Contraction of Heat Shock Protein 70 Genes Uncovers Heat Adaptability of Ostrea denselamellosa
DONG Zhen, LIU Shikai, YU Hong, KONG Lingfeng, and LI Qi
Contraction of Heat Shock Protein 70 Genes Uncovers Heat Adaptability of
DONG Zhen, LIU Shikai, YU Hong, KONG Lingfeng, and LI Qi*
,,,266003,
The milin oysteris a live-bearing species with a sharp decline in the natural population. Unlike other oysters,lives in the subtidal zone and its adaptability to heat, salinity,. is different from most other oysters. Heat shock proteins 70 (HSP70) are a family of conserved ubiquitously expressed heat shock proteins which are produced in re- sponse to stressful conditions, especially heat. In this study, we identifiedgenes through bioinformatic analysis in five species of oyster. Among them,holds the fewest number ofgenes, which may be one of the reasons whycannot tolerate high temperatures. The conserved motifs and gene structures ofsub-family and other types ofwere different from that ofsub-family, which may be due to performing necessary multiple phy- siological functions. Transcription profile analysis forgenes ofindicated that gills play an important role in responding to multiple external challenges. In addition, synteny analysis ofgenes among,andshowed thatgenes in genus ofgenome might have evolved from a common ancestor with genus of. In short, our results lay the foundation for further investigation of the evolution ofgenes and heat adaptability.
; oysters;; heat; adaptability
1 Introduction
Milin oysteris a potential econo- mically important species, which generally inhabits the sub- tidal zone with high salinity along the coasts of China, Ja- pan and Korea (Xu., 2008; Chen., 2011). How- ever, previous studies showed that oysters such asandhad a survival rate of more than 50% at 32℃ (Wang and Li, 2017; Hu., 2020), whilecouldn not survive at 32℃ (Yang., 2003), which means that milin oyster is relatively poorly adapted to high temperatures. Moreover,is a kind of live-bearing oyster, which differs sig- nificantly from oysters,and most other bivalves. During reproduction, the eggs are fertilized and grow to D-shaped larvae in the female mental cavity within 1-3 days (Buroker, 1985; Foighil and Taylor, 2000; Yang., 2001). Previous study showed that species are more likely to be ovoviviviparous when they live in lower- temperature environments, and the female keep the em- bryos in their body to make sure they can grow in a suit- able temperature (Webb., 2006). To date, previous studies onmainly focused on its mito-chondrial genome, seed production and biological charac- teristics (Insua and Thiriot-Quievreux, 1991; Chen., 2011; Yu., 2016; Han., 2022), while the geno- mic-level research on their ecological adaptation remains limited (Xu., 2008).
, also known as, has functions in a wide range of housekeeping and stress-related activities (Rosenzweig., 2019). In previous studies, several members ofhave been cloned inand, and thesegenes were significantly up-regulated when the oysters were under heat stress (Nagata., 2017; Casas and La Peyre, 2020). In addition, with the quick development of genomesequencing, the genome-widegene family has beenstudied in oysters based on their genomes. These genes were found to be expanded in,and, which indicated thatgenes play important roles in adaptation to heat in dynamic environ- ments with a wide variety of stress factors (Zhang., 2012; Powell., 2018; Peng., 2020). Although we have generated the genome assembly of(Dong., 2023),no detailed analysis of thefamily has been performed in.
In this study, in order to provide a comprehensive un- derstanding of thegene family in the,thenumber of gene copies, chromosomal locations, tissue specific expression pattern, structure and motifs were examined based on the genome ofand other oysters. We also analyzed the evolution ofgenes among oysters by performing phylogenetic trees and synteny analyses, and carried out a preliminary study on the ecological adaptability ofby com- parative genomics and gene family enrichment.
2 Material and Methods
2.1 Data Preparation
The genomes, the longest peptides sequences and gff3files of(GCA_902806645.1) (Zhang., 2012),(GCA_002022765.4) (Gomez-Chiarri.,2015) and the genome of(GCA_003671525.1) (Powell., 2018) were downloaded from NCBI website. The longest peptides sequences and gff3 fileofwere obtained from http://soft.bioinfo-minzhao.org/srog/. All these three data sets of(CNA0022698) (Wu., 2022) were obtained from the China National GeneBank DataBase (CNGBdb). In addi- tion, we have generated the genomic data of(Dong., 2023) and(Li., 2023),which can be found in Figshare https://doi.org/10.6084/m9.figshare.19801705 and https://doi.org/10.6084/m9.figshare. 20013503, respectively. To identifygenes in oysters, HSP70 protein sequences fromwere download- ed from UniProt (https://www.uniprot.org/). The RNA-Seq data of.was derived from NCBI web- site with SRA numbers: SRR19238441, SRR19238440, SRR19238438 and SRR19238439.
2.2 Identification of Hsp70 Genes
To identifygenes in,,and,HSP70 protein sequences inwere used as query database. First, basic local alignment search tool algorithm program (BLASTP) (Al- tschul., 1990) was used to get the initial candidategenes ofe?5and iden- tity ≥30%. To make the results more accurate, these can- didate genes were then filtered by conserved domain. The hidden Markov model (HMM) (Eddy, 1996) profile of the HSP70 domain (PF00012) was downloaded from the Pfamprotein family database http://pfam-legacy.xfam.org/family/ HSP70. We filtered the candidate genes of the previous step by running ‘hmmsearch --cut_tc’ algorithm. Finally, we selected the candidate genes with corresponding amino acid length >300 asgenes.
2.3 Sequence Alignment and Phylogenetic Analysis
To investigate the evolutionary relationship offa- mily, thegenes of,andwere used to build a phylogenetic tree. First, the protein sequences of these genes were aligned using MU- SCLE (v3.6) (Edgar, 2004) and then the tree was con-structed using FASTTREE (Price., 2009). The tree was finally decorated and displayed with Interactive Tree of Life (ITOL, https://itol.embl.de/).
2.4 Gene Structure and Conserved Motif
PEPTIDES (2.4.4) (Osorio., 2015) was used to cal- culate the molecular weight and isoelectric point (PI). We used MEME (4.11.2) (Bailey., 2015) with parameters ‘-mod anr -protein -nmotifs 10 -minw 6 -maxw 200’ to de- tect the conserved motif. In addition, the structure of each gene was analyzed based on the coding sequence (CDS) and untranslated region (UTR) data from the gff3 file. Both the conserved motif and gene structure were visualized by TBtools (Chen., 2020).
2.5 Chromosomal Distribution and Synteny Analysis
genes of.were mapped to the chromosomes according to the gff3 file, and the results were processed and visualized by MAPCHART (Voorrips, 2002). To figure out what chromosomes-level changes occurred ingenes in oystersduring evolution, DIAMOND (Bu- chfink., 2015) was used to two-way align the protein sequences of.againstandwith parameters ‘--max-target-seqs 5 --evalue 1e-10’. Then the synteny of genes of these three species was identified based on MCscanX (Wang., 2012). Thecorresponding chromosome was determined by the frac- tion of genes in a block of approximately 25 genes. JCVI (Sleator, 2016) was used to visualize the final result andgenes were marked in red.
2.6 Expression Profiles of OdeHsp70 Genes in Different Tissues
We generated twelve sets of RNA-seq data for tissue specific expression analysis, including gonads, gills, adduc- tor and mantle from each of the three female.in stage of ovulation. These data have already been uploaded to NCBI with SRA numbers SRR19238441, SRR 19238440, SRR19238438 and SRR19238439. First, the RNA-Seq raw data was mapped toge- nome by HISAT2 (v2-2.2.1) (Kim., 2015). Then the quantity of transcriptomes was detected by FEATURE- COUNTS (v2.0.1) (Liao., 2014). The result was stan-dardized using TPM and TMM in order to balance the dif- ferences between tissues and individuals. Based on the stan-dardized quantification results, we plotted a heat map ofgenes ofusing PHEATMAP in R.
3 Results and Discussion
3.1 Identification of Hsp70 Genes
With the strict filtering standard, a total of 401genes were identified, including 59 in, 84 in, 83 in, 84 inand 88 in(Table 1)Compared to other species of oys- ters and previous studies of bivalves, including 133genes in hard calm(Hu., 2022), 61genes in(Cheng., 2016) and 65genes in(Hu., 2019),holds the fewestgenes.
The number ofgenes among bivalves varied great- ly, which might be a reflection of regulatory physiological variation that can help the bivalves adapt to the changing environment, especially temperature (Zhang., 2012).Therefore, the decrease in heat shock protein gene may af- fectto regulate heat-adaptability.
Moreover, we found that thegenes included 6 Heat shock protein70 B2 () genes, which was the same as in, and one copy more than that in; 47 Heat Shock Protein Fa- mily A Member 12 () genes and 6 other types ofgenes. Compared togene family in other spe- cies of oysters and scallops (Cheng., 2016; Hu., 2019), the contraction of thegene family inwas mainly due to the decrease in the number ofgenes (Table 1).
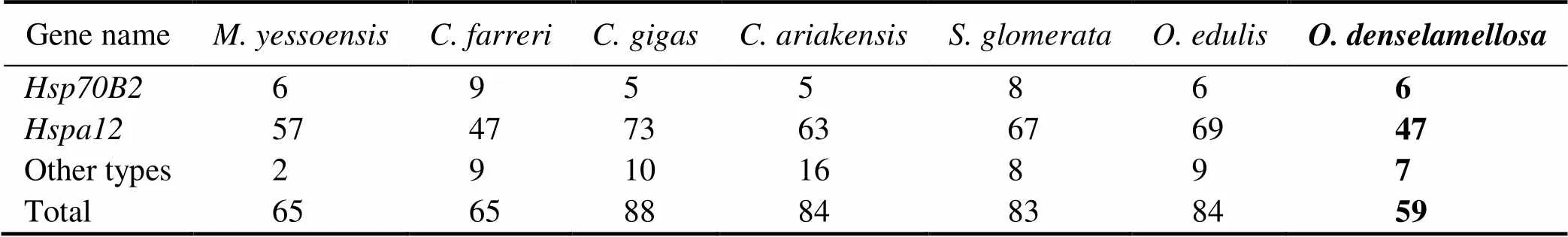
Table 1 Copy numbers of Hsp70 sub-families among mollusk genomes
3.2 Phylogenetic Analysis of the Hsp70 Genes
In order to figure out the evolutionary relationships ofgenes among oysters, four phylogenetic trees with maximum likelihood were constructed based on the long- est protein sequences. Because thegenes ofare well studied and examined, we used its protein se- quences as a reference. These four trees had similar topo- logical structures, and thegene family in the five species of oysters all mainly had two clusters, thesub-family (black branches) and other sub-families (red and green branches), as shown in Fig.1. Thegenes were first clustered together. The other sub-families likeandgene pairs were also clustered together. The other branch consisted of 47sub-family members. Compared with, the copy number ofis significantly reduced (Fig.1). This condition also occurred in a small subset of regions inandFor oysters, not only the total number ofgenes was changing, but the copy number ofsub-family genes was also different. These differences between species may be ex- plained by duplicated genes having independent origins orthere may have existed some genomic rearrangement in this region (Metzger., 2016). This situation inmay be due to the subtidal living environment which is different from the environments of other oysters.
3.3 OdeHsp70 Gene Structure and Conserved Motifs
The information aboutgenes’ location, amino acidlength and MolWts is summarized in https://doi.org/10. 6084/m9.figshare.24152763.v1. The predicted MolWts of theproteins varied from 46.04kDa (ode_0040 25-RB) to 171.14kDa (). In addition,encoded proteins varying from 421aa () to 1508aa (). Compared to other oysters (proteins from 384aa to 2290aa,proteins from 347aa to 2460aa andpro- teins from 316aa to 2595aa), the protein length range ofinwas relatively small. Thismay be due to the evolution ofgenes themselves and the different genome assembly-annotation methods of each species.
A total of ten conserved motifs were detected ingenes. The conserved motifs and gene structures ofsub-family (red) and other types of(green) were different from that ofsub-family. This may be due to performing necessary multiple physiologi- cal functions, according to the conjecture that different mo-tifs may indicate different functions or functional diver- gence (Liu., 2016). Except, the con- served motifs and gene structures of the genes in each of the above three sub-families were highly similar (Fig.2).
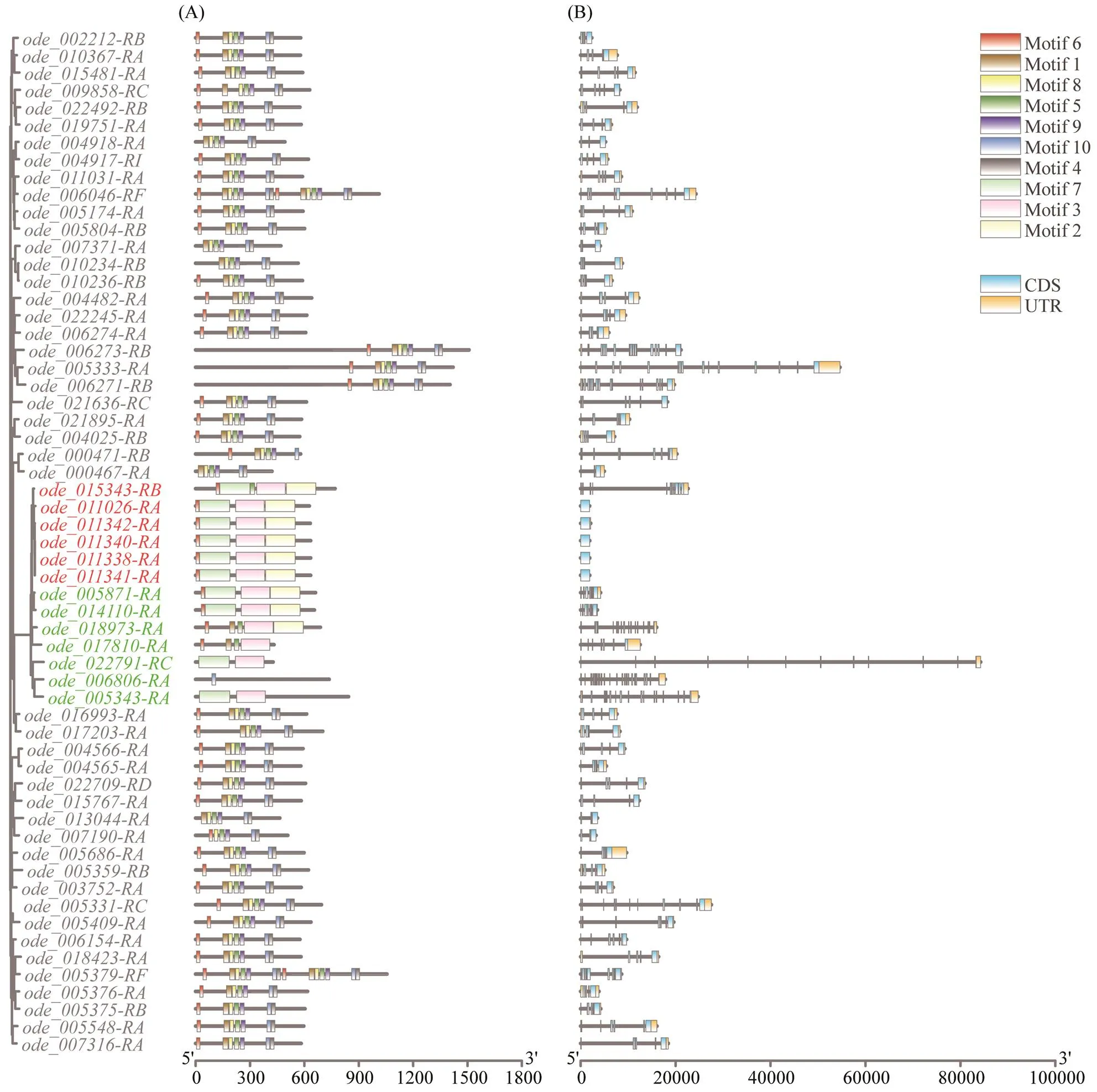
Fig.2 Architecture of conserved protein motifs and gene structure of Hsp70 genes in O. denselamellosa. Different branch colors represent different sub-families, red for Hsp70B2, green for other types, and black for Hspa12. (A) The motifs of Hsp70 proteins in O. denselamellosa. The ten motifs were displayed with different colors. The length of the protein can be estimated using the scale at the bottom. (B) Exons and introns of Hsp70 proteins in O. denselamellosa. Blue boxes in- dicated untranslated regions; orange boxes indicated exons.
3.4 Chromosomal Distribution of OdeHsp70 Genes
A total of 59genes were finally distributedamong 8chromosomes (Fig.3). Among all thesegenes, 28 (48.27%) were located on chromosome 4. The other chromosomes have fewer than 7 genes on each one. Only onegene was observed on chromosome 9. Similarly,genes were alsolocated on 8 chromosomes and 42 of 84 (50.00%) were distributed on chromosome 2 (https://doi.org/10.6084/m9.figshare.24152763.v1). Only onegene was ob- served on chromosome 1 and twogenes were observed on chromosome 6. Unlike the European oyster, in which onegene was distributed on a scaffold, thegenes ofwere all found on chromosomes. It might be a common phenome- non for members of large gene families to be unequally distributed on chromosomes. For example, only19 of 66 transient receptor potential channel genes were located on chromosome 2 in(Fu., 2021).
3.5 Synteny Analysis of OdeHsp70 Genes Between O. edulis and C. ariakensis
Synteny analysis of thegenes was performed among,and. There are many syntenic blocks between the genomes of these oysters, with 616 betweenand, 396 betweenand(Fig.4). Thegenes were mainly distributed on chro- mosome 4 of,chromosome 2 ofandchromosome 5 ofInterestingly, mostgenes located on chromosomes 10 weregenes. For, in addition to chromosomes Ode4 and Oed2,genes were also more or less distributed on other chromosomes, and eight out ten chromosomes of each species have the distribution of this gene family. But the situation was different in, only 6 chromo- somes have the distribution ofgenes, other than chro- mosomes Car3 and Car5. There are distributions on chro- mosomes Car6, Car7, Car8 and Car9, but just in small quan- tities. These results indicated that thegene family may be more conserved and that thegenes widely distributed on more chromosomes ofgenome might have evolved from those of a common an- cestor with.
3.6 Expression Patterns of OdeHsp70 Genes in Different Tissues
RNA-seq data of four tissues, including mantle, gill, ad- ductor muscle and gonad, from an ovulating femalewere used to characterize the expression pro- files ofs. Based on TPM+TMM values, a heat- map ofgenes in various tissues was created(Fig.5).genes were highly expressed in gills and re- latively lowly expressed in mantle and adductor muscle. The same expression pattern has also been reported in hard clamandManila clam(Liu., 2015; Nie., 2017; Hu., 2022). In Bivalve, gills are considered to be sensitive to environ- mental changes, and high expression ofgenes will promote the regulation of the environmental changes re- sponse (Cheng., 2019). Interestingly, somegenes showed highly tissue-specific expression. For exam- ple, fourgenes, including,,and, had high expression in the gills. Genesandshowed high expression levels in the gonad. Genes,were highly ex- pressed in the mantle. Genes,had higher expression in adductor muscle. In addition to a small part of genes like,,and, mostgenes were highly expressed in only one tissue.
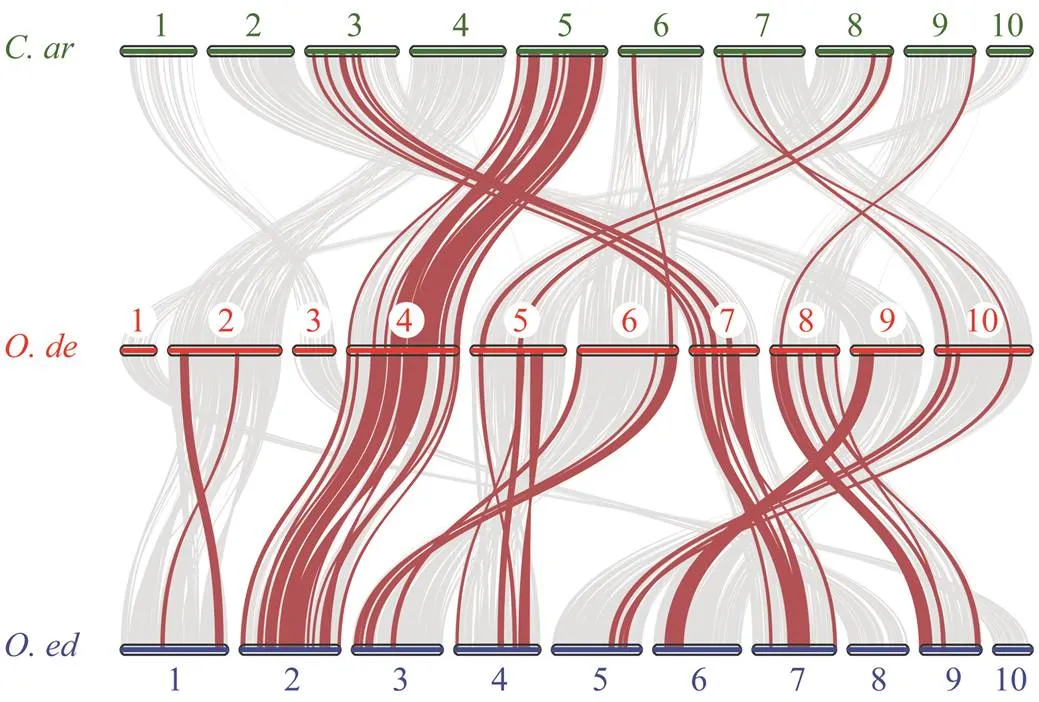
Fig.4 Synteny analyses between the Hsp70 genes of the C. ariakensis (C. ar), O. denselamellosa (O. de), and O. edu- lis (O. ed). Grey lines in the background indicated Syn- teny blocks among these genomes, while red lines high- light syntenic Hsp70 gene pairs.
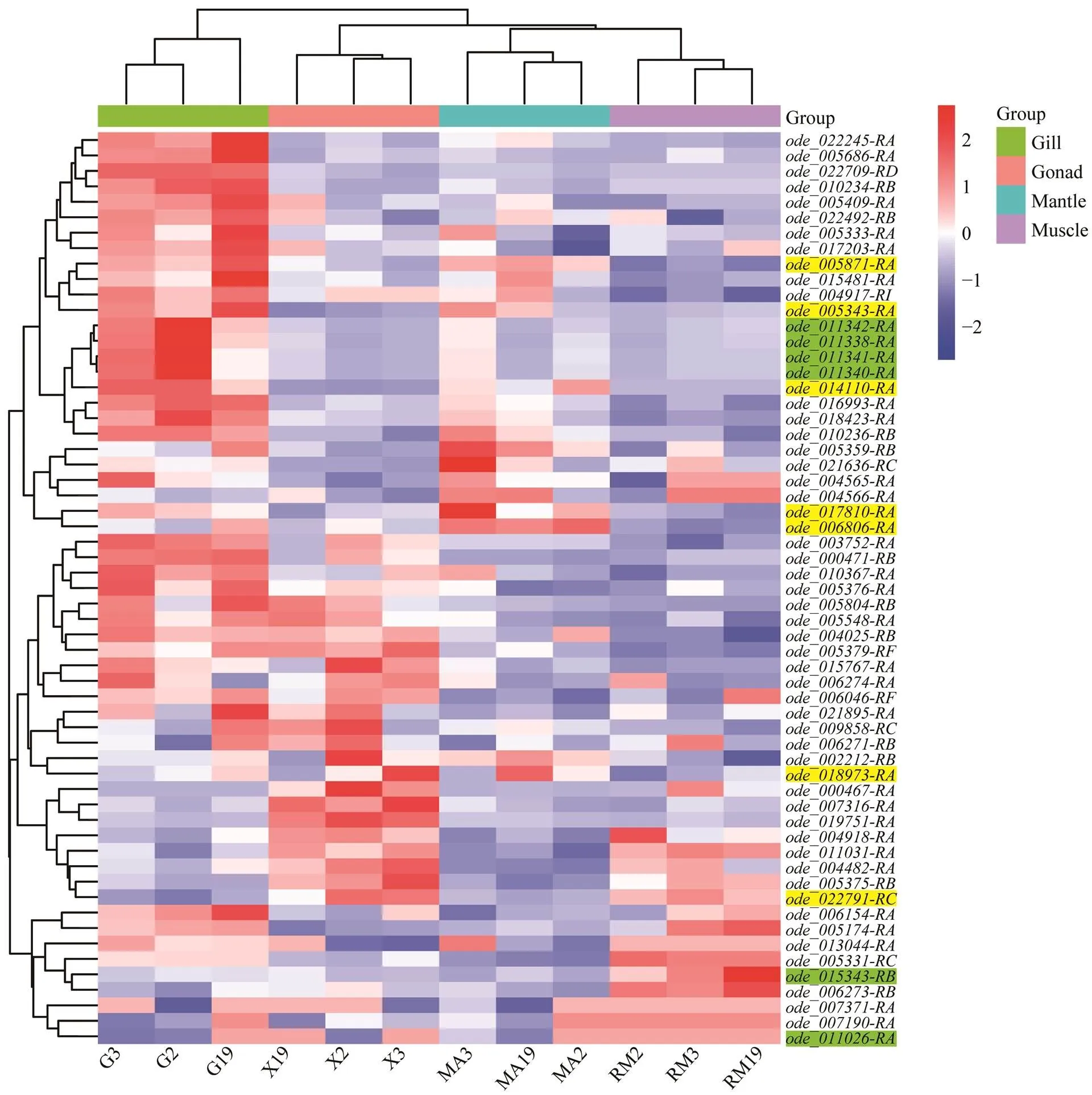
Fig.5 Expression pattern analysis of Hsp70 genes in four tissues of ovulating female O. denselamellosa based on TPM and TMM analyses. G, gill; X, gonad; MA, mantle; RM, muscle. The labels of Hsp70B2 genesare highlighted with green, the labels of Hspa12 genes are not highlighted and the labels of other types genes are highlighted with yellow. The color scale represents Z-score.
4 Conclusions
In summary, a genome-wide analysis ofgene fa- mily in 5 oyster species identified 401 genes including 59 in, 84 in, 83 in, 84 inand 88 in. The fewest num- ber ofgenes inis mainly due to the decrease in the number ofgenes, possibly ex- plaining whycannot tolerate high tem- perature. The gene structure and conserved motif investi- gation revealed that the conserved motifs and gene struc- tures ofgenes and other types ofwere different from that of, which may be due to performing necessary multiple physiological functions. Tran- scription profile analysis forgenes ofsupport that gills play an important role in re- sponding to multiple external challenges. In addition, syn- teny analysis ofgenes betweenanddemonstrated thatgenes inge- nome might have evolved from those of a common an-cestor with. Taken together, the informationobtained in this study lay the foundation for further invest- tigation of the evolution ofgenes and heat adaptability of.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (No. 2022YFD2400305), the China Agriculture Research System Project (No. CARS-49), and the Key R&DProject of Shandong Province (Nos. 2021ZLGX03, 2021LZGC027).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J., 1990. Basic local alignment search tool., 215 (3): 403-410, DOI: 10.1016/S0022-2836(05) 80360-2.
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S., 2015. The MEME Suite., 43 (W1): W39- W49, DOI: 10.1093/nar/gkv416.
Buchfink, B., Xie, C., and Huson, D., 2015. Fast and sensitive protein alignment using DIAMOND., 12 (1): 59-60, DOI: 10.1038/nmeth.3176.
Buroker, N. E., 1985. Evolutionary patterns in the family Ostrei- dae: Larviparity. Oviparity., 90 (3): 233-247, DOI: 10.1016/0022- 0981(85)90169-8.
Casas, S. M., and La Peyre, J. F., 2020. Heat shock protein 70 levels and post-harvest survival of eastern oysters following sublethal heat shock in the laboratory or conditioning in the field., 25 (2): 369-378, DOI: 10.1007/ s12192-019-01056-1.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y.,., 2020. TBtools: An integrative toolkit developed for interactive analyses of big biological data., 13 (8): 1194-1202, DOI: 10.1016/j.molp.2020.06.009.
Chen, L., Li, Q., Wang, Q., Kong, L., and Zheng, X., 2011. Tech- niques of artificial breeding of the oyster., 41 (3): 43-46 (in Chinese with English abstract).
Cheng, D., Liu, H., Zhang, H., Soon, T. K., Ye, T., Li, S.,., 2019. Differential expressions ofgene between golden and brown noble scallopsunder heat stress and bacterial challenge., 94: 924- 933, DOI: 10.1016/j.fsi.2019.10.018.
Cheng, J., Xun, X., Kong, Y., Wang, S., Yang, Z., Li, Y.,., 2016.gene expansions in the scallopand their expression regulation after exposure to the toxic dinoflagellate., 58: 266-273, DOI: 10.1016/j.fsi.2016.09.009.
Dong, Z., Bai, Y., Liu, S., Yu, H., Kong, L., Du, S.,., 2023. Achromosome-level genome assembly ofprovides initial insights into its evolution., 115 (2): 110582, DOI: 10.1016/j.ygeno.2023.110582.
Eddy, S. R., 1996. Hidden Markov models., 6 (3): 361-365, DOI: 10.1016/S0959-440X (96)80056-X.
Edgar, R. C., 2004. MUSCLE: Multiple sequence alignment withhigh accuracy and high throughput., 32 (5): 1792-1797, DOI: 10.1093/nar/gkh340.
Foighil, D. O., and Taylor, D. J., 2000. Evolution of parental careand ovulation behavior in oysters., 15 (2): 301-313, DOI: 10.1006/mpev.1999.0755.
Fu, H., Jiao, Z., Li, Y., Tian, J., Ren, L., Zhang, F.,., 2021. Transient receptor potential (TRP) channels in the Pacific oys- ter (): Genome-wide identification and ex- pression profiling after heat stress betweenand., 22 (6): 3222, DOI: 10.3390/ijms22063222.
Gomez-Chiarri, M., Warren, W., Guo, X., and Proestou, D., 2015. Developing tools for the study of molluscan immunity: The se- quencing of the genome of the eastern oyster,., 46 (1): 2-4, DOI: 10.1016/ j.fsi.2015.05.004.
Han, J., Kim, H. J., Oh, S. Y., and Choi, Y. U., 2022. Reproductive characteristics of the flat oyster(Bival- via, Ostreidae) found on the southern coast of South Korea., 10: 1326, DOI: 10.3390/jmse10091326.
Hu, B., Li, M., Yu, X., Xun, X., Lu, W., Li, X.,., 2019. Di- verse expression regulation ofgenes in scallops after exposure to toxicdinoflagellates., 234: 62-69, DOI: 10.1016/j.chemosphere.2019.06.034.
Hu, Y. M., Li, Q., Liu, S. K., and Kong, L. F., 2020. Effects of acute temperature and salinity stress on the survival and im- mune indexes of Iwagaki oysters,., 27 (3): 286-294 (in Chinese with English abstract).
Hu, Z., Song, H., Feng, J., Zhou, C., Yang, M, J., Shi, P.,., 2022. Massive heat shock protein 70 genes expansion and trans-criptional signatures uncover hard clam adaptations to heat and hypoxia., 9: 898669, DOI: 10. 3389/fmars.2022.898669.
Insua, A., and Thiriot-Quievreux, C., 1991. The characterization of(Mollusca, Bivalvia) chromosomes: Karyotype, constitutive heterochromatin and nucleolus orga- nizer regions., 97 (4): 317-325, DOI: 10.1016/ 0044-8486(91)90324-z.
Kim, D., Landmead, B., and Salzberg, S. L., 2015. HISAT: A fast spliced aligner with low memory requirements., 12 (4): 357-360, DOI: 10.1038/nmeth.3317.
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B., 2017. Time tree: A resource for timelines, timetrees, and divergence times., 34 (7): 1812-1819, DOI: 10.1093/molbev/msx116.
Li, X., Bai, Y., Dong, Z., Xu, C., Liu, S., Yu, H.,., 2023. Chro- mosome-level genome assembly of the European flat oyster () provides insights into its evolution and adap- tation.–, 45 (1): 101045, DOI: 10.1016/j.cbd. 2022.101045.
Liao, Y., Smyth, G. K., and Shi, W., 2014. featureCounts: An ef- ficient general purpose program for assigning sequence reads to genomic features., 30 (7): 923-930, DOI: 10. 1093/bioinformatics/btt656.
Liu, T., Pan, L., Cai, Y., and Miao, J., 2015. Molecular cloning and sequence analysis of heat shock proteins 70 () and 90 () and their expression analysis when exposed to benzo (a) pyrene in the clam., 555 (2): 108-118, DOI: 10.1016/j.gene.2014.10.051.
Liu, X., Tang, S., Jia, G., Schnable, J. C., Su, H., Tang, C.,., 2016. The C-terminal motif of SiAGO1b is required for theregulation of growth, development and stress responses in fox- tail millet ((L.) P. Beauv)., 67 (11): 3237-3249, DOI: 10.1093/jxb/erw135.
Metzger, D. C., Hemmer-Hansen, J., and Schulte, P. M., 2016. Conserved structure and expression ofparalogs in te- leost fishes., 18: 10-20, DOI: 10.1016/j.cbd.2016. 01.007.
Nagata, T., Sameshima, M., Uchikawa, T., Osafune, N., and Ki- tano, T., 2017. Molecular cloning and expression of the heat shock protein 70 gene in the Kumamoto oyster., 83: 273-281, DOI: 10.1007/s12562- 017-1064-6.
Nie, H., Liu, L., Huo, Z., Chen, P., Ding, J., Yang, F.,., 2017. Thegene expression responses to thermal and salinity stress in wild and cultivated Manila clam., 470: 149-156, DOI: 10.1016/j.aquaculture. 2016.12.016.
Osorio, D., Rondon-Villarreal, P., and Torres, R., 2015. Peptides: A package for data mining of antimicrobial peptides., 7 (1): 4-14, DOI: 10.32614/RJ-2015-001.
Peng, J., Li, Q., Xu, L., Wei, P., He, P., Zhang, X.,., 2020. Chromosome-level analysis ofge-nome reveals extensive duplication of immune-related genes in bivalves., 20 (4): 980-994, DOI: 10.1111/1755-0998.13157.
Powell, D., Subramanian, S., Suwansaard, S., Zhao, M., O’Con- nor, W., Raftos, D.,., 2018. The genome of the oysteroffers insight into the environmental resilience of bi- valves., 25 (6): 655-665, DOI: 10.1093/dnares/ dsy032.
Price, M. N., Dehal, P. S., and Arkin, A. P., 2009. FastTree: Com- puting large minimum evolution trees with profiles instead of a distance matrix., 26 (7): 1641-1650, DOI: 10.1093/molbev/msp077.
Rosenzweig, R., Nillegoda, N. B., Mayer, M. P., and Bukau, B., 2019. The Hsp70 chaperone network., 20 (11): 665-680, DOI: 10.1038/s41580- 019-0133-3.
Sleator, R. D., 2016. JCVI-syn3.0–A synthetic genome stripped bare., 7 (2): 53-56, DOI: 10.1080/21655979.2016. 1175847.
Voorrips, R. E., 2002. MapChart: Software for the graphical pre- sentation of linkage maps and QTLs., 93 (1): 77-78, DOI: 10.1093/jhered/93.1.77.
Wang, T., and Li, Q., 2017. Effects of salinity and temperature on growth and survival of juvenile of kumamoto oyster ()., 48 (2): 297-302 (in Chinese with English abstract).
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X.,.,2012. MCScanX: A toolkit for detection and evolutionary ana- lysis of gene synteny and collinearity., 40 (7): e49, DOI: 10.1093/nar/gkr1293.
Webb, J. K., Shine, R., and Christian, K. A., 2006. The adaptive significance of reptilian viviparity in the tropics: Testing the maternal manipulation hypothesis., 60 (1): 115-122, DOI: 10.1111/j.0014-3820.2006.tb01087.x.
Wu, B., Chen, X., Yu, M., Ren, J., Hu, J., Shao, C.,., 2022. Chromosome-level genome and population genomic analysis provide insights into the evolution and environmental adapta- tion of Jinjiang oyster.,22 (4): 1529-1544, DOI: 10.1111/1755-0998. 13556.
Xu, F., and Zhang, S., 2008.. Science Press, Beijing, 336pp (in Chinese).
Yang, M. H., Bong, S. H., and Han, C. H., 2003. Growth and sur- vival rates of flat oyster larvae,, by con- dition of larvae cultivation., 19(2): 133-142.
Yang, M. H., Kim, H. S., Lee, J. Y., and Han, C. H., 2001. Artifi- cial mass culture of flat oyster larvae,, and collection rates according to various spat collection methods., 17 (1): 35-44.
Yu, H., Kong, L., and Li, Q., 2016. Complete mitochondrial ge- nome of(Bivalvia, Ostreidae)., 27 (1): 711-712, DOI: 10.3109/19401 736.2014.913154.
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F.,., 2012. The oyster genome reveals stress adaptation and complexity of shell formation., 490 (7418): 49-54, DOI: 10.1038/ nature11413.
(January 6, 2023;
March 17, 2023;
May 23, 2023)
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2023
Tel: 0086-532-82031622
E-mail: qili66@ouc.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2023年6期
Journal of Ocean University of China2023年6期
- Journal of Ocean University of China的其它文章
- Effects of 5-Azacytidine (AZA) on the Growth, Antioxidant Activities and Germination of Pellicle Cystsof Scrippsiella acuminata (Diophyceae)
- Improving Yolo5 for Real-Time Detection of Small Targets in Side Scan Sonar Images
- Wave Radiation by a Floating Body in Water of Finite Depth Using an Exact DtN Boundary Condition
- Underwater Acoustic Signal Noise Reduction Based on a Fully Convolutional Encoder-Decoder Neural Network
- Revisiting the Seasonal Evolution of the Indian Ocean Dipole from the Perspective of Process-Based Decomposition
- Assessment of Storm Surge and Flood Inundation in Chittagong City of Bangladesh Based on ADCIRC and GIS
