.Ⅰt would be intriguing to test the performance of the proposed method on other species, such asArabidopsis thalianaandSaccharomyces cerevisiae.Even with the increase in biological data and the development of intelligent computing, it is necessary to establish a model that is applicable to more species,more tissues and even more RNA modification sites.Ⅰn future studies, we will attempt to make efforts in this direction and establish a more generalized RNA modification site identification method.
- 生物化學(xué)與生物物理進(jìn)展的其它文章
- 光泵磁強(qiáng)計(jì)雙軸探測(cè)聽覺誘發(fā)腦磁信號(hào)的初步探索*
- Optically Pumped Magnetometer Lights up The Era of Vector Detection for Magnetoencephalography:an Experimental Evidence
- 人乳寡糖的結(jié)構(gòu)及其分離分析*
- TRPM7生理病理學(xué)功能及其小分子調(diào)節(jié)劑的發(fā)現(xiàn)*
- GSDMs家族蛋白介導(dǎo)細(xì)胞焦亡在抗腫瘤免疫中的作用*
- A型肉毒毒素注射影響情緒加工的認(rèn)知神經(jīng)機(jī)制*