Diffusion of nucleotide excision repair protein XPA along DNA by coarse-grained molecular simulations?
Weiwei Zhang(張偉偉) and Jian Zhang(張建)
National Laboratory of Solid State Microstructures,School of Physics,Collaborative Innovation Center of Advanced Microstructures,Nanjing University,Nanjing 210093,China
Keywords: nucleotide excision repair,XPA,one-dimensional diffusion along DNA,molecular simulation
1. Introduction
Nucleotide excision repair(NER)is the central DNA repair pathway widely observed in eubacteria, eukaryotes and archaea. It is responsible for the removal of DNA lesions caused by UV irradiation,chemical mutagens,and other stress damages.[1–5]The abnormalities of NER can lead to the xeroderma pigmentosum(XP)disease,which is featured by sensitivity to sunlight and high risk of developing skin cancers.[6,7]XPA (xeroderma pigmentosum complementation group A) is a key protein in NER,and mutation of XPA gene can lead to severe disease symptoms.[6,8]It has been showed that the XPA may play multiple roles in NER.[9–12]For example,in addition to preferentially binding to various types of DNA lesions and distorted substrates,[10,13]XPA has also been found to interact with several NER related proteins during the damage recognition and verification steps,acting as a scaffold.[11,14]
The human XPA has the length of 273 amino acids and is composed of three domains, including the intrinsically disordered N-terminal region(N-region),the central DNA-binding domain(DBD),and the intrinsically disordered C-terminal region (C-region) (see Fig. 1). The flexibility of the two disordered regions were suggested to promote the binding of XPA to several partners,[15–19]while the central globular DNAbinding domain was found to bind with DNA and interact with numerous NER proteins as well.[20]
Due to its biological importance in NER, XPA has received extensive attention.[7,21]Although previous investigations mostly focused on the interactions of the XPA at the DNA lesion site during the damage recognition and verification steps, recent experimental study showed that detecting the DNA lesions may also involve the one-dimensional diffusion of the XPA at the non-damage site of DNA.[22]Therefore,more detailed investigations for the molecular dynamics of the XPA diffusion along the non-damage site of DNA are important for deeper understanding the biological function of XPA in the NER.
Although all-atom detailed molecular dynamics (MD)simulation has been widely used to directly simulate the details of the inter-molecular interactions involved in the protein-DNA recognitions, it is still extremely timeconsuming to study the whole diffusion process of proteins along DNA.[23–26]Recently, the combination of the atomic-interaction-based coarse-grained model(AICG2+)for proteins[27,28]and the three-site-per-nucleotide (3SPN.2C)model for DNA[29]has shown great success in studying the dynamical and structural features of the protein-DNA interactions.[30–33]Here,based on these models,we performed extensive coarse-grained molecular simulations to study the diffusion dynamics of XPA along the DNA.From our simulations,we found that XPA diffuses along DNA with the DNAbinding residues similar to that identified from crystal structure. It was also found that the diffusion of XPA along DNA is achieved through combination of one-dimensional and threedimensional mechanisms. At low salt concentrations,the onedimensional diffusion with strong rotational coupling is the dominant mechanism. At higher salt concentrations, the diffusion by three-dimensional mechanism becomes more probable. Meanwhile,the one-dimensional diffusion of XPA along DNA displays sub-diffusive behaviour at wide range of salt concentrations. In addition, the simulation showed that the XPA becomes more extended in the presence of DNA or at high salt concentration,suggesting that both DNA binding and increasing salt concentration may promote the binding of other target proteins to XPA by increasing the exposure extent of the binding sites.
2. Model and method
2.1. Coarse-grained model of protein-DNA system.
Throughout this work, to model protein (XPA), the structure-based coarse-grained model was used, where each amino acid in protein was coarse-grained to a single bead located at Cαposition. The interactions between protein beads include AICG2+ potentials and Debye–H¨uckel-type electrostatic interactions. The AICG2+ energy function is given byVAICG2+=Vlocal+VGo+Vexv, whereVlocal,VGO, andVexvare local potentials, non-local structure-based potentials, and excluded volume potentials respectively. The details of these terms can be found in Refs. [28,34,35], and the similar energy function has also been successfully used in the studies of protein folding,enzyme catalysis,chromatin remodeler and nucleosome dynamics in recent works.[27,36–40]Note that the structure-based interactions were omitted for the disordered regions in XPA, and effects of salt concentration are implicitly modeled in the Debye–H¨uckel-type interactions with the details displayed as follows:

whereλDis the Debye length,ε(T,C) is the dielectric constant,andIis ionic strength.
To model dsDNA,the 3SPN.2C model,[29]developed by de Pablo’s group, was used. In this model each nucleotide is simplified to three CG particles, representing the phosphate group (P), sugar (S), and base (B). The interactions among these particles are composed of structure-based local potentials,base pairing,intrastrand base-stacking,interstrand crossstacking, excluded volume effects, and Debye–H¨uckel-type electrostatic interactions. The partial charge of the phosphate beads was set as?0.6e to consider the effect of local counterion condensation effect.[31,41]The 3SPN.2C model model is able to capture several properties of dsDNA and has been widely used to study the DNA-protein interactions.
For the protein-DNA interactions,only the excluded volume effects and the electrostatic interactions were considered. As the electrostatic interaction plays a dominant role in protein-DNA interactions, we paid special attention to this term in the study of protein-DNA binding. For the structured motif in proteins, the partial charges of the CG beads were parametrized by the RESPAC method,[42]which can provide more accurate description of the electrostatic potential. In describing the electrostatic interactions between the protein and DNA,the partial charge of the phosphate bead was set as?1.0,following the work in Ref.[31].
2.2. Simulation details
The model for XPA was built based on the available crystal structure(PDB entry:6j44[8])with missing residues added by modeller9.23.[43]The sequence and structure of XPA are shown in Figs. 1(a), 1(b), and 1(c). For DNA, two random sequences with different lengths were prepared: R100, R150(the sequences and the structures are shown in Figs.1(d)and 1(e)). In our work, the reference structures for DNA were built with the 3DNA package.[44]For the protein-DNA system, the initial structures for DNA and protein in all the simulations were separated and placed randomly. During production runs,the center of mass(COM)of DNA was restrained to the original position and COM distance between XPA(DBD)and DNA was restrained to be smaller than 250 °A.The ionic strength was set to 50, 150, 250, 350, and 450 mM. At each ionic strength,24 simulations were carried out independently,each lasting for 5×108MD steps for XPA-R100 system and over 4×108MD steps for XPA-R150 system,respectively. To characterize the structure features of XPA in the absence of DNA,we also performed several simulations with cumulative length of approximately 1×109MD steps. For all the simulations,we recorded the coordinates of XPA and DNA at every 1×104MD steps. The first 2×107frames were omitted in the calculations of distributions during the posterior analysis.
All the CG simulations were performed by the CafeMol3.2 package.[34]The simulations were conducted by Langevin dynamics with friction coefficientγ=0.02 at temperatureTof 300 K.The structures were visualized by using pymol.[45]
2.3. Analysis
To characterize the search modes of XPA on DNA, we defined a DNA recognition motif consisting of the residues 140, 141, 142, 151, 179, 207, 211,[46]which forms contacts with DNA in the crystal structure. Following the approach used in Ref. [41], the diffusion is considered as sliding if at least five of these residues maintain staying within 15 °A from the DNA.Correspondingly,the diffusion is considered as three-dimensional diffusion if all of the seven residues in the recognition motif leaves the DNA surface by at least 25 °A.In comparison,the protein is considered to perform the hopping dynamics if these residues are 15 °A–25 °A away from DNA.Based on above criteria, we classified both sliding and hopping as one-dimensional diffusion mechanism following the work in Ref[41].
To describe the location of DBD searching site on DNA during one-dimensional diffusion,we used the mean position of the nucleotides that closest to the recognition motif(RM):
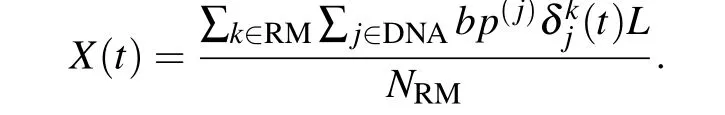
In the above definition,tis simulation time,Lis 3.4 °A (one bp along the strand), RM is the recognition motif, composed of the residues 140,141,142,151,179,207,and 211,NRMis the number of residues in recognition motif,bp(j)is the base pair index of particlejin DNA;δkj=1,ifjis the DNA bead closest to the protein beadk;otherwiseδkj=0.[31]
The protein bead and DNA bead form a contact if their distance is less than 7 °A.Additionally,we defined several local coordinate axes, following the procedure in Ref. [47], to characterize the rotational and translational motions of XPA around DNA.

Fig.1. Sequence and structure of the XPA and DNA studied in this work. (a)Domain architecture of XPA.(b)The sequence of XPA protein with each domain shadowed by the same color as in panel(a). The secondary structure features of DBD domain is also shown. (c)The available crystal sturcture of DBD is shown in cartoon representation and the color scheme for secondary structures is the same as in panel(b).The disorderd regions are represented by dotted lines with the same color as in panel(a). The key DNA-binding residues(residue 140,141,142,151,179,207,211)are shown in ball-and-stick representation and colored differently. The sequences(d)and structures(e)of DNA in this study.
3. Results
3.1. XPA binding and diffusion along DNA
Firstly, to quantify the structural properties of nonspecific interaction interface between XPA and DNA, based on two random DNA sequences with different lengths(R100,R150), we performed MD simulations for the XPA binding to the nonspecific dsDNA site at various salt concentrations and calculated the probabilities of contact formation between the DNA and each residue of the XPA at various salt concentrations. As shown in Fig. 2, there is a striking similarity in the landscape of contact probability between R100 and R150,indicating the conserved binding interface of XPA regardless of properties of DNA.Additionally,for both R100 and R150,one can see that, besides the DBD, residues in the N-region also have high contacting probabilities with DNA,which suggests that the N-region can be involved in the XPA–DNA interactions during the searching process of damage sites. Meanwhile, the results clearly show that increasing salt concentrations tends to decrease the contact probabilities, but without altering the peak locations of the contact probability distributions, which may suggest that the XPA uses the same set of residues for DNA binding at wide range of salt concentrations.Particularly, the residues with high contact probabilities have large overlap with the DNA-binding residues observed in crystal structure of the XPA–DNA complex,[46]demonstrating that the DNA binding surface in the DBD is conserved in the specific and non-specific binding. Such behaviour of maintaining binding interface was also observed in other DNA-binding proteins,[47]and was suggested to facilitate the search of the cognate site on DNA.[48,49]
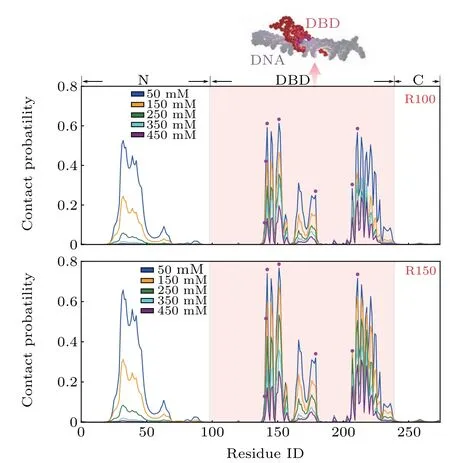
Fig. 2. Contact probabilities of the XPA residues with DNA at various salt concentrations. Residues in the DBD domain are shaded by pink backgound.The key DNA-binding residues in the crystal structure are highlighted by magenta circles. The result with R100(R150)is shown in the upper(lower)panel and a representative structure with DBD bound to R100 is also displayed.
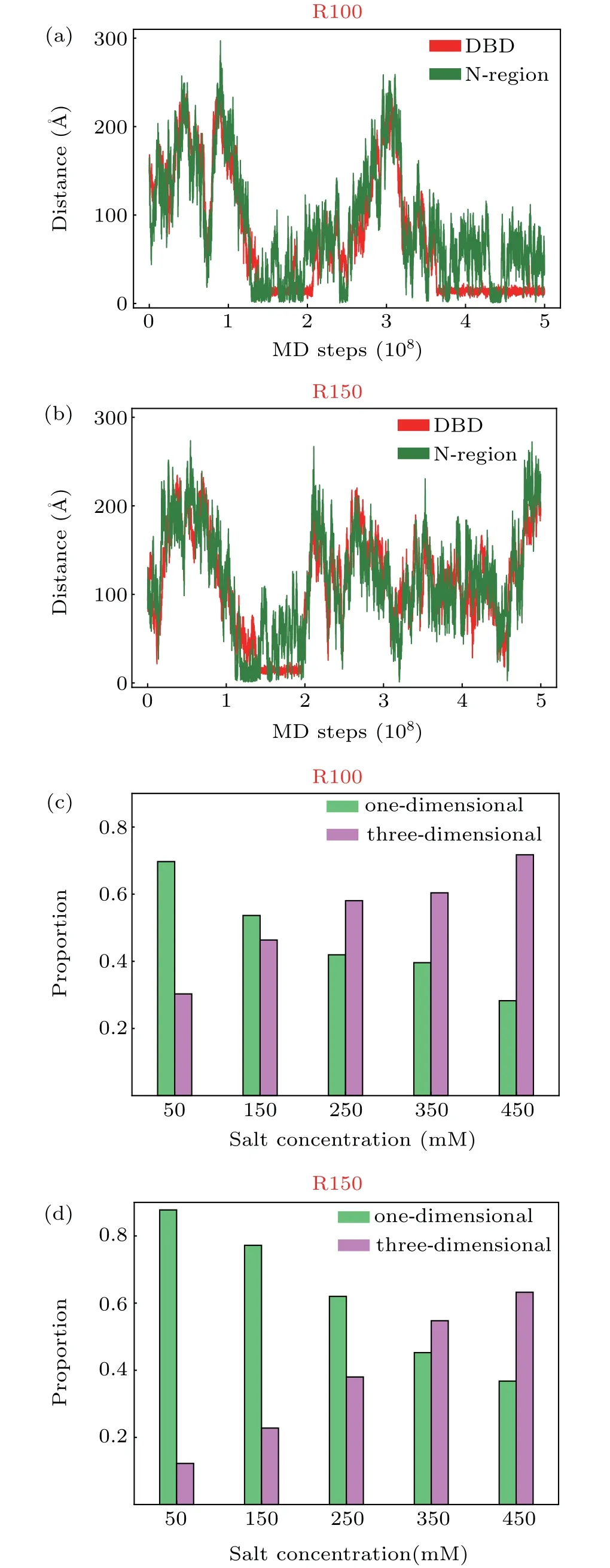
Fig.3. (a)and(b)Time series of the distance between the COM of N-region and DNA surface(green)and that between the COM of DBD and DNA surface (red) at 250 mM with sequence R100 (a) and R150 (b). When calculating the distance between the COM of N-terminal and DNA surface, only the residues ranging from 30 to 50, which have high probabilities of forming contacts with DNA, were considered. (c) and (d) Probabilities of onedimensional diffusion(green)and three-dimensional diffusion(pink)mechanism at different salt concentrations with sequence R100(c)and R150(d).
Besides the properties of binding interface,we also characterized the binding and diffusion process of XPA along DNA. Considering the relatively higher probabilities of contact formation with DNA for the residues in the DBD and the N-region, we plotted the time series of the distances between the above two protein segments and the DNA surface to illustrate the binding dynamics. The centers of mass(COM)of the DBD and that of the residues 30–50 were used to represent the positions of the DBD and the N-region relative to DNA surface. From Figs.3(a)and 3(b), one can clearly see the attachment and detachment processes of the two XPA segments.Such attachment and detachment processes are consistent with experimental observation.[22]However, it is worthy to note that,because of the limited accessible time in our simulation,the repetition of attachment and detachment is only observed sporadically at relative high salt concentration (Fig. 3(a)).Additionally,inspection of trajectory of these two protein segments demonstrated that the attachment/detachment of the DBD and N-region are correlated, lacking of the anchoring and binding mechanism mediated by disordered region as typically observed in other DNA-binding proteins containing disordered regions.[50]After the simultaneous detachments of the N-region and the DBD,the whole XPA performs threedimensional diffusion and then rebinds to the DNA(Fig.3(a)).For many DNA binding proteins, rapidly finding their specific binding sites on DNA in response to external stimulus is vitally important. Thus, in addition to the random threedimensional diffusion driven by thermal diffusion,the protein also uses other searching mechanisms, such as sliding,[51–53]hopping or intersegmental transfer.[54]During sliding,the protein can tightly bind to DNA and diffuse along the DNA helix using the recognition region,[55]in which electrostatic interaction between protein and DNA plays dominant role. Whereas in the hopping diffusion process,the electrostatic interactions are much weaker, which allows the protein to diffuse more freely along the DNA surface and accelerates diffusion by skipping several DNA bases.[41]Unlike three-dimensional diffusion, in which the protein can diffuse far away from DNA,the protein remain staying close to the DNA during the sliding and hopping,which tends to significantly reduce the conformational space during target site search and therefore accelerate the searching of the target site in the DNA.[51]Previous studies showed that the combination of these different search modes is needed to achieve the maximal target-search efficiency.[54,56]Therefore, we also analyzed the probabilities of the one-dimensional diffusion mechanism and threedimensional diffusion mechanism (criteria about classifying different mechanisms can be found in Model and Method) at various salt concentrations for sequence R100 and R150, respectively (Figs. 3(c) and 3(d)). From these results, one can see that, increasing salt concentration can decrease the probability of one-dimensional searching mechanism and increase the three-dimensional searching mechanism for each DNA sequence. As shown in the Debye–Hc¨ukel model, the effect of salt concentration is mainly achieved by changing the length of Debye radius. With the increasing of salt concentrations,the screening effect of the implicit salt ions increases, leading to the decrease of electrostatic interactions. Such results again suggest the dominate role of electrostatic interaction in the XPA–DNA binding.
Considering the important role of one-dimensional diffusion, we then analyzed the diffusion kinetics and calculated the mean square displacement(MSD)of the one-dimensional DBD translational motions along the DNA (MSD(?t) =〈(X(t+?t)?X(t))2〉, where〈〉represents the time average)as a function of time difference ?tat various salt concentrations, and results are shown in Fig.4. The fitting by a power law function MSD(?t) =K?tαwere also shown in the inset by log–log scale. From our results, one can clearly see that the MSD of XPA (DBD) increases upon increasing salt concentration during one-dimensional diffusion for both R100 and R150. The fitting of the diffusion kinetics by the above power law function for R100 and R150 both give theα<1.0,which suggests sub-diffusive motion of DBD along DNA(inset figure in Fig. 4) at wide range of salt concentrations and may facilitate the search of target DNA sequences as suggested by other works.[57–59]Furthermore,this kind of sub-diffusion may be related to the diverse strengths of DNA–XPA interactions,which may be ascribed to the variations of the DNA conformations and the fluctuations of the DNA-protein binding.This kind of connection is consistent with those in many other sub-diffusive dynamics,[59–69]and can be described with continuous time random walk(CTRW)model.[61,69]Note that the exponents of the sub-diffusive dynamics are almost invariant at different salt concentrations for each sequence. This observation suggests that the distributions of the diverse strengths are insensitive to the variations of salt concentrations, since the conformational fluctuations are intrinsic for the the DNAprotein system.This result bring us more insights on the DNAprotein interactions.

Fig.4. Mean square displacement(MSD)of the one-dimensional XPA diffusion along the DNA(R100: upper panel,R150: lower panel)as a function of simulation time at different salt concentrations.The results of power function fitting α are also shown in the inset.
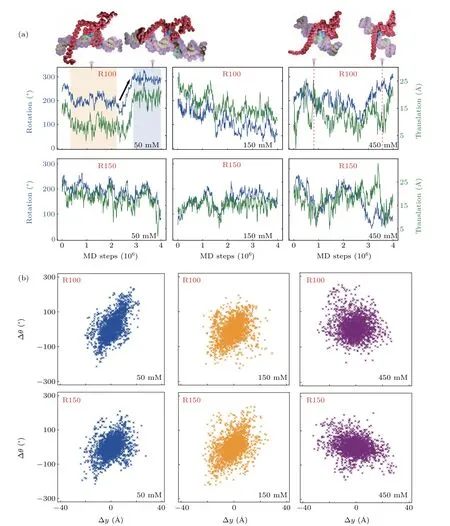
Fig.5. Translational–rotational coupling in the one-dimensional diffusion of XPA along DNA.(a)Representative trajectory fragments of rotation(θ) and translation (y) of XPA along R100 (R150) at the salt concentrations of 50 mM, 150 mM, and 450 mM are shown in upper panel (lower panel). Representive structures with DBD bound to DNA at 50 mM and 450 mM are displayed. (b)The correlation plots between ?θ and ?y of XPA at 50 mM,150 mM,and 450 mM(R100: upper panel,R150: lower panel).
For DNA-interacting proteins, several studies have suggested that diffusion along the DNA helical path is required to achieve efficient target search.[55,70]Meanwhile,the rotationcoupled sliding was proposed to be the general mechanism for the one-dimensional search of protein on DNA.[71]Therefore,we also investigated whether the one-dimensional diffusion of XPA is along helical pitch(rotation-coupled)or not(rotationuncoupled), and the results are shown in Fig. 5. To more appropriately describe this dynamical process,we defined several local axes following the Ref.[47]. The coordinates of the seven residues in the DNA-binding motif mentioned above were used to calculate the rotational angle and translational position along the DNA helical axis. In Fig. 5(a), we plotted the time series of rotational and translational motions of DBD along the R100 and R150 at various salt concentrations during the one-dimensional diffusion. From these trajectories, one can see that the protein can diffuse randomly along DNA at these different salt concentrations. At relatively low salt concentrations(50 mM,150 mM),where the electrostatic interactions are strong, the one-dimensional diffusion occurs dominantly by the rotation-coupled mechanism. While at high salt concentration(450 mM),the electrostatic interactions become weaker, which makes the protein move more freely along the DNA axis, leading to weak correlation between translational and rotational motions. To more clearly characterize the relationship between rotation and translation during one-dimensional diffusion, we also showed the rationaltranslational correlation plot in Fig.5(b)for all trajectories at three different salt concentrations. From these results,one can see an obvious correlation between the rotation and translation at low salt concentrations (50 mM), while it disappears at relatively high salt concentration (450 mM). Particularly,the rotational–translational correlation is still significant at the physiological relevant salt concentration(150 mM).
3.2. Structural properties of XPA
In addition to the diffusion features during this dynamical process, the structural properties of XPA were also studied,since its structure dynamics will surely affect the binding of XPA with other binding partners. The results for these two different DNA sequences are shown in Fig. 6. From our results, one can clearly see that both the DNA binding and varying salt concentration can change the structural properties of XPA and the difference in the effects of R100 and R150 on protein structure at each salt concentration is negligible.In detail, as shown in Fig. 6(a), at low salt concentration(50 mM), due to the strong intra-molecule electrostatic interactions in XPA, these two disordered regions pack against the DBD, leading to small radius of gyration (Rg) of XPA(less than 30 °A) in the absence of DNA, but it increases a lot (around 40 °A) upon binding DNA, which may be caused by the occupancy of intra-molecule interacting sites by DNA.While at high salt concentration (450 mM), the electrostatic interactions between intra and inter-molecule is decreased significantly and consequently the intra and inter molecule motions are more freely, leading to largeRgof XPA with or without DNA, and the effects of DNA binding is much deprecated as well (Fig. 6(b)). What is more, in Figs. 6(c)and 6(d), we also plotted the distribution of contact number between DBD and the disordered regions. In accordance with the above analysis, there are a large number of contacts between N(C)-terminal and DBD,and it decreases much unpon binding DNA at low salt concentration (Fig. 6(c)),while at high salt concentration,the contact number decreases significantly with or without DNA.Notably,the contact number in the absence of DNA is still less compared with binding DNA.From the analysis of the contact number,it becomes clear that both increasing salt concentrations and DNA binding can lead to the decrease of contact number,indicating more extended conformation of XPA. Based these observations, considering the dominate role of electrostatic interactions in describing the XPA–DNA interactioins and N(C)–DBD interactions in XPA, we can conclude that, both the DNA and salt concentration can act as modulating factors of the XPA structure.
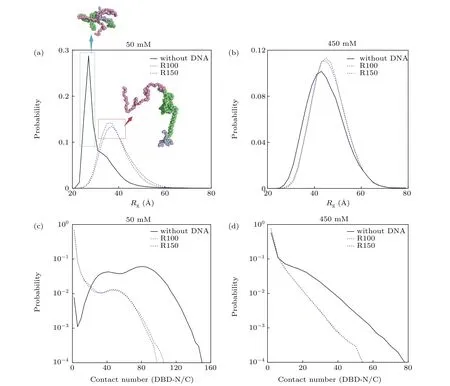
Fig.6. Effect of DNA binding and salt concentration on the conformation of XPA.(a)and(b)Rg distribution of XPA with(R100: blue,R150:purple)and without(black)DNA binding at the salt concentrations of 50 mM(a)and 450 mM(b). The representative structures of XPA are also shown,and the color scheme for domains is the same as in Fig.1(a). (c)and(d)Distribution of contact number formed between the DBD domain and the two disordered regions of XPA with(R100: blue,R150: purple)and without(black)DNA binding at the salt concentrations of 50 mM(c)and 450 mM(d).
Nowdays, more and more studies have found that the conformation of protein is very sensitive to external stimulus, which play an important role in modulating the conformation of protein involed in various biological activities, like a cascade of reactions or signal transduction in cells,etc.[72–76]Interestingly, previously studies have also found that both salt concentration and binding DNA could affect the protein structure.[77–82]Hence,toghter with the findings that the XPA needs to cooperate with other binding partners during the NER pathway, and multiple binding sites for these partners are located at both DBD and disordered regions(N-terminal and Cterminal),[16,17,20,83–86]we can speculate that both the DNA itself and the salt concentration play a role in modulating the NER and at high salt concentration,the protein predominately adopt an extended conformation, ready for interacting with other partners,while at relative low salt concentration,the disordered regions tend to pack tightly with the DBD without DNA, however, upon DNA binding, the N(C)–DBD contacts are much reduced,which can lead to increased exposure of the binding sites in XPA for recruiting other partner proteins.
4. Conclusion
Here, we used a coarse-grained model to study the dynamical process of XPA diffusion along two different DNAs.Our results showed that the diffusion properties of XPA along these two DNAs are pretty similar,which indicates the robustness of dynamical behaviour of XPA along DNA.From more detailed perspective,our results showed that the XPA diffuses along the DNA with interface similar to the recognition motif found in the crystal structure, which may promote the search efficiency for the DNA target. Additionally, we also characterized the diffusion process of XPA along DNA. The XPA diffuses along DNA using combination of one-dimensional and three-dimensional search mechanisms,and increasing salt concentration can decrease the proportion of one-dimensional mechanism. During one-dimensional diffusion, the protein displays subdiffusive motions, which has also been observed in several biological system. Additionally, the correlation analysis of rotational and translational motions of DBD along DNA also showed that at relatively low salt concentration,the one-dimensional DBD translational motion is coupled to rotation,that is to say,sliding along helical pitch. Whereas at high salt concentration,the correlation disappears. All these results suggest that, like many other DNA binding proteins,[54,87,88]XPA maintains its position and orientation with respect to targeted DNA base pairs during one-dimensional diffusion at relatively low salt concentration, ensuring the efficient target search.
In addition to binding to DNA, XPA has been found to act a key scaffold protein, interacting with many proteins involved in NER.[86]Therefore,we analyzed the structural features of XPA at various salt concentrations,in the presence and absence of DNA, respectively. The results showed that both DNA and salt concentration can modulate the protein structure. At low concentration,DNA binding can significantly increase theRgof protein and decrease the contact number between DBD and disordered regions,indicating more extended conformation of XPA.While at high concentration, the weak interaction between protein and DNA and intra-molecular interaction will let the protein to be in the extensible state as well. Based on these discussions, we can speculate that even at relatively low salt concentrations,when XPA bind to DNA,the contacts between DBD and other disordered regions are reduced, which tends to increase the exposure extent of the binding sites for other repair proteins,promoting the scaffold function of the XPA.
Acknowledgment
The authors thank the insightful discussions with Jun Wang,Wenfei Li,and Wei Wang.
- Chinese Physics B的其它文章
- Physical properties of relativistic electron beam during long-range propagation in space plasma environment?
- Heterogeneous traffic flow modeling with drivers’timid and aggressive characteristics?
- Optimized monogamy and polygamy inequalities for multipartite qubit entanglement?
- CO2 emission control in new CM car-following model with feedback control of the optimal estimation of velocity difference under V2X environment?
- Non-peripherally octaalkyl-substituted nickel phthalocyanines used as non-dopant hole transport materials in perovskite solar cells?
- Dual mechanisms of Bcl-2 regulation in IP3-receptor-mediated Ca2+release: A computational study?

