Sensitive and specific detection of circuIating tumor ceIIs promotes precision medicine for cancer
Qin-Qin Huang, Xing-Xiang Chen, Wei Jiang, Shui-Ling Jin, Xing-Yu Wang, Wei Liu, Shi-Shang Guo,Jian-Cheng Guo, Xing-Zhong Zhao
1Department of Otolaryngology, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou 450014, China.
2Center for Precision Medicine of Zhengzhou University, Zhengzhou 450052, China.
3Division of Life Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong.
4Department of oncology, the first affiliated hospital of Zhengzhou university, Zhengzhou 450052, China.
5Faculty of Health Science, University of Macau, Avenida da Universidade, Taipa, Macau, China.
6Key Laboratory of Artificial Micro- and Nano-Structures of Ministry of Education, School of Physics and Technology, Wuhan University, Wuhan 430072, China.
Abstract Circulating tumor cells (CTCs) have the potential to provide genetic information for heterogeneous tumors, which may be useful for monitoring disease progression and developing personalized therapies. However, the isolation of CTCs for molecular analysis is challenging due to their extreme rarity and phenotypic heterogeneity, which hinders the transformation of CTCs into traditional clinical applications. In order to achieve clinically significant CTC detection,devices utilizing novel microfluidics and nanotechnology have been developed to achieve high sensitivity and specificity capture of CTCs. In this review, we discuss these newly developed devices for CTC capture and molecular characterization for early diagnosis and determining ideal treatment regimen to better manage these cancers clinically.In addition, the potential prognostic values of CTCs as treatment guidelines and that ultimately contribute to realize personalized treatment are also discussed.
Keywords: Circulating tumor cells, sensitivity, treatment, precision medicine
INTRODUCTION
The main cause of cancer-related mortality is cancer metastasis. During this greatly complicated and multistage disseminative process, tumor cells (the seeds) detach from primary roots, shed into blood and lymph circulation, undergo the immune attack and shear stress, travel to preferable metastasis soil, and eventually seed and proliferate to develop metastases. On their way to the potential organs, these circulating tumor cells (CTCs) undergo epithelial-mesenchymal transition (EMT)[1], thereby resulting in enhanced motility and migratory ability that facilitates vasculature invasion. Upon reaching a suitable niche, the CTCs undergo mesenchymal-epithelial transition (MET), subsequently reacquiring the stem cell properties and reactivating proliferative capability to colonize at metastatic sites[2]. In order to prevent and surveil the development of metastasis disease, especially metastasic carcinoma, the detection and characterization of CTCs are of great interest to scientists. CTCs were first detected in cancer patient in 1869 by Australian physician namedThomas Ashworth. In the past couple of decades, numerous studies have suggested that the presence of CTCs in the blood of cancer patients has the clinical potential as a noninvasive diagnosis marker and a prognosis indicator known as a “l(fā)iquid biopsy” to replace traditional invasive biopsy, whilst also facilitating technical advances for detection of CTCs.
CTC analysis has a variety of clinical applications, including real-time non-invasive monitoring of CTCs as biomarkers for new cancer therapies as well as identifying new potential therapeutic targets that directly inhibit cancer metastasis. Although the potential applications of CTC analysis appear to be very promising,due to the rarity (one CTC per billion hematologic cells) and heterogeneity (e.g., differences in morphology and gene expression) of CTCs in the blood of cancer patients, there are few commercially available techniques for clinical use. High sensitivity and specificity of CTC detection methods thus have a great impact on improving patient outcomes. Therefore, currently available technologies for CTC detection have become increasingly more sensitive and reliable, with the goal of early cancer detection and thus successful cancer treatment. An important new direction in this field is the development of devices and materials that provide information beyond CTC enumeration. Integrated devices allow for the separation of heterogeneous CTCs to facilitate a more in-depth characterization of these cells (e.g., phenotypic and molecular profiling) to develop a personalized treatment plan. Nanomaterials and microfuidic-based nanotechnologies may be the most promising strategies for implementing ideal CTC capture devices to replace traditional tools, primarily relying on their small size, high throughput capacity and large surface-to-volume ratio to solve the problem of CTC heterogeneity[3]. In this review, we will provide an overview of current CTC isolation strategies and molecular characterization with brief insights into the potential clinical implications of CTC capture and characterization.
SENSITIVE CTC ISOLATION METHODS
CTCs may have the potential to predict the disease progression in patients with early-stage or advanced cancer,even before the formation of primary tumor. However, the extreme rarity of CTCs in blood poses a challenge for detecting CTCs from blood; for example, one study indicated that only 1.43% of 350 metastasis cancer patients had ≥ 500 CTCs/7.5 mL blood[4]. Inability to draw large volume of blood from patients highlights the need for improved CTC isolation methods to achieve sensitive and specific CTC detection in small sample volumes. CTC isolation methods have been developed based on either biological (surface antigen, cytoplasmic protein, invasion capacity,et al.) or physical (size, density, deformability and charge,et al.) properties of tumor cells. We discuss the most popular technologies and latest advances in the following sections [Figure 1].
Detection of CTCs based on their biological properties
Immunomagnetic beads-based isolation
The most widely used enrichment method is a positive selection method based on the epithelial cell adhesion molecule (EpCAM) antibodies[5-7]. So far, CellSearch System (Menarini Silicon Biosystems, Italy)is the first and also the only one being up to the standard of US Food and Drug Administration (FDA),which consists of the CellTracks Autoprep and the CellSearch Epithelial Cell kit, integrating EpCAM based immunomagnetically enrichment, 4’,2-diamidino-2-phenylindole (DAPI) based cell nuclei staining, CD45-Allophycocyan specified leukocyte negative selection and cytokeratin 8,18,19-Phycoerythrin specified epithelial cells positive selection into an objective indicator (EpCAM+, DAPI+, CD45-, cytokeratin+) of CTC counts. In 2004, it was cleared for monitoring the outcome of therapies and optimizing clinical decision for breast cancer; later, it was also cleared for use in prostate and colorectal cancers. Through the CellSearch,which has become the benchmark for all other CTCs isolation methods, CTC counts have been associated with prognosis for progression-free survival (PFS) and overall survival (OS) in these three kinds of metastatic cancer[8-10].
Although clinical correlations have been identified, methods for large scale isolation of CTCs from peripheral blood are lacking, therefore, efforts have been focused on improving the isolation sensitivity and efficiency. Talasazet al.[11], reported a magnetic sweeping device (MagSweeper, Stanford University, Stanford)consisting of a nonadherent plastic sheath covered magnetic rod with anti-EpCAM antibody functionalized beads, allowing for a ~60% capture efficiency to target cells and a purity of 100% for HLA-A2 cells. Ephesia technology integrated anti-EpCAM functionalized self-assembled magnetic beads with microfuidics,demonstrating a capture efficiency > 94%[12]. Similar immunomagnetic platforms [Figure 2A] also included the Magnetic Sifter with magnetic pores incorporated into a microfuidic chip[13]. Moreover, compared with CellSearch system, Adna Test (Qiagen, Hannover), a highly specific immunomagnetic cell-isolation system, with its improved antibody cocktails provided an effective approach to increase the efficiency of CTCs capture and complements the CellSearch for detection of CTCs[14]. Mayoet al.[15]utilized MACS cell separation platform (Miltenyi Biotec) based on a mixture of cytokeratin (CK) coated magnetic beads to isolate CTCs in lung cancer patients.
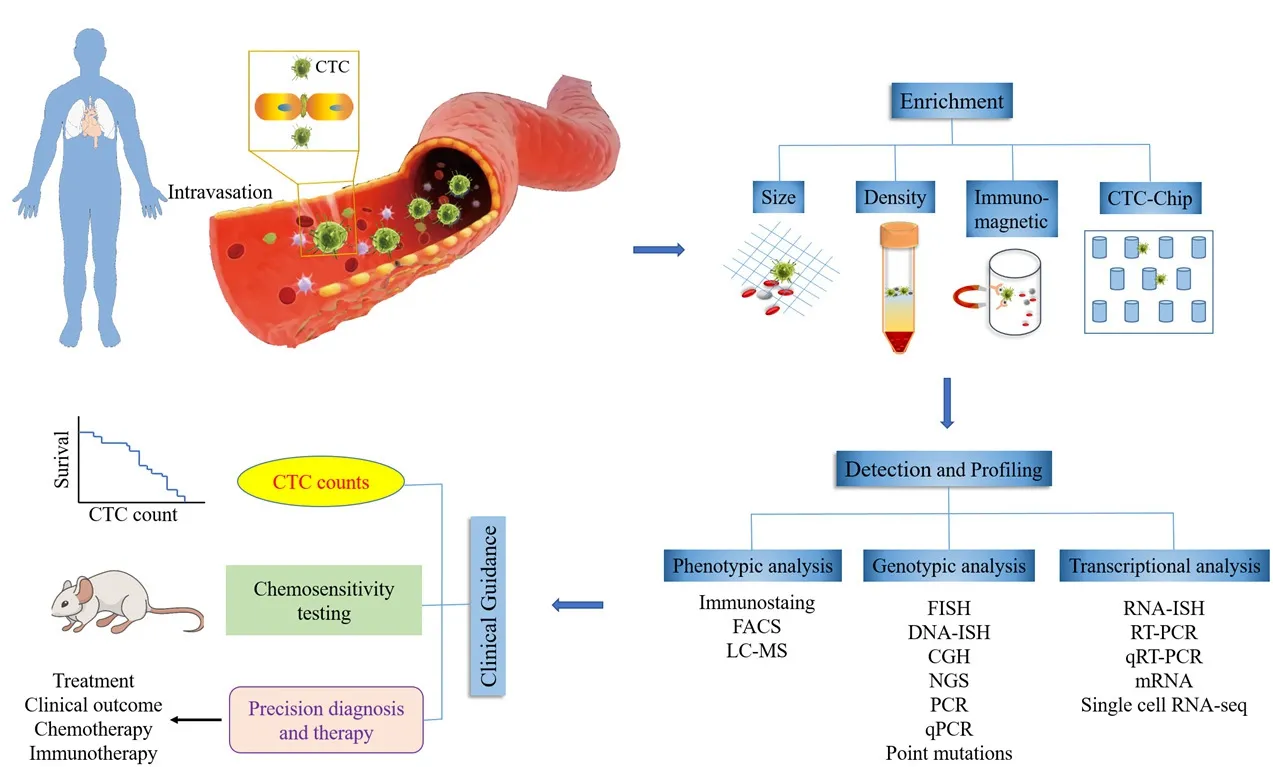
Figure 1. Illustration of current applications of CTC technologies. The CTCs exit the primary tumor and intravasate into the bloodstream.CTCs are enriched through various CTC isolation technologies such as size, density, immunomagnetic and CTC-Chip. Detection methods are utilized to detect CTCs based on phenotypic, genotypic and transcriptional analysis. The clinical applications of isolated and detected CTCs.The CTC count is associated with the potential of patient’s survival. CTCs can be good chemotherapy monitoring markers for predicting drug sensitivity/resistance in preclinical and clinical settings. CTC: circulating tumor cells; FACS: fluorescence activated cell sorting; LC-MS: liquid chromatograph-mass spectrometry; FISH: fluorescence in situ hybridization; CGH: comparative genomic hybridization; NGS: next-generation sequencing; PCR: polymerase chain reaction; qRT-PCR: quantitative reverse transcription-polymerase chain reaction
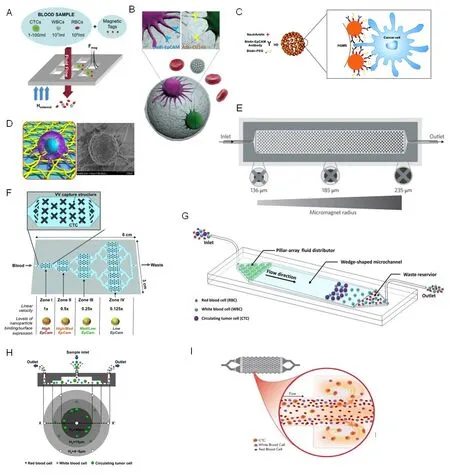
Figure 2. Strategies for CTC enrichment. A: A magnetic sifter device for CTC isolation when a magnetic field is applied. Magnetically labeled CTCs are captured at the edges of the pores, while unlabeled cells pass through the pores under fluid flow; B: dual antibodies (anti-EpCAM and anti-CD146) and biodegradable gelatin nanoparticle-coated microbeads for the capture of mesenchymal CTCs; C: a microfluidic device embedded a wedge-shaped microchamber for cell separation based on multiple biophysical properties; D: a 3D bionic cytosensor with PLGA nanofibers for CTC capture; E: vortex technology exploited for CTC isolating; F: a multizone velocity valley device for isolating heterogeneous CTCs in four different regions of varying linear velocities; G: the hollow glass microspheres with nanotopographical structures (NSHGMS) for excellent CTC isolation; H: a microfluidic device embedded a pyramid-shaped microchamber for size-based CTC separation; I: a MagRC approach for separating and in-line profiling of heterogeneous CTCs. A: Copyright Royal Society of Chemistry,2013. Reproduced with permission from reference[13]; B: Copyright Ivyspring International Publisher, 2013. Reproduced with permission from reference[27]; C: Copyright Royal Society of Chemistry, 2018. Reproduced with permission from reference[42]; D: Copyright BioMed Central, 2018. Reproduced with permission from reference[43]; E: Copyright Nature, 2017. Reproduced with permission from reference[45];F: Copyright Wiley, 2015. Reproduced with permission from reference[46]; G: Copyright Institute Of Electrical And Electronics Engineers,2018. Reproduced with permission from reference[52]; H: Copyright Springer, 2018. Reproduced with permission from reference[53]; I:Copyright Wiley, 2018. Reproduced with permission from reference[54]
However, a number of studies have shown that the levels of CTCs estimated by EpCAM-based methods including CellSearch, is uncorrelated with prognosis in patients with some types of carcinomas. Most of the evidence attributes this inconsistency to the large degree of heterogeneity in CTCs. Specifically, CTCs might undergo full (or partial) EMT during dissemination, resulting in several phenotypes including epithelial,mesenchymal or hybrid (epithelial/mesenchymal) CTCs. These subpopulations of cells may insufficiently bind to antibodies, thereby evading detection[16,17]. Therefore, a lack of sensitive and specific biomarkers still hinders the isolation and detection of CTCs. Recent studies provide some probabilities. Here are some examples of successful markers. Glycan sialyl-Tn (STn) is often associated with cancer metastasis and expressed in metastatic colorectal and bladder tumors. Neveset al.[18]fabricated a STn affinity-based microfuidic device for specifically isolating STn+ CTCs, following an enzyme-based method to recover viable CTCs for downstream analyses. It showed greatly higher isolation efficiency from the blood of patients with advanced bladder and colorectal cancers. Plastin3 (PLS3) is expressed in metastatic cancer cells but absent in normal cells[19]. Similarly, telomerase which is expressed at high levels in almost all the cancer cells, but not in normal cells, plays an important role in cancer immortality by replenishing chromosome ends[20]. Green fuorescent protein (GFP) fused adenoviral was employed as a probe to target telomerase in cancer cells, and this strategy was applied to detect and isolate CTCs in Non-Small Cell Lung Cancer (NSCLC) to evaluate response to radiation therapy and to potentially detect recurrence and progression of disease. Oncofetal chondroitin sulfate (ofCS) is expressed in both epithelial and mesenchymal tumor cells, as well as the cells that have undergone EMT, suggesting that it may be an ideal candidate for isolating and analyzing CTCs[21,22].Agerb?ket al.[23]employed recombinant VAR2CA (rVAR2) to efficiently target ofCS expressed CTCs from patients with hepatic, prostate, pancreatic or lung cancer, allowing for isolation of a larger and more diverse population of CTCs compared to anti-EpCAM-antibody approaches. More recently, Dinget al.[24]detected Folate receptor (FR) positive CTCs in peripheral blood from 200 patients with lung adenocarcinoma, and further determined that FR+ CTC number could be used for screening solitary pulmonary nodules (SPNs) in patients and diagnosing early-stage lung cancer with sensitivity of 70.2% and specificity of 79.3%. Meanwhile,more specific biomarker for specific subgroup of CTCs is of interest. Cyclooxygenase-2 (COX-2) has been implicated in transforming growth factor-β (TGF-β1) mediated EMT progress[25]and has a higher level of expression in subpopulations of mesenchymal CTCs correlated with distant metastases[26]. These results suggested FR might be a novel biomarker for isolation and therapy targets. A subpopulation of tumor cells can express cluster of differentiation 146 (CD146) during EMT process, during which EpCAM expression is reduced. Therefore, Huanget al.[27][Figure 2B] designed dual antibodies (anti-EpCAM and anti-CD146)and biodegradable gelatin nanoparticle-coated microbeads to improve the capture of mesenchymal CTCs,achieving high efficiency ( > 80%) and high cell viability (92.5%).
All aforementionedex vivodetection systems require substantial quantities of blood. The GILUPI CellCollector (NANOMEDIZIN), approved by Conformite Europeenne in 2012, is another commercial EpCAM positive based selection device and is the first developedin vivoCTC isolation system to overcome the limitations of blood sample volume[28-30].
Except those EpCAM-based positive selection, CD45 negative selection is applied to deplete the CD45+ cells,mostly using RosetteSep system (Stem Cell Technology, Vancouver), and to analyze the EpCAM-negative CTCs in combination with EPISPOT (Epithelial Immunospot assay, France). Ramirezet al.[31]first evaluated the EPISPOT assay on a large cohort of metastatic breast cancer patients with a positive rate of 59% compared with the 48% positive rate using CellSearch, demonstrating its clinical prognostic relevance.
Microfluidic and nanotechnology-based CTC devices
Microfuidic devices enable efficient processing of complex blood samples with minimal damage to target cells.Owing to the synergistic benefits of the microfuidic devices and immunomagnetic separation, microchipbased immunomagnetic technologies are also commonly used for CTC detection. The most representative microfuidic device based on anti-EpCAM for CTCs isolation is a microscope slide sized CTC-Chip with a mass of geometrically distributed microposts coated with anti-EpCAM[32]. A 98% viability of captured CTCs was reported with minimal preprocessing and low fow stress[33]. Ozkumuret al.[34]developed an automated platform, termed “CTC-iChip”, combining the strengths of microfuidics and the benefits of magnetic-based cell isolation for single-cell separation. This CTC-iChip was able to detect theEML4-ALKgene fusion in lung cancer, suggesting that it could be a promising tool for clinical diagnosis. Then Stottet al.[35]developed a microvortex-generating herringbone (HB)-Chip for effective capture of CTCs. The micromixer device was fabricated to enhance the cell-surface interaction. Subsequently, improvement was achieved by employing nanostructured substrates and chaotic micromixers, increasing the recovery rate up to 95%[36].
For the sake of increasing the sensitivity of capturing exceedingly rare CTCs, many efforts have been made to fabricate nanostructures into the microfuidics to increase the interaction between ligands and cells;such devices include electropolymerized polymer nanodots[37], electrospun TiO2 nanofiber[38], and silicon nanowires[39-41]. More recently, Donget al.[42][Figure 2C] utilized a nanotopographical surface (NSHGMS),based on the CTC isolation technology of anti-EpCAM antibody modified Self-foating hollow glass microspheres (HGMS), to achieve excellent capture performance (93.6% ± 4.9% efficiency and 30 cells/mL detection limit in 20 min). A preferable method was based on a combination of advantages of different approaches. Wuet al.[43][Figure 2D] tactfully fabricated a 3D bionic cytosensor with electrospun polymers(PLGA) nanofibers crosswise stacked on Ni micropillars for better CTC filopodia climbing, subsequently coupled with immuno-selection by anti-EpCAM quantum dots, demonstrating a sensitive detection range and limit of 101-105cells/mL and 8 cells/mL, respectively, as well as a recovery range of 93.5%-105%.
However, fabricating these nanoscale substrates is time-consuming. Shenget al.[44]developed a microfuidic device combined with DNA aptamer modified gold nanoparticles (AuNPs) to enhance the capture performance without elaborate establishment for nanostructure. When compared with aptamer on the surface alone, the binding efficiency of AuNPs-aptamer showed a 39-fold increase and the capture efficiency rose from 49% to 92%. In order to profile the dynamic phenotypes of rare CTCs, Poudinehet al.[45][Figure 2E]fabricated a magnetic nanoparticles-enabled ranking cytometry (MagRC) approach to separate and in-line profile heterogeneous CTCs based on the longitudinal profile of magnetic field gradients. They demonstrated that this device was capable of profiling CTCs with higher sensitivity at a single-cell resolution in unprocessed blood from cancer patients compared to other previously developed magnetic sorting techniques. Similarly,an immunomagnetic nanoparticle-mediated binning and profiling approach [Figure 2F] was developed to separate CTCs with different phenotypes based on the differential expression of surface markers[46]. The CTC subpopulations could be spatially sorted in different compartments of a fuidic chip, providing a powerful means to sort heterogeneous CTCs and investigate EMT in patient CTCs.
Detection of CTCs based on their physical properties
CTCs undergo cellular processes (EMT, MET,et al.) during dissemination, resulting in a number of phenotypes. Thus, it is important to determine which CTC fractions possess greater metastatic potential and/or stronger resistance to immune surveillance and medical treatment. In this case, CTCs would have better prognostic and therapeutic values. The aforementioned methods depend on specific markers of interest for isolation; however, subpopulations of CTCs lacking the markers may be unintentionally overlooked.Therefore, additional methods that could serve as complements to protein markers are urgently needed.
Size-based CTC isolation
Alternative methods that isolate CTCs dependent on physical properties have been developed to replace or complement the antibody-based isolation methods. Most of the CTCs are believed to be larger than normal blood cells (leukocytes, erythrocytes). And pores with ≈ 8 μm in diameter have been shown to be appropriate for CTC detainment.
Thanks to various advantages, such as retention of cell morphology, antigen independence, and high sensitivity and specificity, membranous filter devices, for example, isolation by size of epithelial tumor cells(ISET) (Rarecells Diagnostics, Paris, France), have caught more attention in CTC researches. In a comparative study, Baiet al.[47]estimated the clinical effect of CTCs by using CellSearch system and ISET devices among patients with renal cell carcinoma (RCC), discovering that ISET was more appropriate for RCC patients.CTCBIOPSY (Wuhan YZY Medical Science and Technology Co., Ltd., China) is a commercial one-step ISET device which could complete automatic detection and identification within 10 min[48]. The Parsortix technology (ANGLE plc) incorporated a microscope slide sized cassette for CTC separation based on cell size and compressibility[49]. Owing to the excellent capture performance and the advantage of easy retrieval of viable CTCs for downstream analysis, the FDA clearance process of this device for diagnose is underway.The ClearCell FX system (Clearbridge BioMedics, Singapore), one of the first automated cell separation and retrieval systems, is a new label-free and size-based technology with extremely high recovery rates by dean fow fractionation[50]. These devices, together with other similar size exclusion platforms, such as ScreenCell (ScreenCell, France) based on microporous membrane filter[51], CellSieveTM (Creatv Microtech),and MetaCell (Ostrava, Czech Republic), constitute the next generation label-free CTCs enrichment technologies, demonstrating CTC isolation and detection with high efficiency, purity and viability.
Moreover, size difference can be combined with other physical features to improve capture yield. For example,a recent wedge-shaped microfuidic device [Figure 2G] based on the difference in size, as well as rigidity and nuclear/cytoplasmic ratio between CTCs and normal blood cells, was fabricated to enhance CTC isolation,exhibiting excellent capture performance with ≥ 85% capture efficiency[52]. Similarly, benefiting from those multiple biophysical properties, Liuet al.[53][Figure 2H] developed a pyramid-shaped microchamber to achieve a more than 85% capture efficiency and a 93% recovery yield. In addition, vortex technology has been exploited and validated for isolating CTCs based on differences in size, shape and deformability by inertial microfuidics and laminar micro-vortices. VTX-1 liquid biopsy system [Figure 2I] was developed for fully automated isolation and enumeration of CTCs with either high recovery mode or high purity mode in the vortex microfuidic chip[54].
Density-based CTC isolation
The density of nucleated CTCs lies between plasma and red blood cells, and within the scope of white blood cells. Quantitative buffy coat analysis by centrifuging for separation was established by Stephen C. Wardlaw in 1983[55]. AccuCyte separation based on this principle is the first step of the commercial RareCyte Platform(RareCyte, Inc. Seattle)[56], coupled with fuorescence analyzing (CyteFinder system) and picking (CytePiker)to count and retrieve cells for downstream single-cell characterization, overcoming the limitation of capture methods which are dependent on sizes that might miss the small sized-CTCs and immunomarkers that might not be expressed on some subpopulations. Some commercial density gradient solutions, such as Ficoll-Paque(GE Healthcare) and Percoll (GE Healthcare), provide simple-to-use and inexpensive methods for separating CTCs in the mononucleocyte layer from granulocytes and erythrocytes. OncoQuick (Greiner Bio-One/Hexal Gentech, Germany) consists of a sterile tube with a porous barrier inserted above separation medium,allowing the simple, rapid and highly efficient enrichment of CTCs through density-based centrifugation and size-based separation.
Dielectrophoresis based CTC isolation
The overlap of size or density between CTCs and normal cells may affect the efficiency of these size-/densitybased approaches. Electrical properties of CTCs have been applied to discriminate them from other normal cells using dielectrophoresis (DEP). Based on conventional DEP devices, microchips are used to manipulate electric fields to achieve higher capture efficiency and recovery rate. Nguyenet al.[57]fabricated a microchip to guide target lung CTCs to sensing electrodes by DEP and hydrodynamic forces, achieving a LOD of 3 cells and an efficiency over 90% at 50 kHz electric field intensity within 10 minutes. The commercial DEPArray TM cartridge (Menarini Silicon Biosystems, Italy) that contains an array of electrodes embedded with detection sensors is based on the same principle for isolating target cells for subsequent analysis.
MOLECULAR CHARACTERIZATION OF CTCS
Not all the CTCs are detectable and not all detected CTCs have the potential for metastases, indicating that CTC enumeration alone may not be an effective marker of progressive disease. A commonly used chemotherapy agent, isosfamide, was reported to decrease the number of lung cancer nodules but also increase CTC frequency in a pre-clinical model of osteosarcoma[58]. Although a large number of clinical trials have suggested that CTC presence is associated with poor survival in patients with some metastatic cancers[59].Their characterization, including phenotyping and genotyping, could lead to a better understanding of heterogeneity of metastatic tumor and further facilitate the management of patients for individualized treatment.
Protein analysis of CTCs
Most CTC detection assays are compatible with identification systems for numeration and follow-up characterization. The most common procedure consists of morphological analysis (size, shape, nuclear cytoplasmic ratio, cell shrinkage), immunohistochemical analysis[4][Figure 3A], and fuorescence immunocytochemistry (ICC). There are various markers that are useful for ICC analysis. For instance, DAPI(nuclear counterstaining), pan-keratin (positive marker), and CD45 (negative marker) are applied in the CellSearch system. Fluorescence channels and specific antibodies are now accessible for users to define the detection of more established markers, for example, epidermal growth factor receptor (EGFR)[60], androgen receptor (AR)[61], folate receptor (FR)[62], and several EMT-related markers (such as vimentin) etc., which are not only the identification markers but also therapy-associated targets. Furthermore, the ELISPOT assay combines cell culture and an immunospot test to quantitatively and qualitatively detect single viable cells expressing-cancer associated marker proteins.
One of the more reliable analytical approaches is protein profiling of captured CTCs, which can shed light on the roles of CTCs in tumor metastases and disease progression. At present, proteomic analyses mostly rely on mass spectrometry and the sensitivity of this method is improved with appropriate sample preparation and targeted cell enrichment. High-resolution porous layer open tube based liquid chromatograph-mass spectrometry (PLOT-LC-MS) lead to identification of approximately 4000 proteins of 100-200 MCF-7 cells with zeptomole detection sensitivity. Recently, Zhuet al.[63]incorporated the nanodroplet processing platform(nanoPOTS) with ultrasensitive LC-MS, allowing identification of 1500-3000 proteins from 10-140 cells. He subsequently combined CD45 negative selection and laser capture microdissection-based purification with nanoPOTS-LC-MS for studying protein expression of rare or single CTCs, identifying 164 and 607 protein groups of 1 and 5 spiked LNCaP cells, respectively[64].
Because of these analytical advances, a wealth of information regarding disease metastasis and progression is being discovered. For example, a recent report about profiling of single live CTC protease activity demonstrated the increased expression of matrix metalloproteases (MMPs) secreted by CTCs relative to normal cells is capable of triggering proteolytic processes that assist in invasion and immune evasion[65].
Gene analysis of CTCs
Fluorescencein situhybridization (FISH) technology is highly commercial and easily accessible for analysis of genetic alterations (deletion, insertion, translocation and rearrangement) at the chromosomal level, and is widely used in CTC identification and characterization. For example, RNA-ISH has been utilized for detection of onco-miRNA (such as miRNA-21)[66], while DNA-ISH has been used for quanitifying copy number of cancer-related genes (such as HER2/neugene)[13]. Based on an optimized FISH method, Frithiofet al.[67][Figure 3B] performed a CellSearch-based CTC separation assay to quantitatively measure HER-2 amplification in breast cancer CTCs. They validated that FISH was superior to protein evaluation ofHER-2status in predicting breast cancer patients' response toHER-2targeted immunotherapy and found that one in six patients underwent CTCHER-2amplification during the treatment of metastatic disease.
Hybridization analyses, much like ICC analyses, are limited by the availability of both antibodies and microscope filters. Multiple polymerase chain reaction (PCR) targeting associated RNA and DNA for detecting assorted genetic mutations may overcome these limitations. Real-time PCR (RT-PCR) can be used to determine the differences of gene expression between CTCs and normal cells. And quantitative reverse transcription PCR(qRT-PCR) takes advantage of a reverse transcriptase reaction to convert RNA into cDNA before regular RTPCR. Global or specific gene expression profiling of CTCs has been generated by real-time reverse transcription-PCR analysis, providing insights about mechanism of cancer and development of novel diagnostic biomarkers and therapeutic targets[68]. These technologies following CTC isolation has been used for identification of various gene markers, such as ALDH1 (stem cell marker), phosphoinositide kinase-3 (PI3Kα), TWIST1, TP53, and Akt2(stem cell markers)[69]. Mostertet al.[70]performed a CTC isolation assay and mRNA expression profiling using the CellSearch technique in 142 metastatic colorectal cancer (mCRC) patients. They measured 95 mRNAs by RT-qPCR and found that 34 CTC-specific mRNAs were higher in patients with ≥ 3 CTCs compared with healthy donors. This CTC-specific gene panel for mCRC patients, such asKRT19,KRT20andAGR2, may aid in characterizing how CTCs with different expression profiles contribute to malignancy, thereby furthering the realization of individualized cancer treatment. Camptonet al.[71]developed a comprehensive and sensitive platform, named as AccuCyte?-CyteFinder? system, for identification and characterization of individual CTCs. Using the whole genome amplification (WGA) product, they confirmed that the TP53 gene, which is known to contain the R175H mutation in SKBR3, enables personalized, molecularly-guided cancer treatment.
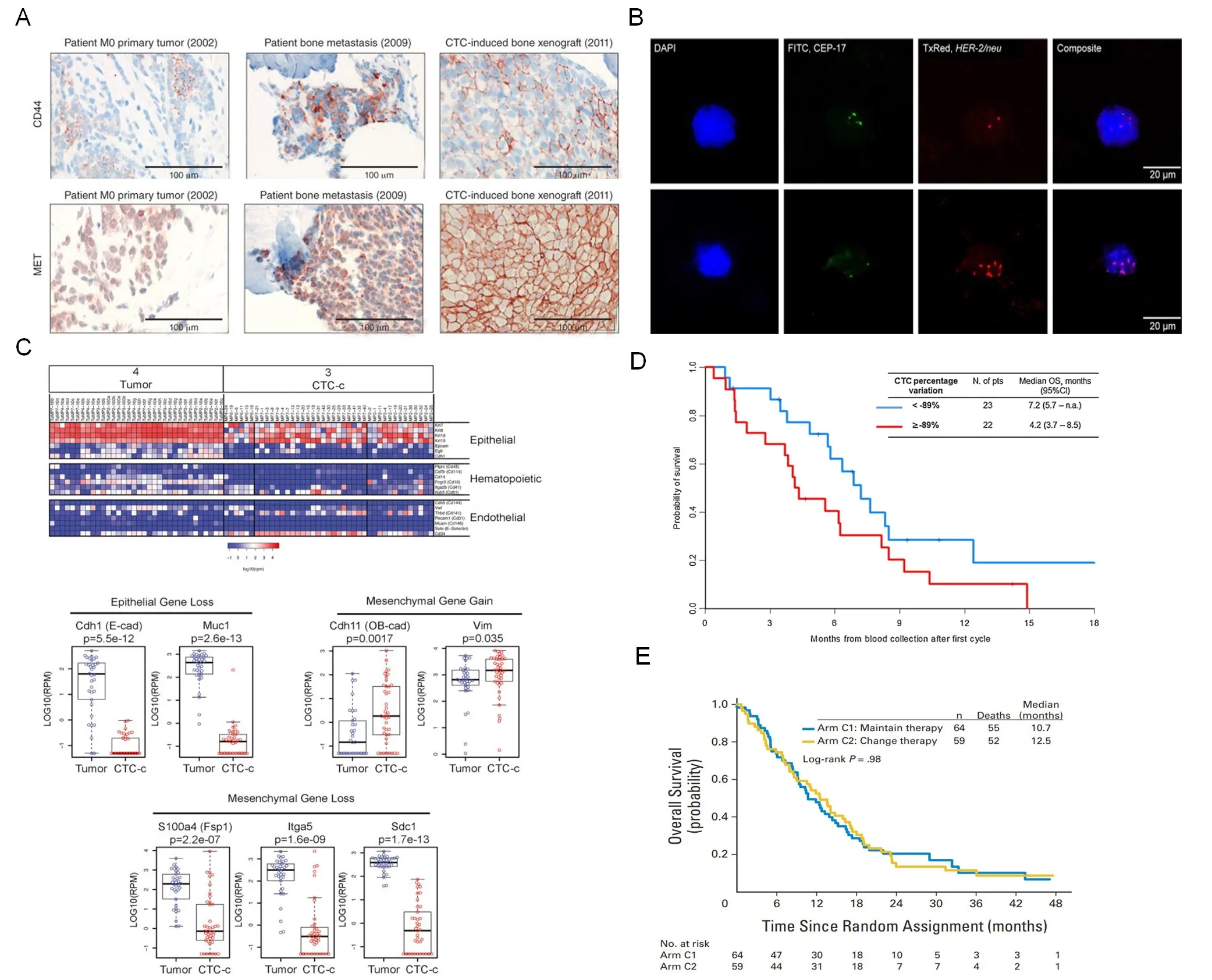
Figure 3. Molecular profiling and clinical application of CTCs. A: Immunohistochemical analysis of the expression of CD44 and MET in primary tumor (M0, nonmetastatic stage), bone metastasis and CTC-induced bone xenograft after transplantation of sorted CTCs; B:FISH images of a patient with metastatic breast cancer have no detectable HER-2 amplification (top panel), and the lower panel is HER-2 amplified CTC; C: the expression heatmap of epithelial, hematopoietic, and endothelial markers in primary tumors and classical epithelial CTCs (CTC-c). Epithelial and mesenchymal genes differentially expressed in CTCs vs. tumors; D: Kaplan-Meier curves of overall survival according to CTC change after one cycle of chemotherapy; E: overall survival (OS) and PFS in patients with metastatic breast cancer for whom therapy failed to reduce CTCs at first follow-up (approximately 21 days after first dose of chemotherapy), or randomly assigned to maintain the original chemotherapy (arm C1) or to switch to an alternative chemotherapy (arm C2). A: Copyright Nature Publishing Group, 2013. Reproduced with permission from reference[4]; B: Copyright Dove Medical Press, 2016. Reproduced with permission from reference[67]; C: Copyright Cell Press, 2014. Reproduced with permission from reference[75]; D: Copyright Elsevier, 2014. Reproduced with permission from reference[79]; E: Copyright American Society of Clinical, 2014. Reproduced with permission from reference[80]
Ampli1 TM (Menarini Silicon Biosystem), a product developed for single-cell WGA, can be used to amplify DNA for downstream genotyping analysis, including comparative genomic hybridization (CGH) and next-generation sequencing (NGS). Upon its development in 1992, CGH technology opened a new avenue in genomic investigation and, more particularly, in cancer gene analysis. In the past, CGH was applied for analysis of tumor tissues, but many studies have suggested that data from primary tumors alone is insufficient. Array CGH, which exploits ordered arrays of genomic DNA sequences, is widely used for analysis of CTCs, including identification of genomic alterations which include insertion/deletion, singlenucleotide variations, copy number variations (CNVs)[72]; identification of candidate oncogenes or tumor suppressors; identification of novel biomarkers involved in metastasis, cancer progression and therapy response; and identification of subgroups of CTCs[73]. High-throughput NGS is another strong technology to analyze heterogeneity of CTCs and reveal the mechanisms of metastasis, which might be the Achilles’heel in disease progression. Bertucciet al.[74]utilized aCGH and NGS to compare DNA copy number and mutational profiles of 365 cancer-related genes between primary tumors and metastases and discovered a degree of divergence for actionable driver genes that might be extremely relevant with cancer metastasis.Profiting from whole genome amplification technology, the limited amount of single-cell genomic DNA sample can be amplified indistinguishable for sequencing.
Single-cell RNA sequencing was used to detect the heterogeneity of CTCs, unveiling the mechanism of drug resistance of androgen receptor (AR) inhibitors in prostate cancer[61]. The technique was also used to identify conduct a transcriptomic analysis in pancreatic CTCs, finding increased expression of stromalderived extracellular matrix (ECM) proteins, which facilitate cell migration and invasiveness, in CTCs from mice and humans with pancreatic cancer[75][Figure 3C].
Single-cell exome sequencing of isolated CTCs from cancer patient, revealed insertion/deletion and singlenucleotide variation in CTCs after whole genome amplification. The results showed cancer-type specific CNVs that are reproducible within cells of the same patient, or even between patients with the same type of cancer[76].
CLINICAL IMPLICATIONS
Prognostic and diagnostic value of CTCs
Several studies have highlighted the correlation between CTC burden and treatment effect, indicating the prognostic value for patients receiving chemotherapy or surgery and the potential of surveillance of disease recurrence or metastasis [Table 1]. Several years after chemotherapy, patients with high-risk breast cancer with elevated CTC counts in their peripheral blood were reported to have worst survival prospect[77].Similarly, patients with colorectal cancer with elevated CTC counts were more likely to have recurrence after 3 years of curative resection, showing the relation between post-operative CTCs and poor prognosis[78].The prognostic value of CTCs was demonstrated by Nicolaet al.[79][Figure 3D] in 60 patients with extensive SCLC. After assessment with the CellSearch system, the group isolated and analyzed CTCs in 90% (54/60)of patients at baseline and demonstrated that CTC count was significantly associated with the number of organs involved. A reduction in CTC count of more than 89% after chemotherapy significantly improved the prognosis accuracy and was associated with a better outcome. They concluded that only the change of CTC count after the first chemotherapy cycle provided clinically relevant information. However, there were still some clinical trials (such as: gov NCT00382018) that failed to observe improved PFS or OS for cancer patients with decreased CTCs after therapies[80][Figure 3E]. Compared to either CTCs levels or cancer specific antigen levels alone, the combination of both two biomarkers provided a notably better predictive indicator for patients with advanced cervical cancer[81].
Paired CTCs are correlated strongly with origin tumor cells, thus characterization of this subset of genes of CTCs might help to predict primary tumor origin. And CTCs detached from different parts within same tumor or even from different tumors are originally heterogeneous in nature. Gene expression profiling of CTCs from patients with different metastatic cancers showed the different but unique gene expression patterns of those cancer types, providing novel noninvasive diagnostic tools[68]and essential information for personalized treatment.
On the other hand, CTCs undergoing certain transitions, such as EMT and MET, might generate new genetic alterations which are absent in the primary tumor, but related with potential distant tumor. Characterization of these subgroups of CTCs might assist in localizing specific distant metastatic sites. Additionally, some subgroup of CTCs may harbor changes that are undetectable in the tumor of origin, but are related to drugresistance and management of treatment. Revealing the qualitative and quantitative divergence between the CTCs and the primary tumor or within CTCs by genotyping and phenotypic analysis is crucial for future studies of individualized medicine in metastatic disease. However, some changes present in CTCs homogenously take place within primary tumor, suggesting CTCs, to some extent, might be a noninvasive and real-time indicator for following cancer progression and monitoring therapeutic response.
CTCs predict therapy outcome and provide personalized therapeutic targets
CTC enumeration can be a good marker for predicting drug sensitivity/resistance in preclinical and clinical settings. CTC quantity reasonably correlates well with clinical and instrumental tumor response. To investigate the clinical significance of CTCs in predicting the tumor response to chemotherapy, Wuet al.[82]detected CTCs at baseline and during chemotherapy in 453 eligible lung cancer patients, indicating that disease control rate (DCR) of CTC-negative patients was significantly higher than that of CTC-positive patients; more importantly, patients also showed higher OS. The CTC status has been reported to be related to prognosis and is altered in response to chemotherapy in many other tumor types, such as colorectal cancer[10], breast cancer[83], osteosarcoma[58]. Smerageet al.[80]confirmed the prognostic significance of CTCs in patients with metastatic breast cancer, demonstrating that an increased number of CTCs was associated with poor prognosis. Early conversion to alternate cytotoxic therapies was ineffective in prolonging OS in patients with increased CTCs after receiving 21 days of first-line chemotherapy. For this population, a more effective treatment than standard chemotherapy is needed.
CTC characterization can generate predictions of drug potency and therapeutic efficacy before or during treatment according to analysis of targeted protein expression and signaling pathway activity. The development of immune check point inhibitors for cancer therapy, for example, anti-PD-L1, have achieved much success due to increased efficacy and decreased toxicity[84]. However, PD-L1 expression detection for prediction of therapeutic response using tumor biopsies prior to treatment is invasive and insufficiently precise to guide treatment planning, resulting in some cancer patients being treated with an expensive but ineffective and toxic therapy.Instead, serial monitoring of patients treated with immune checkpoint inhibitors showed that a decrease of CTC score correlated with improved PFS and OS, and further demonstrated that RNA-based CTCs score during the immunotherapy has the potential to be a predictive biomarker for immunotherapeutic outcome[85].
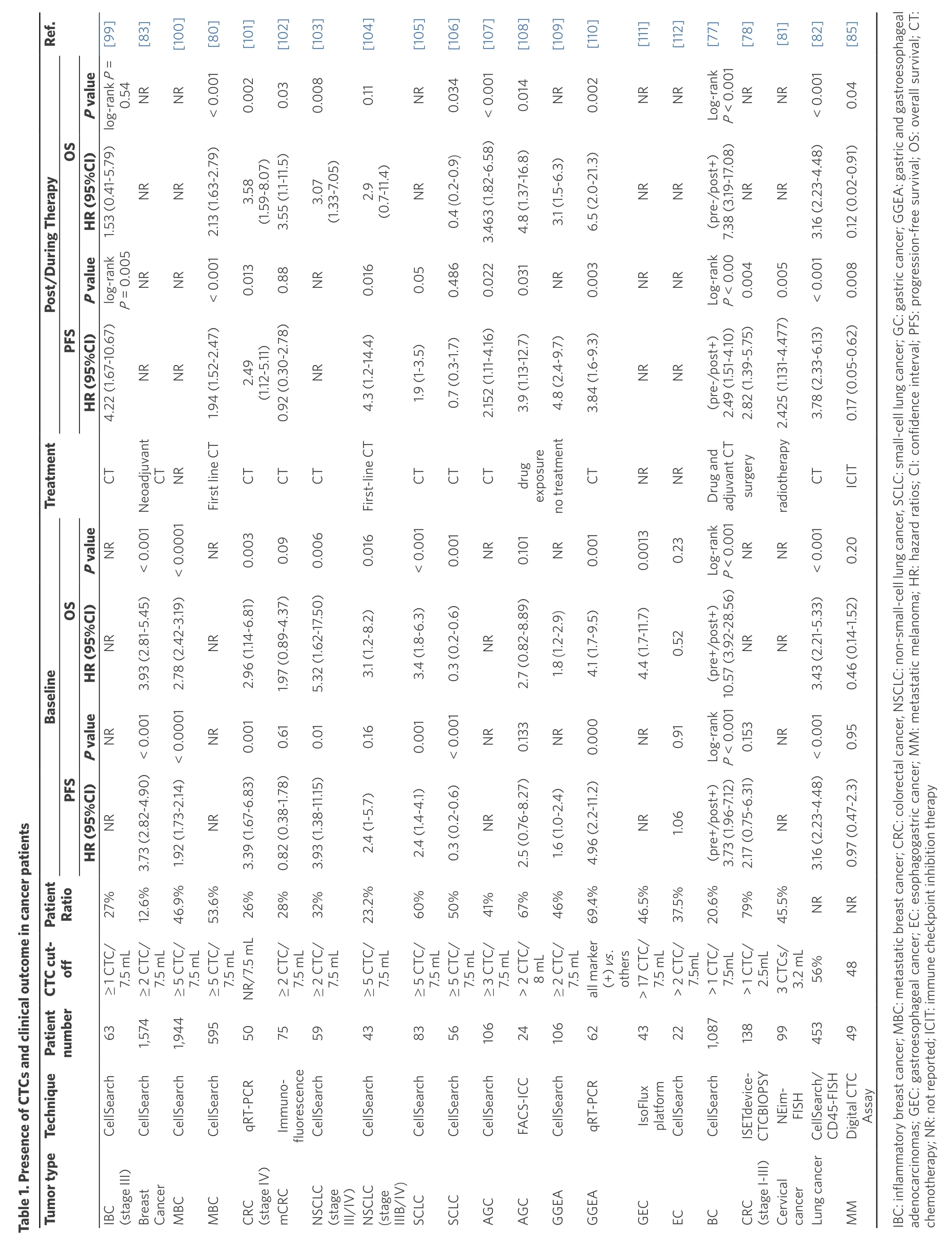
Table 1. Presen ce of CT Cs an d clin ical ou tcom e in cancer pa tien tsimages/BZ_149_204_298_2193_3186.png IBadch C: inflamm atory b reast can cer; M BC: m etastatic b reast canphceagog r; C RC: colorectal canM: m cer, N SC LC: n on-small-cell lun g can cer, SCLC: sm en ocarcino mas; G EC: gastroeso ph ageal can cer; EC: esot inh astric can cer; M etastatic m elan om a; H R: h azard ratio s; C I: c em otherapy; N R: n ot rep orted; ICIT: imm un e checkpo in ib itio n therapy
Beyond enumeration alone, CTCs could provide crucial information of tumor malignancy via molecular characterization, leading to better treatment monitoring and molecular-/cancer cell-targeted therapies. The genotypic changes in CTCs provided the best suitable targeted therapy and enabled assessment treatment regimen efficacy over time. Epidermal growth factor receptor (EGFR) on the CTC surface has been verified as extremely significant in the process of tumor growth and progression. EGFR inhibitors (HER2 inhibitors,tyrosine kinase inhibitors, TKI and monoclonal antibodies) have been licensed for treatment of cancers caused by EGFR up-regulation, such as NSCL, breast, renal cell, squamous cell, colon and pancreatic cancers.However, in some cases, EGFR-targeted inhibitors are not effective due to the emergence of drug-resistance mutations. Mutation screening analysis of EGFR in CTCs may provide an explanation for drug-resistance mechanism and also reveal possibilities for diagnostic and therapeutic interventions[60]. Inhibitors of other therapeutic molecular targets including mTOR, such as temsirolimus, and phosphoinositide 3-kinase(PI3K), such as ZSTK474, LY294002, have shown to have anti-proliferation in clinical trials[86,87]. Most cell populations of the immune system play an important role in survival and seeding, or even enhancing the growth of tumorigenic subpopulations of CTCs. The molecular characterization of CTCs might assist in unveiling intercellular interaction mechanisms and providing potential therapeutic targets. For instance,the extracellular surface interacting protein, PD-L1, is one such target that is currently generating much interest[84]. In consideration of costs and toxicity of anti-PD-L1 therapy, predictive biomarkers able to distinguish responders from non-responders are in urgent demand. Real-time CTC analysis provides significant information on drug resistance[88]. In addition, CTCs are now regarded as a new cellular therapeutic target. Photodynamic therapy was used to selectively kill GFP-expressing CTCs by energy transfer between expressed GFP and pre-accumulated rose bengal (RB) in cells, demonstrating that clearance of CTCs could reduce metastasis and extend survival[89].
CONCLUSION AND OUTLOOK
The potential clinical value of CTCs has been established. Advances in CTC isolation and molecular characterization offer the possibility for early detection and diagnosis, improve the satisfaction of therapies,as well as expand our knowledge about underlying mechanisms of cancer dissemination and progression.
Although the tremendous technical advances in CTC isolation and detection make it possible to analyze extremely rare CTCs, there are still many hurdles. First, a criterion to standardize different kinds of detection assays is urgently needed in clinical applications. Secondly, while the emergence of new predictive biomarkers leads to clearer recognition about tumor metastases and disease progression, novel targets for prognosis and treatment need to be further validated and standardized. The next frontier of CTCs detection lies in thorough characterization, which might rely on developing single-cell multi-omic technologies, including genomics,proteomics, transcriptomics etc.. Finally, research findings provide arguments in favor of the hypothesis that only a subpopulation (metastasis-initiating cells, MICs) of CTCs in patient blood is responsible for initiating carcinoma metastasis. A majority of cancer cells may never develop into metastatic phase, but instead maintain a dormant state or die from the anoikis, immune attacks and physical shear stress in the vasculature. However, our understanding about the requirements for CTCs being activated from latency into overt metastases is far from complete. Apart from CTCs, other noninvasive “l(fā)iquid biopsies” might provide more supplementary information, including some cell-free components such as circulating tumor DNA (ctDNA)[90], microRNAs (miRNA), exosomes, as well as long-coding RNA (lncRNA)[91-94]. Recently, due to the success in immunotherapy of cancers, immune checkpoint blocker programmed death-ligand 1 (PDL1), whose expression in CTCs correlate with tumor status[95], has gained interest as a potential independent prognostic marker for PFS and OS[96], extending the spectrum of noninvasive liquid biopsies[97]. And, CTC detection may also have the potential to monitor the efficacy of anti-PD-L1 therapy[98]. Comprehensive and systemic liquid biopsy analyses may contribute to thorough understanding of metastatic malignancy and better management of cancer patients.
DECLARATIONS
Authors’ contributions
Huang QQ and Chen XX contributed equally to this work.
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work was supported by the National Natural Science Foundation of China (31800085), National Natural Science Foundation for Major Research Instruments (81527801).
Conflicts of interest
Both authors declared that there are no conficts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
? The Author(s) 2019.
REFERENCE
1. Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013;19:1438-49.
2. Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, et al. Metastatic colonization requires the repression of the epithelialmesenchymal transition inducer Prrx1. Cancer Cell 2012;22:709-24.
3. Huang Q, Wang Y, Chen X, Wang Y, Li Z, et al. Nanotechnology-Based Strategies for Early Cancer Diagnosis Using Circulating Tumor Cells as a Liquid Biopsy. Nanotheranostics 2018;2:21-41.
4. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539-44.
5. Kling J. Beyond counting tumor cells. Nat Biotechnol 2012;30:578-80.
6. Huang Q, Chen B, He R, He Z, Cai B, et al. Capture and release of cancer cells based on sacrificeable transparent MnO2 nanospheres thin film. Adv Healthc Mater 2014;3:1420-5.
7. Huang Q, Cai B, Chen B, Rao L, He Z, et al. Efficient Purification and Release of Circulating Tumor Cells by Synergistic Effect of Biomarker and SiO2 @Gel-Microbead-Based Size Difference Amplification. Adv Healthc Mater 2016;5:1554-9.
8. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9.
9. Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 2006;12:4218-24.
10. Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, et al. Relationship of circulating tumor cells to tumor response, progressionfree survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21.
11. Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A 2009;106:3970-5.
12. Saliba AE, Saias L, Psychari E, Minc N, Simon D, et al. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc Natl Acad Sci U S A 2010;107:14524-9.
13. Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip 2014;14:78-88.
14. Fina E, Callari M, Reduzzi C, D’Aiuto F, Mariani G, et al. Gene expression profiling of circulating tumor cells in breast cancer. Clin Chem 2015;61:278-89.
15. Mayo C, Ortega FG, Gimenez-Capitan A, Molina-Vila MA, Serrano MJ, et al. CK-coated magnetic-based beads as a tool to isolate circulating tumor cells (CTCs) in human tumors. Transl Lung Cancer Res 2013;2:65-71.
16. Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8.
17. Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science 2013;341:1186-8.
18. Neves M, Azevedo R, Lima L, Oliveira MI, Peixoto A, et al. Exploring sialyl-Tn expression in microfluidic-isolated circulating tumour cells: A novel biomarker and an analytical tool for precision oncology applications. N Biotechnol 2019;49:77-87.
19. Yokobori T, Iinuma H, Shimamura T, Imoto S, Sugimachi K, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res 2013;73:2059-69.
20. Takakura M, Matsumoto T, Nakamura M, Mizumoto Y, Myojyo S, et al. Detection of circulating tumor cells in cervical cancer using a conditionally replicative adenovirus targeting telomerase-positive cells. Cancer Sci 2018;109:231-40.
21. Dorsey JF, Kao GD, MacArthur KM, Ju M, Steinmetz D, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: Pilot study results. Cancer 2015;121:139-49.
22. Frick MA, Kao GD, Aguarin L, Chinniah C, Swisher-McClure S, et al. Circulating Tumor Cell Assessment in Presumed Early Stage Non-Small Cell Lung Cancer Patients Treated with Stereotactic Body Radiation Therapy: A Prospective Pilot Study. Int J Radiat Oncol Biol Phys 2018;102:536-42.
23. Agerb?k MO, Bang-Christensen SR, Yang MH, Clausen TM, Pereira MA, et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat Commun 2018;9:3279.
24. Ding C, Zhou X, Xu C, Chen J, Ju S, et al. Circulating tumor cell levels and carcinoembryonic antigen: An improved diagnostic method for lung adenocarcinoma. Thorac Cancer 2018;9:1413-20.
25. Neil JR, Johnson KM, Nemenoff RA, Schiemann WP. Cox-2 inactivates Smad signaling and enhances EMT stimulated by TGF-beta through a PGE2-dependent mechanisms. Carcinogenesis 2008;29:2227-35.
26. Cai J, Huang L, Huang J, Kang L, Lin H, et al. Associations between the cyclooxygenase-2 expression in circulating tumor cells and the clinicopathological features of patients with colorectal cancer. J Cell Biochem 201810.1002/jcb.27768:1-7.
27. Huang Q, Wang FB, Yuan CH, He Z, Rao L, et al. Gelatin Nanoparticle-Coated Silicon Beads for Density-Selective Capture and Release of Heterogeneous Circulating Tumor Cells with High Purity. Theranostics 2018;8:1624-35.
28. He Y, Shi J, Shi G, Xu X, Liu Q, et al. Using the New CellCollector to Capture Circulating Tumor Cells from Blood in Different Groups of Pulmonary Disease: A Cohort Study. Sci Rep 2017;7:9542.
29. Gallerani G, Cocchi C, Bocchini M, Piccinini F, Fabbri F. Characterization of tumor cells using a medical wire for capturing circulating tumor cells: A 3D approach based on immunofluorescence and DNA FISH. J Vis Exp 2017; doi: 10.3791/56936:e56936.
30. Markou A, Lazaridou M, Paraskevopoulos P, Chen S, Swierczewska M, et al. Multiplex Gene Expression Profiling of In Vivo Isolated Circulating Tumor Cells in High-Risk Prostate Cancer Patients. Clin Chem 2018;64:297-306.
31. Ramirez JM, Fehm T, Orsini M, Cayrefourcq L, Maudelonde T, et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem 2014;60:214-21.
32. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9.
33. Sequist LV, Nagrath S, Toner M, Haber DA, Lynch TJ. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. J Thorac Oncol 2009;4:281-3.
34. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179-47.
35. Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A 2010;107:18392-7.
36. Wang S, Liu K, Liu J, Yu ZT, Xu X, et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl 2011;50:3084-8.
37. Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, et al. Functionalized conducting polymer nanodots for enhanced cell capturing: the synergistic effect of capture agents and nanostructures. Adv Mater 2011;23:4788-92.
38. Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater 2012;24:2756-60.
39. Shen Q, Xu L, Zhao L, Wu D, Fan Y, et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates.Adv Mater 2013;25:2368-73.
40. Hou S, Zhao L, Shen Q, Yu J, Ng C, et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed Engl 2013;52:3379-83.
41. Hou S, Zhao H, Zhao L, Shen Q, Wei KS, et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater 2013;25:1547-51.
42. Dong Z, Yu D, Liu Q, Ding Z, Lyons VJ, et al. Enhanced capture and release of circulating tumor cells using hollow glass microspheres with a nanostructured surface. Nanoscale 2018;10:16795-804.
43. Wu X, Xiao T, Luo Z, He R, Cao Y, et al. A micro-/nano-chip and quantum dots-based 3D cytosensor for quantitative analysis of circulating tumor cells. J Nanobiotechnology 2018;16:65.
44. Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013;7:7067-76.
45. Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol 2017;12:274-81.
46. Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L,et al. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chem Int Ed Engl 2015;54:139-43.
47. Bai M, Zou B, Wang Z, Li P, Wang H, et al. Comparison of two detection systems for circulating tumor cells among patients with renal cell carcinoma. Int Urol Nephrol 2018;50:1801-9.
48. Chen F, Wang S, Fang Y, Zheng L, Zhi X,et al. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application.Oncotarget 2017;8:3029-41.
49. Obermayr E, Maritschnegg E, Agreiter C, Pecha N, Speiser P, et al. Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget 2018;9:812-23.
50. Lee Y, Guan G, Bhagat AA. ClearCell(R) FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells.Cytometry A 2018;93:1251-4.
51. Yanagita M, Luke JJ, Hodi FS, Janne PA, Paweletz CP. Isolation and characterization of circulating melanoma cells by size filtration and fluorescent in-situ hybridization. Melanoma Res 2018;28:89-95.
52. Qin L, Zhou W, Zhang S, Cheng B, Wang S, et al. Highly efficient isolation of circulating tumor cells using a simple wedge-shaped microfluicdic device. IEEE Trans Biomed Eng 201810.1109/tbme.2018.2875361.
53. Liu F, Wang S, Lu Z, Sun Y, Yang C, et al. A simple pyramid-shaped microchamber towards highly efficient isolation of circulating tumor cells from breast cancer patients. Biomed Microdevices 2018;20:83.
54. Sollier-Christen E, Renier C, Kaplan T, Kfir E, Crouse SC. VTX-1 Liquid Biopsy System for Fully-Automated and Label-Free Isolation of Circulating Tumor Cells with Automated Enumeration by BioView Platform. Cytometry A 2018;93:1240-5.
55. Wardlaw SC, Levine RA. Quantitative buffy coat analysis. A new laboratory tool functioning as a screening complete blood cell count.JAMA 1983;249:617-20.
56. Kaldjian EP, Ramirez AB, Sun Y, Campton DE, Werbin JL, et al. The RareCyte(R) platform for next-generation analysis of circulating tumor cells. Cytometry A 2018;93:1220-5.10.
57. Nguyen NV, Jen CP. Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens Bioelectron 2018;121:10-8.
58. Chalopin A, Tellez-Gabriel M, Brown HK, Vallette F, Heymann MF, et al. Isolation of circulating tumor cells in a preclinical model of osteosarcoma: effect of chemotherapy. J Bone Oncol 2018;12:83-90.
59. Bahnassy AA, Saber MM, Mahmoud MG, Abdellateif MS, Abd El-Mooti Samra M, et al. The role of circulating tumor cells in metastatic breast cancer: prognostic and predictive value. Mol Biol Rep 2018;45:2025-35.
60. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77.
61. Alama A, Truini A, Coco S, Genova C, Grossi F. Prognostic and predictive relevance of circulating tumor cells in patients with non-smallcell lung cancer. Drug Discov Today 2014;19:1671-6.
62. Ding C, Zhou X, Xu C, Chen J, Ju S, et al. Circulating tumor cell levels and carcinoembryonic antigen: An improved diagnostic method for lung adenocarcinoma. Thorac Cancer 2018;9:1413-20.
63. Zhu Y, Piehowski PD, Zhao R, Chen J, Shen Y, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells. Nat Commun 2018;9:882.
64. Zhu Y, Podolak J, Zhao R, Shukla AK, Moore RJ, et al. Proteome Profiling of 1 to 5 Spiked Circulating Tumor Cells Isolated from Whole Blood Using Immunodensity Enrichment, Laser Capture Microdissection, Nanodroplet Sample Processing, and Ultrasensitive nanoLCMS. Anal Chem 2018;90:11756-9.
65. Dhar M, Lam JN, Walser T, Dubinett SM, Rettig MB, et al. Functional profiling of circulating tumor cells with an integrated vortex capture and single-cell protease activity assay. Proc Natl Acad Sci U S A 2018;115:9986-91.
66. Ortega FG, Lorente JA, Garcia Puche JL, Ruiz MP, Sanchez-Martin RM, et al. miRNA in situ hybridization in circulating tumor cells--MishCTC. Sci Rep 2015;5:9207.
67. Frithiof H, Aaltonen K, Ryden L. A FISH-based method for assessment of HER-2 amplification status in breast cancer circulating tumor cells following CellSearch isolation. Onco Targets Ther 2016;9:7095-103.
68. Smirnov DA, Zweitzig DR, Foulk BW, Miller MC, Doyle GV,et al. Global gene expression profiling of circulating tumor cells. Cancer Res 2005;65:4993-7.
69. Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res 2012;14:R15.
70. Mostert B, Sieuwerts AM, Bolt-de Vries J, Kraan J, Lalmahomed Z, et al. mRNA expression profiles in circulating tumor cells of metastatic colorectal cancer patients. Mol Oncol 2015;9:920-32.
71. Campton DE, Ramirez AB, Nordberg JJ, Drovetto N, Clein AC,et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining.Bmc Cancer 2015;15:360.
72. Kaveh F, Baumbusch LO, Nebdal D, Borresen-Dale AL, Lingjaerde OC, et al. A systematic comparison of copy number alterations in four types of female cancer. BMC Cancer 2016;16:913.
73. Van Beers EH, Nederlof PM. Array-CGH and breast cancer. Breast Cancer Res 2006;8:210.
74. Bertucci F, Finetti P, Guille A, Adelaide J, Garnier S, et al. Comparative genomic analysis of primary tumors and metastases in breast cancer. Oncotarget 2016;7:27208-19.
75. Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 2014;8:1905-18.
76. Ni X, Zhuo M, Su Z, Duan J, Gao Y, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 2013;110:21083-8.
77. Trapp E, Janni W, Schindlbeck C, Juckstock J, Andergassen U, et al. Presence of Circulating Tumor Cells in High-Risk Early Breast Cancer During Follow-Up and Prognosis. J Natl Cancer Inst 2019;111:380-7.
78. Yang C, Shi D, Wang S, Wei C, Zhang C, et al. Prognostic value of pre- and post-operative circulating tumor cells detection in colorectal cancer patients treated with curative resection: a prospective cohort study based on ISET device. Cancer Manag Res 2018;10:4135-44.
79. Nicola N, Antonio R, Alessandro M, Simona S, Simona B, et al. Prognostic value of circulating tumor cells’ reduction in patients with extensive small-cell lung cancer. Lung Cancer 2014;85:314-9.
80. Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol 2014;32:3483-9.
81. Wen YF, Cheng TT, Chen XL, Huang WJ, Peng HH, et al. Elevated circulating tumor cells and squamous cell carcinoma antigen levels predict poor survival for patients with locally advanced cervical cancer treated with radiotherapy. PLoS One 2018;13:e0204334.
82. Wu ZX, Liu Z, Jiang HL, Pan HM, Han WD. Circulating tumor cells predict survival benefit from chemotherapy in patients with lung cancer. Oncotarget 2016;7:67586-96.
83. Bidard FC, Michiels S, Riethdorf S, Mueller V, Esserman LJ, et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J Natl Cancer Inst 2018;110:560-7.
84. Swaika A, Hammond WA, Joseph RW. Current state of anti-PD-L1 and anti-PD-1 agents in cancer therapy. Mol Immunol 2015;67:4-17.
85. Hong X, Sullivan RJ, Kalinich M, Kwan TT, Giobbie-Hurder A, et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc Natl Acad Sci U S A 2018;115:2467-72.
86. Namatame N, Tamaki N, Yoshizawa Y, Okamura M, Nishimura Y, et al. Antitumor profile of the PI3K inhibitor ZSTK474 in human sarcoma cell lines. Oncotarget 2018;9:35141-61.
87. Grunwald V, Keilholz U, Boehm A, Guntinas-Lichius O, Hennemann B, et al. TEMHEAD: a single-arm multicentre phase II study of temsirolimus in platin- and cetuximab refractory recurrent and/or metastatic squamous cell carcinoma of the head and neck (SCCHN) of the German SCCHN Group (AIO). Ann Oncol 2015;26:561-7.
88. Mazel M, Jacot W, Pantel K, Bartkowiak K, Topart D,et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol Oncol 2015;9:1773-82.
89. Kim YR, Yoo JK, Jeong CW, Choi JW. Selective killing of circulating tumor cells prevents metastasis and extends survival. J Hematol Oncol 2018;11:114.
90. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10.
91. Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, et al. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer 2016;15:48.
92. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST,et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82.
93. Halvorsen AR, Sandhu V, Sprauten M, Flote VG, Kure EH, et al. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol 2018;57:1225-31.
94. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50.
95. Ilie M, Szafer-Glusman E, Hofman V, Chamorey E, Lalvee S, et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann Oncol 2018;29:193-9.
96. Strati A, Koutsodontis G, Papaxoinis G, Angelidis I, Zavridou M, et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann Oncol 2017;28:1923-33.
97. Boffa DJ, Graf RP, Salazar MC, Hoag J, Lu D,et al. Cellular Expression of PD-L1 in the Peripheral Blood of Lung Cancer Patients is Associated with Worse Survival. Cancer Epidemiol Biomarkers Prev 2017;26:1139-45.
98. Kulasinghe A, Perry C, Kenny L, Warkiani ME, Nelson C, et al. PD-L1 expressing circulating tumour cells in head and neck cancers.BMC Cancer 2017;17:333.
99. Hall CS, Karhade M, Laubacher BA, Kuerer HM, Krishnamurthy S, et al. Circulating Tumor Cells and Recurrence After Primary Systemic Therapy in Stage III Inflammatory Breast Cancer. J Natl Cancer Inst 2015;107.
100. Bidard F-C, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. The Lancet Oncology 2014;15:406-14.
101. Barbazan J, Muinelo-Romay L, Vieito M, Candamio S, Diaz-Lopez A, et al. A multimarker panel for circulating tumor cells detection predicts patient outcome and therapy response in metastatic colorectal cancer. Int J Cancer 2014;135:2633-43.
102. Romiti A, Raffa S, Di Rocco R, Roberto M, Milano A, et al. Circulating tumor cells count predicts survival in colorectal cancer patients. J Gastrointestin Liver Dis 2014;23:279-84.
103. Zhou J, Dong F, Cui F, Xu R, Tang X. The role of circulating tumor cells in evaluation of prognosis and treatment response in advanced non-small-cell lung cancer. Cancer Chemother Pharmacol 2017;79:825-33.
104. Muinelo-Romay L, Vieito M, Abalo A, Nocelo MA, Baron F, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel)2014;6:153-65.
105. Messaritakis I, Politaki E, Kotsakis A, Dermitzaki EK, Koinis F, et al. Phenotypic characterization of circulating tumor cells in the peripheral blood of patients with small cell lung cancer. PLoS One 2017;12:e0181211.
106. Messaritakis I, Politaki E, Plataki M, Karavassilis V, Kentepozidis N, et al. Heterogeneity of circulating tumor cells (CTCs) in patients with recurrent small cell lung cancer (SCLC) treated with pazopanib. Lung Cancer 2017;104:16-23.
107. Li Y, Gong J, Zhang Q, Lu Z, Gao J, et al. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer 2016;114:138-45.
108. Meulendijks D, de Groot JW, Los M, Boers JE, Beerepoot LV, et al. Bevacizumab combined with docetaxel, oxaliplatin, and capecitabine,followed by maintenance with capecitabine and bevacizumab, as first-line treatment of patients with advanced HER2-negative gastric cancer: A multicenter phase 2 study. Cancer 2016;122:1434-43.
109. Pernot S, Badoual C, Terme M, Castan F, Cazes A, et al. Dynamic evaluation of circulating tumour cells in patients with advanced gastric and oesogastric junction adenocarcinoma: Prognostic value and early assessment of therapeutic effects. Eur J Cancer 2017;79:15-22.
110. Kubisch I, de Albuquerque A, Schuppan D, Kaul S, Schaich M, et al. Prognostic Role of a Multimarker Analysis of Circulating Tumor Cells in Advanced Gastric and Gastroesophageal Adenocarcinomas. Oncology 2015;89:294-303.
111. Brungs D, Lynch D, Luk AW, Minaei E, Ranson M, et al. Cryopreservation for delayed circulating tumor cell isolation is a valid strategy for prognostic association of circulating tumor cells in gastroesophageal cancer. World J Gastroenterol 2018;24:810-8.
112. Sclafani F, Smyth E, Cunningham D, Chau I, Turner A, et al. A pilot study assessing the incidence and clinical significance of circulating tumor cells in esophagogastric cancers. Clin Colorectal Cancer 2014;13:94-9.
 Journal of Cancer Metastasis and Treatment2019年4期
Journal of Cancer Metastasis and Treatment2019年4期
- Journal of Cancer Metastasis and Treatment的其它文章
- AUTHOR INSTRUCTIONS
- Cancer stem cells in liver metastasis from colon adenocarcinoma express components of the reninangiotensin system
- Breast cancer, metastasis, and the microenvironment: disabling the tumor cell-tostroma communication network
- Peripheral biomarkers for pediatric brain tumors:current advancements and future challenges
- Targeting autophagy with small molecules for cancer therapy
- Breast cancer metastasis to the stomach
