Identification of candidate genes for aphid resistance in upland cotton by QTL mapping and expression analysis
Qiushung An,Zhenyun Pn,Nurimnguli Aini,Peng Hn,Yunlong Wu,Chunyun You,Xinhui Nie,
a Key Laboratory of Oasis Ecology Agricultural of Xinjiang Production and Construction Corps,Agricultural College,Shihezi University,Shihezi 832003,Xinjiang,China
b Cotton Research Institute of the Shihezi Academy of Agriculture Science,Shihezi 832011,Xinjiang,China
Keywords:GhLAC4-3 Lignin Gossypium hirsutum Aphid resistance
ABSTRACT Lignin is one of the main components of cell walls and is essential for resistance to insect pests in plants.Cotton plants are damaged by aphid(Aphis gossypii)worldwide but resistant breeding is undeveloped due to scarce knowledge on resistance genes and the mechanism.This study reported a lignin biosynthesisrelated gene identified in the F2 population derived from the cross between cotton cultivars Xinluzao 61(resistant to aphid) and Xinluzao 50 (susceptible to aphid).A quantitative trait locus was mapped on chromosome D04 with a logarithm of odds (LOD) score of 5.99 and phenotypic effect of 27%.RNA-seq analysis of candidate intervals showed that the expression level of GH_D04G1418 was higher in the resistant cultivar than in the susceptible cultivar.This locus is close to AtLAC4 in the phylogenetic tree and contains a conserved laccase domain.Hence,it was designated GhLAC4-3.Silencing of GhLAC4-3 in Xinluzao 61 via virus-induced gene silencing (VIGS) resulted in decreased lignin content and increased susceptibility to aphids.These results suggest that GhLAC4-3 might enhance aphid resistance by regulating lignin biosynthesis in cotton.
1.Introduction
Aphid (Aphis gossypiiGlover) is a serious insect pest in most cotton-producing areas in China.As a piercing-sucking insect,aphids cause severe losses in yield and quality in cotton production.Chemical controls for this pest are being used less frequently because the wide use of pesticides has direct or indirect adverse effects on non-target insects,humans,and the environment [1].Therefore,understanding the genetic mechanism of aphid resistance in cotton is the most fundamental way to improve cotton quality and reduce environmental pollution.
Lignin,a complex phenolic polymer deposited in cell walls of higher plants,proves to be involved in host resistance to pathogens and insect pests in addition to the functions of plant mechanical support and transport of mineral and water [2].Lignin is mainly composed of three monomers: guaiacyl(G),syringyl(S),and trace ρ-hydroxyphenyl(H) [3].In apoplast,they are oxidized by laccase to produce lignin [4].
Laccase is a glycoprotein containing copper ions,first isolated from the juice of the Japanese lacquer tree(Rhus vernicifera).There are 17 known laccase family members inArabidopsis thaliana[5].Only four laccase genes,AtLAC4,AtLAC11,AtLAC15,andAtLAC17,were reportedly involved in lignin biosynthesis.In cotton,Zhang et al.[6] found that the total lignin content and verticillium wilt resistance of transgenicArabidopsis thalianawere also significantly enhanced with the increase ofGhLAC15expression.Hu et al.[7]also found that with the increase and decrease ofGhLAC1expression,the degree of lignification enhancement of plant cell wall and the resistance to biotic and abiotic stresses also increased and decreased.Therefore,we hypothesize that plant laccase (LACs) is involved in lignin biosynthesis to regulate plant pest resistance.This study was conducted using QTL mapping and expression analysis to identify candidate genes for aphid resistance in upland cotton.
2.Materials and methods
2.1.Mapping population and evaluation of aphid resistance
The F2population,containing 600 individuals,was derived from the cross between upland cotton cultivars Xinluzao 61 (resistant male parent) and Xinluzao 50 (susceptible female parent).It was used for genetic analysis and mapping of aphid resistance QTL.Seeds of Xinluzao 61 were germinated and grown in greenhouse under conditions of 28/25 °C,16 h light/8 h dark cycle,and 60%humidity.
Aphids were collected from the farm fields of Shihezi University in 2019 summer.The pests were fed with a susceptible cotton line to reproduce offspring until the infesting experiment.Non-choice tests of aphids in greenhouses and fields were conducted to evaluate the aphid resistance.The parents and the F2seeds were planted on the farm of Shihezi University and five adult aphids were transferred to the back of young leaves of each plant at the four-leaf stage of cotton.The quantity of aphid on individual plants was investigated 14 d after infestation.Resistance was scored from 1 to 5 with the criteria of 1 ≤n<50,51 ≤n<100,101 ≤n<150,151 ≤n<200,andn≥200 aphids per plant,which was modified from the method of Liang et al.[8].
2.2.Restriction-site-associated DNA (ddRAD) sequencing and genetic map construction
Genomic DNA was extracted from young leaves of the resistant(n=41) and susceptible (n=42) plants from 83 F2individuals derived from cross Xinluzao 61 × Xinluzao 50 and both parents using a genomic DNA extraction kit (Tiangen,Beijing,China) and double-digested withMseI andSacI (New England Biolabs,Beijing,China) to construct sequencing libraries.The 85 samples were sequenced using an Illumina HiSeq 2000 platform (Illumina Trading,Shanghai,China).Then,the Haplotype Caller and Genotype GVCFs module in the GATK software (version 3.7,https://software.broadinstitute.org/gatk/) were used to detect the variation in all samples according to the following criteria:1)the percentage of genotypic deletions in offspring was ≤50%;2) the frequency of minor alleles was ≥ 20%;3) the heterozygosity of offspring was ≤ 60%;and 4) the relative heterozygosity of offspring was ≤75%.To ensure the quality of subsequent analysis,we only selected the offspring whose heterozygosity rate and genotype deletion rate did not exceed 15% and 80%,respectively,for subsequent analysis.
A genetic linkage map was constructed as described by Xie et al.[9].The MEM algorithm of the BWA software (version 0.7.15-r1140) was used to compare the sequencing data with the reference genome (TM-1_ZJU_v2.1) [10].We used QTL Cartographer(version 1.17j)software(NCSU,Raleigh,NC,USA)for QTL analysis.The Composite Interval Mapping (CIM) method with 95% confidence intervals and 1.0 cM walking speed precision was chosen for QTL mapping.Putative QTL were declared after 1000 permutation trials using a log-likelihood(LOD)threshold of 2.5.Peaks with LOD values higher than the LOD cut-off were considered to be significant (1000 times,P=0.05).
2.3.Library construction of RNA-seq data
In the four-leaf stage of cotton,we used young leaves at the top of cotton plants for the RNA-seq analysis.The RN38 EASYspin plus Plant RNA kit (Aidlab Biotech,Beijing,China) was used to extract the total RNA from each sample.Libraries were sequenced by Novogene (Novogene,Tianjin,China) using an Illumina HiSeq system(Illumina,San Diego,CA,USA).To quantify the gene expression level,we calculated the Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) of each gene according to the length of the gene and calculated the reading mapped to the gene.Then,DESeq2 software(version 1.20.0,https://www.bioconductor.org/packages/release/bioc/html/DESeq2.)was used to compare the differential expression between the two combinations.We referred to the methods of Benjamin and Hochberg [11] to adjust the obtainedP-value to control the error detection rate.|log2-FoldChange|>1 and FDR <0.05 were set as the threshold of significant difference expression.
2.4.Real-time quantitative PCR and RT-PCR
To determine the expression level ofGhLAC4-3in VIGS plants,RT-PCR and qRT-PCR were performed.RNA was extracted from young cotton leaves of the silent plants and the control plants at the four-leaf stage.The M-MLV method was used to reverse transcribe total RNA (2 μg) into a cDNA template (TaKaRa,Dalian,Liaoning,China).This cDNA was used as a template for reverse transcription PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR).All specific primers (Table S1) were designed using NCBI online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index).RTPCR was performed with a program of an initial denaturation step at 95 °C for 5 min,followed by 34 cycles of denaturation at 95 °C for 30 s,annealing at 60 °C for 30 s,and extension at 72 °C for 30 s.The SYBR Green qPCR Super Mix (Transgen Biotech,Beijing,China) was used to conduct qRT-PCR of candidate genes on Light Cycler 480II (Roche,Mannheim,Germany).The qRT-PCR was run under the following conditions: an initial denaturation step at 95°C for 30 s,45 cycles of denaturation at 95°C for 5 s,annealing at 60°C for 15 s,and extension at 60°C for 15 s.The cottonGhUBQ7gene was used as an internal reference.The relative expression was analyzed using the 2-ΔΔCTmethod[12].The average value of each sample was determined from the measured value of at least five treated or untreated plants using three experimental replicates.
2.5.VIGS assays
A virus-induced gene silencing (VIGS) assay was performed on Xinluzao 61 according to the method described by Gao et al.[13].GhCLA1,a gene encoding 1-deoxyxylose 5-phosphate synthase,was used as the positive control.Approximately 300-400 bp cDNA fragments ofGhLAC4-3were amplified from Xinluzao 61 using gene-specific primers (Table S1).The resulting products were cloned into pTRV2 withEcoRI andKpnI(New England Biolabs,Beijing,China) to produce recombinant vectors pTRV:GhCLA1,pTRV:00,pTRV1,and pTRV:GhLAC4-3.Then,the vectors were introduced intoA.tumefaciensstrainGV3101and mixed with the strains carrying constructs pTRV:00,pTRV:GhCLA1 or pTRV:GhLAC4-3 (1:1,V/V),respectively.Finally,the mixed strains were injected separately into the flat cotyledon of each seedling.Seedlings were incubated for 24 h in the dark.After the appearance of the albino phenotype,the silencing effect ofGhLAC4-3was detected using the RNA derived from the cotton leaves.
2.6.Choice and no-choice assays of cotton aphid
Leaves of control plants and silent plants were placed in 29×44 cm plastic boxes or 9 cm round glass dishes for the selective and non-selective tests of cotton aphid [7].The experiment was performed in triplicate for each treatment and control,involving eight leaves per replicate.
2.7.Histochemical staining and content determination of lignin
To observe xylem development and lignin deposition in plants,sections of stems and young leaves were chemically stained using Wiesner′s reagent [14].In short,sections of stems and young leaves were put in phloroglucinol solution (2%,v/v,95% ethanol)for 10 min,transferred to 18% HCl solution for 5 min,and then immediately photographed under the fluorescence microscope(DM2500;Leica,Wetzlar,Germany).
Lignin contents of the first internodes and young leaves of different plants were determined using the Klason method [15].Briefly,after removing other compounds from the stems and young leaves with a phosphate buffer,Triton X-100,NaCl and acetone,the lignin content was determined via the lignin-thioglycolic acid(LTGA) reaction.The experiment was performed in triplicate for each treatment and control,involving 10 leaves or stem per replicate.
2.8.Statistical analysis
Thet-test in SPSS (version 26.0) statistical software (SPSS,Chicago,IL,USA)was used for statistical analysis.The Origin2021 software (OriginLab,Northampton,MA,USA) was used for plotting.The values used in the figure are mean ± SD.
3.Results
3.1.Four cotton aphid-resistant QTL were identified via ddRAD-seq
Aphid bioassays for two cotton cultivars with Xinluzao 61 or Xinluzao 50 to aphid infection were performed in greenhouse and field settings for two years.We observed that the number of aphids per plant of the Xinluzao 61 was significantly lower than that of the Xinluzao 50 (Fig.1A).Further,frequency distribution analysis performed on the 600 F2population (Xinluzao 61 × Xinluzao 50) individuals indicated that aphid resistance of cotton traits was normally distributed (Fig.S1).These data suggest that the aphid resistance of cotton is a quantitative trait.
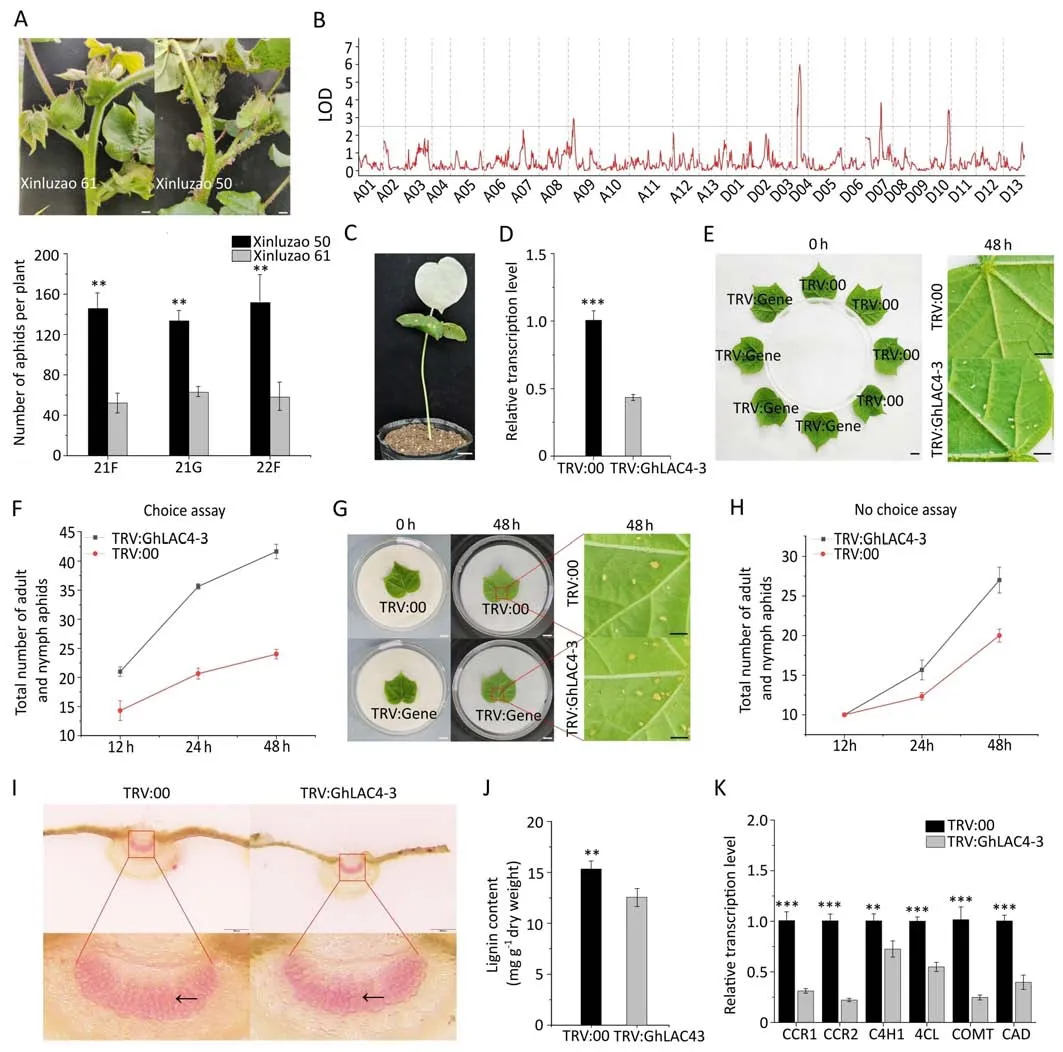
Fig.1.Mining and identification of the aphid-resistant gene GhLAC4-3.(A)Representative images of the resistant Xinluzao 61 and susceptible Xinluzao 50 cultivars following aphid infestation(upper panel),and the number of aphids on Xinluzao 61 and Xinluzao 50 plants in the field(21F and 22F)and greenhouse(21G)(lower panel).(B)Logarithm of odds (LOD) scores calculated from the phenotype data of 83 individuals derived from F2 for a genome-wide scan.Thresholds for identifying QTL were determined by permutation(1000 times,P=0.05)and are shown with gray horizontal lines.(C)Twelve days after TRV:GhCLA1 was injected into cotton cotyledon,the newly developed true leaves and stems of cotton showed bleaching,which proved that the VIGS system was effective.(D) qRT-PCR analysis showed that the relative expression of GhLAC4-3 was significantly decreased in TRV:GhLAC4-3 plants compared to TRV:00 plants.(E,F)Choice feeding assay for cotton aphids.(G,H)Non-choice feeding assay for cotton aphids.For aphid choice and non-choice feeding experiments,the number of aphids on leaves of each plant was recorded at 12,24,and 48 h.Photos were taken with a microscope(DM2500,Leica)at 0 h before and 48 h after aphid introduction.(I)Cross sections of young leaves from TRV:GhLAC4-3 and TRV:00 plants stained with phloroglucol HCl.The lignin in the red box in the upper picture is magnified and shown in the lower picture,with the black arrow pointing to the lignin fiber.Scale bar,2000 μm.(J)Determination of lignin content in young leaves of TRV:GhLAC4-3 and TRV:00 plants.(K)Relative expression levels of genes involved in lignin biosynthesis in young leaves of TRV:GhLAC4-3 and TRV:00 plants.In A,D,F,H,J,and K,values are means ± SD; n=3.*, P <0.05;**, P <0.01;***, P <0.001.Scale bars,1 cm in (A,C,E,and G).
To study the QTL controlling cotton aphid resistance,83 extreme individuals derived from the F2population were selected from grades 1 and 5,and the double digestion restriction site associated DNA sequencing(ddRAD-seq) was performed [16].Approximately 40.7 million clean reads were obtained.The average value of the unique mapping of all samples and the average value (%) of the mapping were 2,953,190 and 61.93%,respectively,and 94.78%of the bases were of high quality,with the GC content ranging from 38.57% to 39.32% (Table S2).The variation of all samples showed that 15,866 single nucleotide polymorphisms (SNPs) and 1052 InDels were distributed on 26 chromosomes (Table S3).These results indicate that the quantity and quality of the data are sufficient for further analysis.
Four unique QTL,qRA-A09.1,qRA-D04.1,qRA-D07.1,and qRAD10.1,were detected on chromosomes A09,D04,D07,and D10,respectively,by the genome-wide linkage analysis (Fig.1B).The calculation results showed that these QTL conferring aphid resistance explained 72% of the phenotypic variation,of which the LOD score of qRA-D04.1 was 5.99 and theR2was 27% (Table S4).Therefore,a 9.47 Mb region spanning 40.67-50.14 Mb on chromosome D04 was defined as the primary target region associated with aphid resistance.Consulting the reference genome (TM-1_ZJU_v2.1) revealed that there were 460 genes in this region.
3.2.Expression pattern of candidate genes was detected after aphid invasion
The leaves that were infected with aphids for two weeks were collected for transcriptome sequencing to elucidate the changes in gene expression of aphid-resistant and susceptible cotton lines.RNA-seq analysis showed that the expression of 18 candidate genes derived from the 460 genes revealed by ddRAD-seq differed significantly between aphid-resistant and susceptible cotton plants.Moreover,the expression levels ofGH_D04G1280,GH_D04G1369,GH_D04G1126,GH_D04G1418,andGH_D04G1459were significantly higher in Xinluzao 61 cultivars than in Xinluzao 50 cultivars following aphid infection(Table S5).Furthermore,the sequencing analysis of these candidate genes detected a 10 bp insertion inGH_D04G1418genomic DNA of Xinluzao 50 in exon-4 of the predicted cDNA.Also,a frameshift that could result inGH_D04G1418′s loss-of-function mutation was generated(Fig.S2A).Comparison of amino acid sequences revealed that the 280th amino acid ofGH_D04G1418in Xinluzao 50 changed and the translation ended in S300 (Fig.S2B).However,structural variation and SNPs of the other four candidate genes were not found between the DNA sequences of Xinluzao 61 and Xinluzao 50(Figs.S3-S6).Therefore,GH_D04G1418can be used as a candidate gene for further research.
Sequence alignment and conservative domain prediction of proteins showed thatGH_D04G1418is a homolog of theArabidopsisgeneAtLAC4,which has the same conserved domain as 17Arabidopsislaccase genes.Phylogenetic tree analysis also demonstrated thatGH_D04G1418andAtLAC4grouped together(Fig.S2C).In addition,tissue expression analysis revealed thatGH_D04G1418was mainly expressed in stems (Fig.S2D).Previous studies have shown thatGH_D04G1418is systematically namedGhLAC4-3[17].Therefore,we have adopted this nomenclature.
3.3.GhLAC4-3 positively correlates with resistance to aphid in upland cotton via expression analysis
To examine the function ofGhLAC4-3in cotton’s defense against aphids,VIGS was performed on Xinluzao 61.Twelve days after TRV:GhCLA1 was injected into cotton cotyledons,the newly developed true leaves and stems of cotton showed albinism,which showed that the VIGS system was effective (Fig.1C).In parallel experiments,the expression level of the target geneGhLAC4-3in the VIGS-treated plants was assessed using RT-PCR and qRT-PCR.It was found that the relative expression level ofGhLAC4-3was reduced by about 60% in the silenced plants compared to the control plants (Figs.1D,S7).
To determine the tolerance ofGhLAC4-3to pests,we conducted aphid selection and non-selective feeding experiments on young leaves of the VIGS plants.The results showed that the number of aphids on the silent plant was higher than that on the control plant(Fig.1E-H).These findings indicate that a reduction inGhLAC4-3gene expression level increases the susceptibility of cotton to aphids.
3.4.GhLAC4-3 participates in lignin biosynthesis of upland cotton
To determine whetherGhLAC4-3is involved in lignin biosynthesis,phloroglucinol HCl staining and content determination of lignin were performed on the stems and young leaves of the VIGS plants.It was found that the intensity of red staining in young leaves and stems of TRV:GhLAC4-3 plants was higher than that of TRV:00 plants(Figs.1I,S8A),and the same result was obtained using lignin content determination (Figs.1J,S8B).Based on these results,we detected the expression level of genes related to lignin biosynthesis.The expression levels of six lignin biosynthesis-related genes,namelyGhCCR1,GhCCR2,GhC4H1,Gh4CL,GhCOMT,andGhCAD,located upstream ofGhLAC4-3,were decreased significantly in TRV:GhLAC4-3compared to in TRV:00 plants (Fig.1K).These results indicate thatGhLAC4-3participates in the biosynthesis of plant lignin by regulating the expression level of genes related to lignin biosynthesis.
4.Discussion
Aphid is one of the main pests in cotton production.Therefore,understanding the genetic mechanism of aphid resistance is the most effective and sustainable way to reduce aphid damage and increase cotton yield long term.Compared with whole genome sequencing,ddRAD-seq is a technique that can capture numerous SNPs in a short time and at a lower cost [18].It has the advantage of rapidly discovering SNPs in a large number of individuals and generating genotype data[19].In this study,to detect the genomic regions associated with aphid resistance in cotton,ddRAD-seq was performed on 83 extreme samples of the F2population,and a main effect QTL on ChrD04,including 460 genes,was obtained by genome-wide linkage analysis.Then,five genes were identified as candidate genes associated with aphid resistance by further screening by RNA-seq and gene function annotations.The expression levels of these genes in resistant plants were significantly higher than those in sensitive plants under aphid infestation.These suggest that they may play an important role in cotton defense against aphids.
Cell wall modifying enzymes present in aphid saliva can help the mouthparts absorb plant nutrients by disrupting cell wall polymers during feeding.Lignin in the cell wall is difficult to degrade and is considered a vital part of the plants’ defense response because it is difficult for aphid mouthparts to penetrate the cell wall[20,21].For example,inChrysanthemum morifolium,CmMYB15andCmMYB19combine with the promoter element AC of the lignin biosynthesis gene to regulate that gene,thereby enhancing the resistance of chrysanthemums to aphids [22,23].In wheat (Triticum aestivumL.),the activities of apoplastic peroxidase and β-1,3-glucanase in resistant cultivars were significantly higher than those in susceptible cultivars once infected by wheat aphids.However,the peroxidase activity was associated with lignin crosslinking in the cell wall [24].In this study,we identified a new insect-resistance gene,GhLAC4-3,which belongs to the laccase gene family.The VIGS analysis showed thatGhLAC4-3affected lignin accumulation in the cell wall by regulating the expression of critical genes involved in lignin synthesis.Therefore,GhLAC4-3has been potentially associated with aphid resistance.Furthermore,these findings provide unprecedented insights into the defense mechanisms of cotton laccase genes against aphids.
CRediT authorship contribution statement
Qiushuang An:Formal analysis,Writing-original draft,Project administration.Zhenyuan Pan:Writing -review &editing.Nurimanguli Aini:Investigation.Peng Han:Visualization.Yuanlong Wu:Writing-review&editing.Chunyuan You:Supervision,Resources.Xinhui Nie:Conceptualization,Writing-review&editing,Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Corps Science and Technology Innovation Talent Plan(2021CB028),the Shihezi Science and Technology Research Key Field Science and Technology Research Project (2022NY01),the Fifth Division Science and Technology Plan Project (2021NY02),and the Young and Middle-aged Leading Talent Plan (2020CB017).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.03.006.
- The Crop Journal的其它文章
- Genomic location of Gb1, a unique gene conferring wheat resistance to greenbug biotype F
- iBP-seq: An efficient and low-cost multiplex targeted genotyping and epigenotyping system
- RPNet: Rice plant counting after tillering stage based on plant attention and multiple supervision network
- SimET: An open-source tool for estimating crop evapotranspiration and soil water balance for plants with multiple growth cycles
- SPP-extractor: Automatic phenotype extraction for densely grown soybean plants
- Improving maize growth and development simulation by integrating temperature compensatory effect under plastic film mulching into the AquaCrop model

