Prognostic implications of epidermal growth factor receptor variant III expression and nuclear translocation in Chinese human gliomas
Kaiyuan Yang, Xiaohui Ren, Liyuan Tao, Peipei Wang, Haihui Jiang, Li Shen,Yiming Zhao, Yong Cui, Mingxiao Li, Song Lin
1Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing 100070, China; 2China National Clinical Research Center for Neurological Diseases, Beijing 100070, China; 3Beijing Neurosurgical Institution, Capital Medical University, Beijing 100050, China; 4Research Center of Clinical Epidemiology, Peking University Third Hospital, Beijing 100191, China; 5Department of Cell Biology, Peking University Health Science Center, Beijing 100191, China; 6Peking University Stem Cell Research Center, Beijing 100191, China.
*These authors contributed equally to this work.
Correspondence to: Song Lin, MD. Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing 100070, China. Email:linsong2005@126.com.
Abstract Objective: To determine the prognostic implications and clinical significance of epidermal growth factor receptor variant III (EGFRvIII) expression and EGFRvIII nuclear translocation in Chinese human gliomas.Methods: We retrospectively examined EGFRvIII expression and EGFRvIII nuclear translocation using immunohistochemistry in specimens of 240 Chinese patients with glioma, including 84 World Health Organization(WHO) II gliomas, 84 WHO III gliomas and 72 glioblastomas (WHO IV). Factors that correlated with EGFRvIII and EGFRvIII nuclear translocation expression were analyzed by the Chi-square test. Kaplan-Meier methodology and Cox regression were used for the survival analysis.Results: Log-rank tests showed that patient age, Karnofsky performance scale (KPS) score, tumor grade,EGFRvIII expression, EGFRvIII nuclear translocation, 1p/19q codeletion, isocitrate dehydrogenase (IDH)mutation, Ki-67 labeling index and O6-methylguanine-DNA methyltransferase (MGMT) status (P<0.05) were significantly correlated with overall survival (OS) time. Multivariate Cox regression analysis revealed that patient age, tumor grade, EGFRvIII nuclear translocation, 1p/19q codeletion, and IDH mutation (P<0.05) were significantly correlated with OS. Patients with a high level of EGFRvIII nuclear translocation (≥7%) had both significantly shorter OS [hazard ratio (HR): 1.920, 95% confidence interval (95% CI): 1.228-3.003, P=0.004] and progression-free survival (PFS) times (HR: 1.661, 95% CI: 1.116-2.471, P=0.012) than those with a low level of EGFRvIII nuclear translocation (<7%).Conclusions: A high level of EGFRvIII nuclear translocation in glioma is an independent factor indicating a poor prognosis, but EGFRvIII expression is not an independent clinical prognostic factor. The level of EGFRvIII nuclear translocation maybe a novel and crucial prognostic biomarker in glioma.
Keywords: Glioma; EGFRvIII expression; EGFRvIII nuclear translocation; biomarker; prognosis
Glioma is the most common intracranial tumor with a high degree of malignancy and strong invasiveness, accounting for approximately 80% of intracranial malignant tumors(1), of which glioblastoma multiform (GBM) exhibits the highest degree of malignancy. In recent years, despite continuous progress in surgical treatment, radiation therapy, chemotherapy, targeted therapy and comprehensive individualized treatment measures, the treatment effect and prognosis of glioma have remained poor, with a median overall survival (OS) of GBM patients of only 18 months despite undergoing surgery and postoperative radiotherapy and chemotherapy (2), and a five-year survival rate of less than 10% (3). Therefore, it is very important to actively explore the pathogenic mechanism of glioma and to discover new therapeutic targets.
Tumor cells have the ability to proliferate rapidly.During the process of rapid proliferation, if DNA repair and amplification disorders produce new gene mutations,tumor cells can further increase the growth rate and proliferative ability of tumor cells and thus increase their malignancy (4). Previous studies have confirmed that 57%of GBM patients contain epidermal growth factor receptor(EGFR) gene mutations, amplification, rearrangements and other genetic mutations (5), which are associated with various EGFR mutations. EGFR is a macromolecular transmembrane glycoprotein with a molecular weight of 170 kDa, which consists of an extracellular ligand junction and region, transmembrane hydrophobic region and intracellular kinase district 3 domains. EGFR and ErbB2(HER-2/neu), ErbB3, and ErbB4 four receptors together constitute the tyrosine kinase receptor family. Among them, epidermal growth factor receptor variant III(EGFRvIII) is the most common form of EGFR mutant,and compared to wild-type EGFR, EGFRvIII contains a partially deleted extracellular ligand junction, as well as mRNA deletion of exons 2 to 7 (6), leading to the deletion of a total of 801 base pairs in the extracellular domain. The changed extracellular structure of bases 6-273 in the domain creates a new binding site that differs from the wild-type EGFR. It can directly and sustainably activate EGFR-mediated activation without a corresponding EGFR ligand over a series of processes of downstream multiple effectors, such as the Ras-MAP kinase pathway, PI3K/AKT pathway and STAT-3 signal transduction pathway, which may participate in regulating cell growth and apoptosis (7),thereby promoting the abnormal proliferation of tumor cells and inhibiting apoptosis (8). EGFRvIII expression is increased in various malignant tumors such as breast cancer,non-small lung cell carcinoma, prostate cancer and glioma,but it is not expressed in normal human tissues (9-11). It is also closely related to the proliferation, invasion, migration,apoptosis and tumor-related angiogenesis of tumor cells (6).
Previous laboratory studies have shown that EGFRvIII may play a key role in the proliferation and invasion of GBM and some other tumors mainly in two ways: on the one hand, as a transmembrane protein without liganddependent activation, EGFRvIII can directly activate downstream signaling molecules, which could promote tumor proliferation, migration and invasion (7); on the other hand, exposure of the membrane protein EGFRvIII nuclear positioning signal during the process of endocytosis causes EGFRvIII translocation from the cell membrane to the nucleus (12), potentially activating many signaling pathways related to tumorigenesis and migration and indicating a poor prognosis in patients with GBM and some other tumors (13-15). However, the clinical prognostic significance of EGFR and EGFRvIII in glioma patients remains very controversial (16-20).
Thus, the purpose of this study was to identify the clinicopathological factors and prognostic implications associated with EGFRvIII overexpression and EGFRvIII nuclear translocation in patients with glioma.
Materials and methods
Patient selection and tumor specimens
During December 2011 and September 2015, 240 cases of glioma patients who underwent tumor removal in the neurosurgical department of Beijing Tiantan Hospital were included in the study. Two hundred and forty specimens,including 84 cases of World Health Organization (WHO)II glioma, 84 cases of WHO III glioma and 72 cases of WHO IV glioma, were obtained from the tumor resection.All tumor samples were independently histologically examined and graded by three experienced neuropathologists based on the 2007 WHO Classification of Tumors of the Central Nervous System (21). Specifically,the histological diagnoses of the tumor specimens were reviewed and confirmed by a third senior neuropathologist.If the first two pathologists did not agree on the diagnosis,a third senior neuropathologist would resolve the judgment. If the three neuropathologists could not reach an agreement, the case was submitted to the pathological committee of Beijing Neurosurgical Institute and Beijing Tiantan Hospital for a final diagnosis. All patients provided written informed consent to participate in the present study, which was approved by the Medical Ethics Committee of Capital Medical University.
The radiological, clinical, operative and pathological records of all patients were retrospectively evaluated. After treatment, all the patients were closely followed every month for the first year, every 3 months for the second year, and every 6 months thereafter, including the records of neuro-imaging, adjuvant therapies, OS and PFS. The adjuvant therapies after the operation included radiotherapy and chemotherapy. Radiotherapy consisted of a total dose of 60 Gy, which was separated into 30 daily fractions of 2 Gy each time. Chemotherapy regimens included temozolomide-based protocols (temozolomide 75 mg/m2during radiotherapy, and/or 150-200 mg/m2at 5 d/cycle after radiotherapy) (22) and nimustine (ACNU)-based protocols (ACNU 90 mg/m2, d 1, and teniposide(VM26) 60 mg/m2, d 1-3) (23). The OS was defined as the duration from the date of surgery to the date of death or last known follow-up, and the PFS was defined as the duration from the date of surgery to the date of recurrence as demonstrated by radiology.
Molecular detection
Molecular markers, including 1p/19q codeletion, isocitrate dehydrogenase1/2 (IDH1/2) mutation, and O6-methylguanine-DNA methyltransferase (MGMT)promotor methylation, were studied. Chromosomes 1 and 19 were analyzed by the fluorescence in situ hybridization(FISH) method, and the IDH1/2 mutation was detected by sequence analysis, both using a previously described protocol (24). MGMT promoter methylation was assessed by methylation-specific PCR (MSP) as described previously by our team (25).
Immunohistochemical staining
Immunohistochemical staining was performed using antibodies against Ki-67 and EGFRvIII as reported previously (16,26). Briefly, specimens were fixed in formalin and sectioned at a thickness of 4 μm. For antigen retrieval, the slides were boiled in 10 mmol/L citrate buffer(pH 6.0) for 2 min after deparaffinization and rehydration.Endogenous peroxidase was then blocked with 3% aqueous hydrogen peroxide. The sections were incubated with primary antibody at 4 ℃ overnight. Next, the sections were washed five times with phosphate buffer solution (PBS) and incubated with the secondary antibody at 37 ℃ for 30 min.Then, the antibodies were detected using diaminobenzidine as a chromogen, and the slides were counterstained with hematoxylin. Primary antibodies were diluted in PBS with 1% bovine serum albumin (BSA) at the following concentrations: mouse monoclonal anti-human Ki-67, 1:400, was purchased from Santa Cruz Biotechnology (Dallas, TX, USA); ready-to-use mouse monoclonal anti-EGFRvIII antibody (cat#HTA0001) was purchased from Beijing Cellonis Biotechnologies Co.,LTD (Beijing, China), which exclusively detects EGFRvIII protein and does not cross-react with wild type EGFR (27).
Evaluation of Ki-67 labeling index and EGFRvIII expression
The Ki-67 and EGFRvIII immunohistochemical staining results were semiquantitatively scored as reported previously (17). The staining intensity was calculated by two experienced pathologists without knowledge of the patients' clinical information. Ki-67 with a brownish brown or brown nucleus showed positive staining, and five randomly selected high magnification (×400) fields were counted. The expression levels considered a positive rate of 5% as the dividing line for Ki-67. EGFRvIII staining of brown granules in the cell membrane/cytoplasm and/or nuclear was positive. Under a high-power field (×400),positive cells from 5 randomly selected fields were counted.Positive cell counts ≤4.0% were given a score of 1, between 5%-29% a score of 2, between 30%-59% a score of 3, and≥60% a score of 4, according to the staining strength. A colorless count was given a score of 0, pale brown a score of 1, medium brown a score of 2, and brown a score of 3.Based on the product of the two scores, the immunoreactivity for EGFRvIII was finally scored as follows: 0, negative; 1-4, weakly positive; 6-8, moderately positive; 9-12, strongly positive. According to the maximally selected log-rank statistic, an EGFRvIII score <4 was regarded as low expression of EGFRvIII, and an EGFRvIII score ≥4 was regarded as high expression of EGFRvIII. Controls without positive control tissues and primary antibody were included in all cases to guarantee the quality of staining. In the case of any contradiction, the two observers reviewed the slides simultaneously to reach an agreement.
Evaluation of EGFRvIII nuclear translocation
The EGFRvIII-positive (“weakly positive”, “moderately positive” and “strongly positive”) expression tumor specimens were independently assessed for nuclear staining by two experienced pathologists who were blinded to the patient clinical information. At high magnification (×400),positive cells from 5 randomly selected fields were counted(at least 200 tumor cells per field were counted), as previously reported (28). According to the maximally selected log-rank statistic, the EGFRvIII-positive expression tumor specimens were categorized into 2 groups based on the proportion of nuclear translocation. When the proportion of labeled nuclear/all nuclear tumor was 7% or more, the nuclear translocation of the specimen was regarded as “high level”; when the proportion was less than 7% or EGFRvIII expression was exclusively membrane/cytoplasmic, the nuclear translocation of the specimen was considered “l(fā)ow level”.
Analysis of extent of resection and tumor size
Tumor size and extent of resection were assessed with magnetic resonance imaging (MRI) performed within 3 d of surgery. We used Neusoft PACS/RIS image diagnostic workstation V5.5.5.70613 (Neusoft Medical Systems Co.,Ltd., Shenyang, China) for semiautomatic volumetry to measure the extent of resection, as gross-total resection(GTR) (≥98% resection) and no GTR (<98%) (29). Tumor size was determined by the maximal diameter of the tumor in the axial, coronary and/or sagittal planes. We determined the tumor size and extent of resection using the contrast-enhanced region on the contrast-enhanced T1-weighted images of high-grade (WHO III-IV) gliomas; the tumor size and extent of resection of low-grade (WHO II)gliomas were measured using T2-weighted/Flair images.
Statistical analysis
All data were analyzed using IBM SPSS Statistics (Version 25.0; IBM Corp., New York, USA) and R software (version 3.5.0; R Foundation for Statistical Computing, Vienna,Austria). The maximally selected log-rank statistic was used to dichotomize EGFRvIII expression and EGFRvIII nuclear translocation for OS. A minimum P value approach was used to perform a cutoff point analysis. The maximally selected log-rank statistic was calculated using the “maxstat(version 0.7-25)” package in R. The Chi-square test (or Fisher's exact test), t-test and nonparametric test were used to compare categorical and continuous variables between groups as appropriate. Kaplan-Meier (log-rank test) and Cox regression analyses were used to evaluate independent factors for progression-free survival (PFS) and OS. Because of the similarity between EGFRvIII expression and EGFRvIII nuclear translocation in nature and because EGFRvIII expression was highly correlated with EGFRvIII nuclear translocation (P<0.001), EGFRvIII expression and EGFRvIII nuclear translocation were analyzed separately in the Cox regression analysis to prevent multicollinearity. All statistical tests were two-sided using a 0.05 significance level.
Results
Overall characteristics of study patients
This study cohort included 240 Chinese patients with glioma ranging from WHO II-IV. The clinical details of all the patients are shown in Table 1. The patient ages ranged from 14 to 72 years with a mean of 42±11 years.One hundred forty-three (59.6%) patients were male, and 97 (40.4%) were female. The median preoperative Karnofsky performance scale (KPS) score was 80 (range,20-100). Tumor sizes ranged from 2.0 to 10.0 cm, with a mean size of 5.7±1.6 cm. One hundred sixty-four (68.3%)patients received gross total resection (GTR) of the tumor,and 76 (31.7%) patients received non-GTR resection. One hundred ninety-eight patients (82.5%) received postoperative chemotherapy, and 178 patients (74.2%)received postoperative radiotherapy.
Until the last follow-up, 107 of 240 (44.6%) patients experienced tumor progression, and 85 of 240 (35.4%)patients died. The mean PFS and OS was 35.3 (95% CI,33.9-36.7) months and 40.2 (95% CI, 39.0-41.4) months,respectively. Only four patients were lost to follow-up and were considered censored observations in the survival analysis.
EGFRvIII overexpression in glioma tissues
Immunohistochemistry revealed EGFRvIII staining in the cytomembrane/cytoplasm and nuclei of glioma cells(Figure 1). Cases with EGFRvIII expression (“moderately positive” and “strongly positive”) were detected in 73 of 240 (30.4%) gliomas. The high expression rates of EGFRvIII cases were found in all WHO grades of gliomas analyzed: WHO grade II, III and IV was 10/84 (11.9%),20/84 (23.8%) and 43/72 (59.7%), respectively (Table 1).
EGFRvIII nuclear translocation in glioma tissues with EGFRvIII-positive expression
EGFRvIII nuclear translocation proportion analysis wasperformed in glioma specimens with EGFRvIII-positive(“weakly positive”, “moderately positive” and “strongly positive”) expression (Figure 1). Cases with EGFRvIII“high level” nuclear translocation (the proportion of labeled nuclear/all tumor nuclear was 7% or more) were detected in 95 of 240 (39.6%) gliomas. A “high level”nuclear translocation of EGFRvIII cases was found in all WHO grades of gliomas analyzed: WHO grade II, III and IV were 26/84 (31.0%), 20/84 (23.8%) and 49/72 (68.1%),respectively (Table 1).
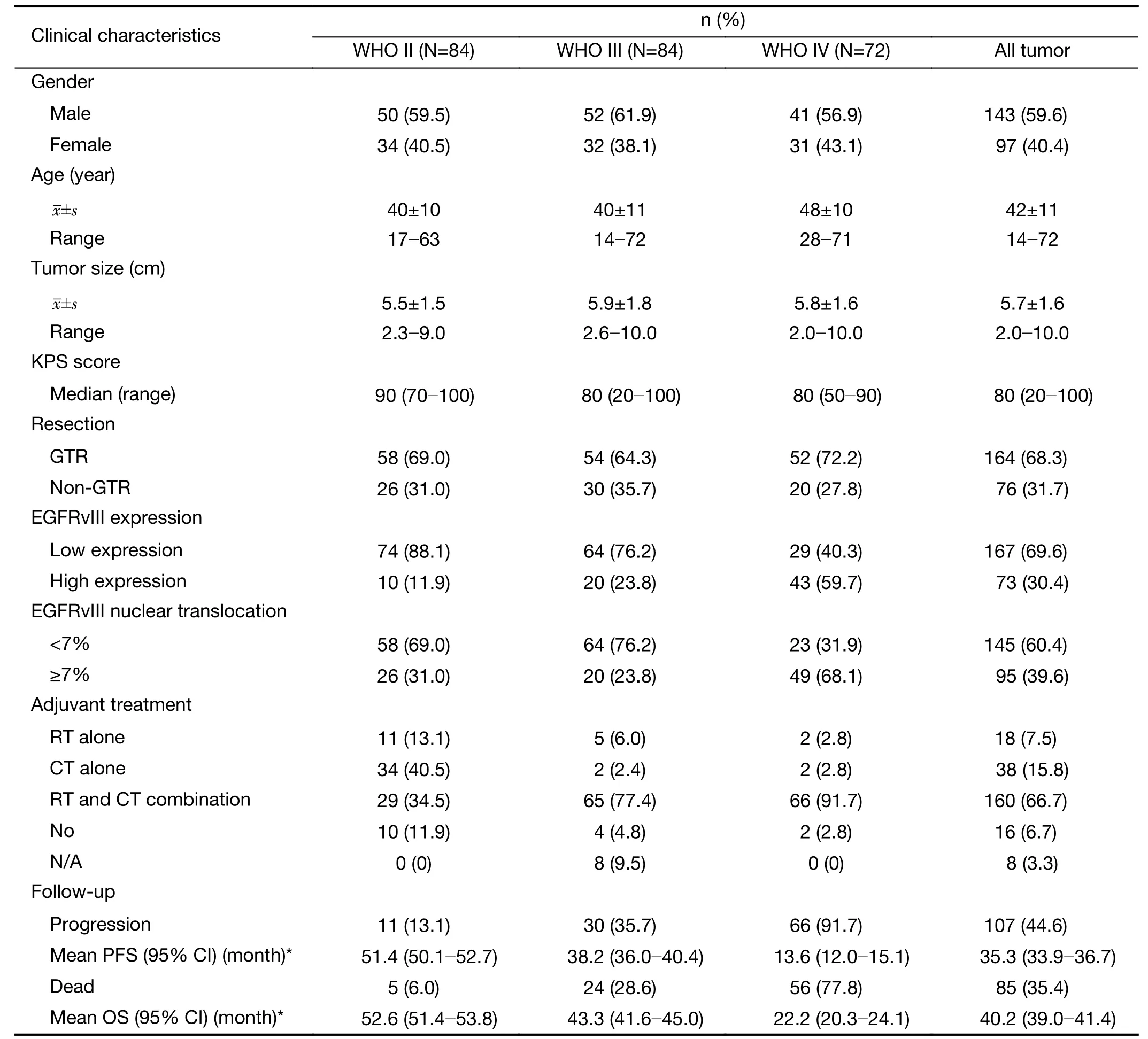
Table 1 Baseline characteristics for all patients (N=240)
Factors correlated with EGFRvIII expression and EGFRvIII nuclear translocation
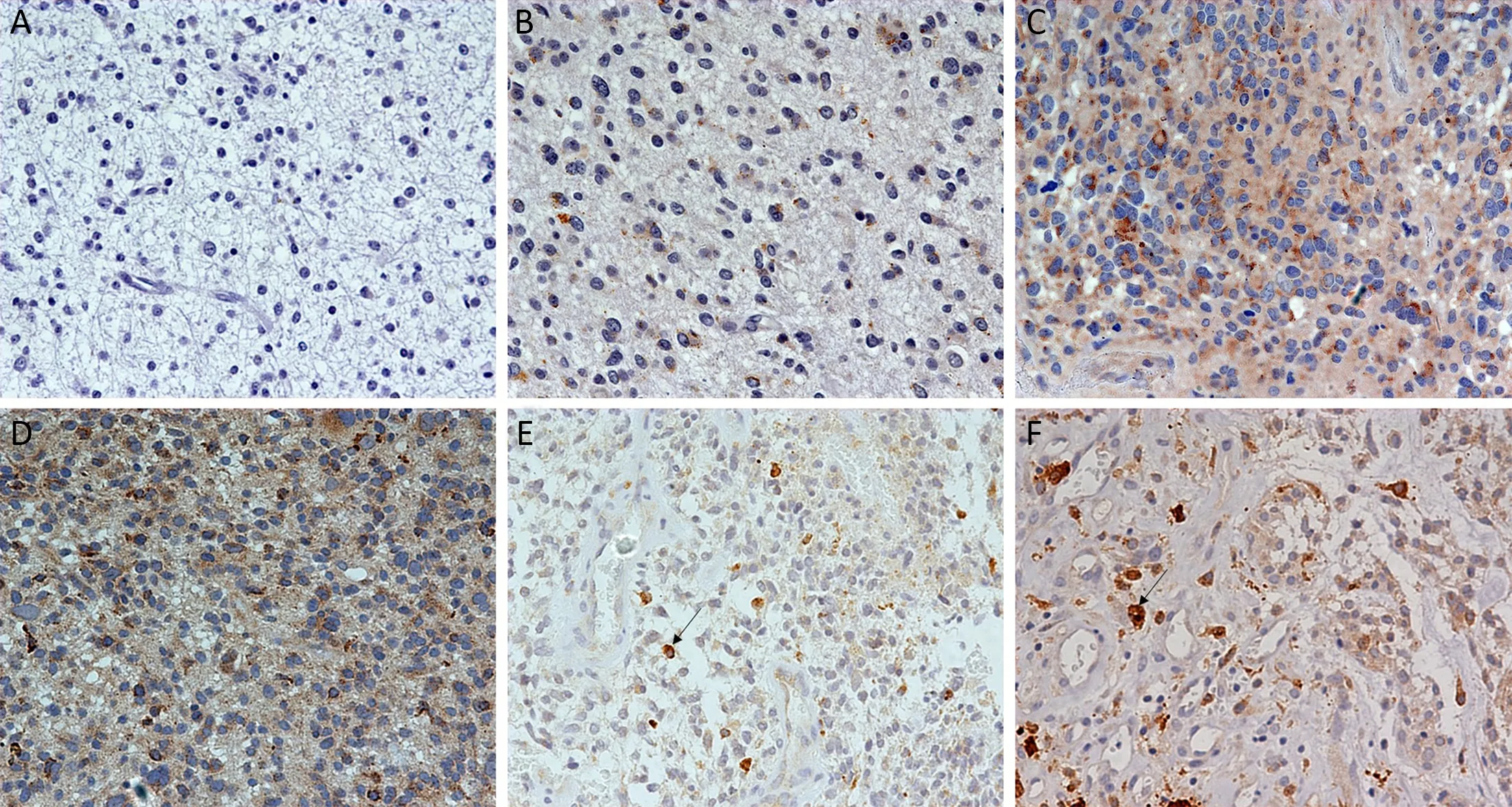
Figure 1 Immunohistochemical staining for epidermal growth factor receptor variant III (EGFRvIII) expression and EGFRvIII nuclear translocation in glioma. (A) Negative EGFRvIII expression; (B) Weakly positive EGFRvIII expression; (C) Moderately positive EGFRvIII expression; (D) Strongly positive EGFRvIII expression; (E) Proportion of labeled nuclei/all tumor nuclei less than 7% (low-level EGFRvIII nuclear translocation); (F) Proportion of labeled nuclei/all tumor nuclei greater than 7% (high-level of EGFRvIII nuclear translocation).Original magnification (×400).
The factors associated with EGFRvIII expression were analyzed by the Chi-square test, including patient age,gender, KPS score, tumor size, tumor grade, IDH1/2 mutation, 1p/19q codeletion, tumor origin, MGMT methylation, Ki-67 labeling index and EGFRvIII nuclear translocation. Univariate analysis revealed that patient age(P=0.001), KPS score (P=0.037), tumor grade (P<0.001),IDH1/2 mutation (P<0.001), Ki-67 labeling index(P=0.020) and EGFRvIII nuclear translocation (P<0.001)were correlated with EGFRvIII expression (Table 2).
The factors associated with EGFRvIII nuclear translocation were analyzed by the Chi-square test, including patient age, gender, KPS score, tumor size, tumor grade,IDH1/2 mutation, 1p/19q codeletion, MGMT methylation, Ki-67 labeling index and EGFRvIII expression. Univariate analysis revealed that patient age(P=0.030), KPS score (P=0.019), tumor grade (P=0.045),IDH1/2 mutation (P=0.038) and EGFRvIII expression(P<0.001) were correlated with EGFRvIII nuclear translocation (Table 3).
Factors correlated with survival by univariate analysis
The clinical prognostic factors associated with PFS and OS were analyzed by Kaplan-Meier survival analyses (Table 4).According to the log-rank analysis, the prognostic factors that correlated with a longer PFS included patient age <50 years (P<0.001), KPS score >80 (P<0.001), lower EGFRvIII expression (Figure 2A, P=0.001), a low level of EGFRvIII nuclear translocation (Figure 2C, P<0.001), Ki-67 labeling index ≤5% (P=0.001), lower tumor grade (P<0.001),1p/19qcodeletion (P<0.001), MGMT methylation(P<0.001) and IDH1/2 mutation (P<0.001), as shown in Table 4.
The factors that correlated with a longer OS included patient age <50 years (P<0.001), KPS score >80 (P<0.001),lower EGFRvIII expression (Figure 2B, P=0.002), a low level of EGFRvIII nuclear translocation (Figure 2D,P<0.001), Ki-67 labeling index ≤5% (P=0.001), lower tumor grade (P<0.001), 1p/19q codeletion (P<0.001),MGMT methylation (P<0.001) and IDH1/2 mutation(P<0.001), as shown in Table 4.
EGFRvIII nuclear translocation independently predicts shorter survival in glioma patients by multivariate analysis
Patient age, KPS score, tumor grade, 1p/19q codeletion,MGMT methylation, IDH1/2 mutation, Ki-67 labeling index, EGFRvIII expression and EGFRvIII nuclear translocation were included in the Cox regression analysis(Table 5, 6).
In the multivariate Cox regression analysis for PFS, the factors that independently correlated with PFS were age ≥50 years (HR: 1.758, 95% CI: 1.139-2.712, P=0.011),higher tumor grade (HR: 4.839, 95% CI: 2.516-9.038,P<0.001), 1p/19q codeletion (HR: 0.426, 95% CI:0.228-0.794, P=0.007), IDH mutation (HR: 0.513, 95%CI: 0.312-0.843, P=0.008), KPS score >80 (HR: 0.524,95% CI: 0.351-0.782, P=0.002) and a high level of EGFRvIII nuclear translocation (HR: 1.661, 95% CI:1.116-2.471, P=0.012) (Table 5).

Table 2 Clinical factors associated with EGFRvIII expression level by univariate analysis
In the multivariate Cox regression analysis for OS, the factors that independently correlated with OS were age ≥50 years (HR: 1.733, 95% CI: 1.079-2.781, P=0.023), higher tumor grade (HR: 7.972, 95% CI: 3.127-20.324, P<0.001),1p/19q codeletion (HR: 0.423, 95% CI: 0.205-0.873,P=0.020), IDH mutation (HR: 0.575, 95% CI:0.334-0.991, P=0.046) and a high level of EGFRvIII nuclear translocation (HR: 1.920, 95% CI: 1.228-3.003,P=0.004) (Table 6).
Discussion
In recent years, molecular targeted therapy for cancer hasbeen increasing (30,31). Its greatest advantage is that it can specifically act on the corresponding molecular targets without obvious side effects on other tissues. Therefore,actively exploring specific and effective molecular targets for the treatment of glioma is of great significance.Currently, the prognostic significance of EGFR/EGFRvIII in patients with glioma has attracted extensive attention.Several laboratory studies have confirmed that EGFRvIII overexpression is closely related to proliferation, invasion,migration, apoptosis and tumor-related angiogenesis of glioblastoma cells (6,32-34). Considering the role of EGFRvIII in tumor proliferation and exclusive expression on tumor cells, EGFRvIII has become an ideal target for identifying and developing novel therapeutic approaches.Various targeting strategies for EGFRvIII in GBM have been described to date, including anti-EGFR antibodybased approaches (35), therapeutic vaccines (36,37),chimeric antigen receptor (CAR) T-cell therapy (38) and Bispecific T Cell Engager (39).
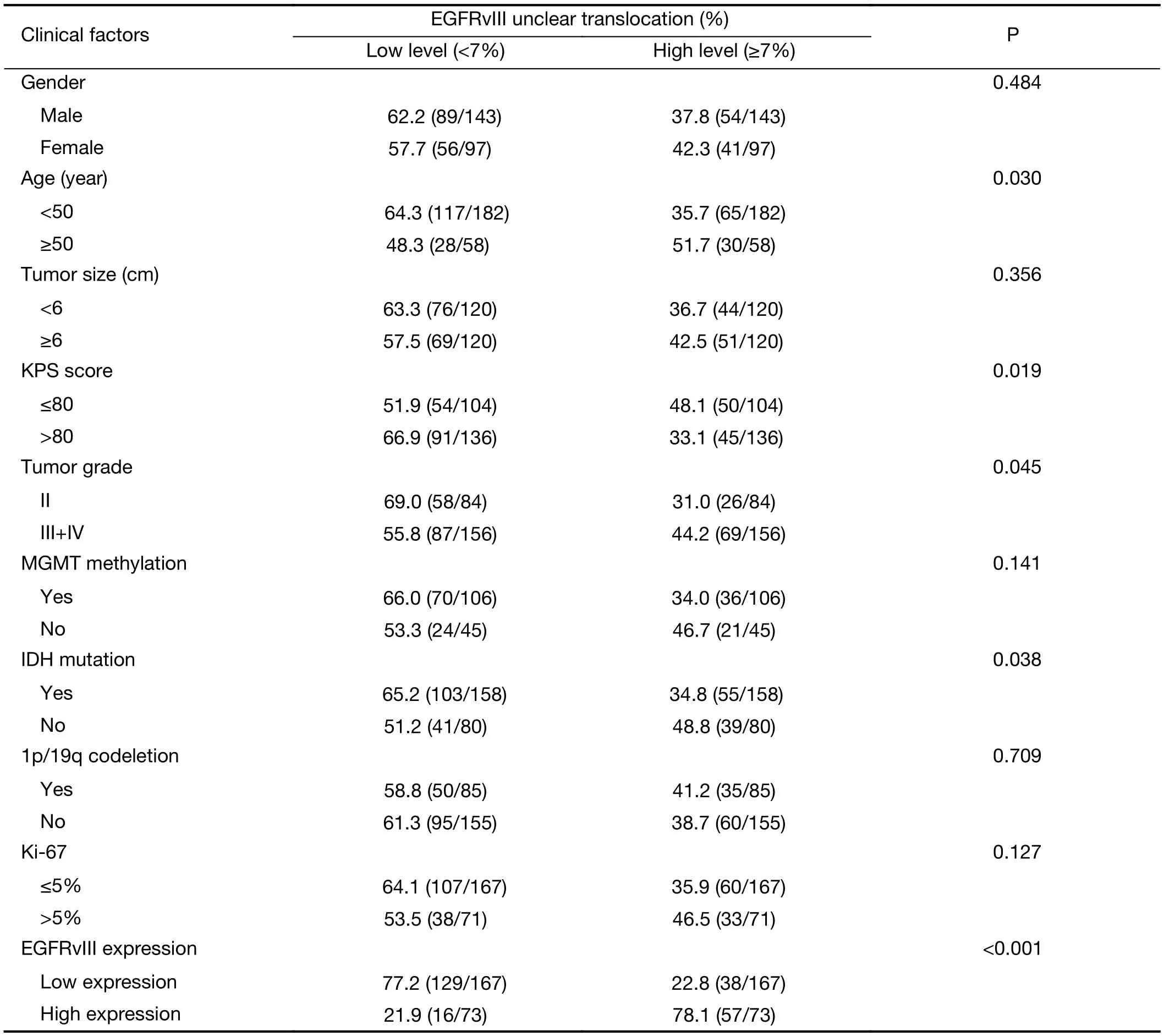
Table 3 Clinical factors associated with EGFRvIII nuclear translocation level by univariate analysis
However, the prognostic role of EGFRvIII in patients with glioma has been highly controversial. Weller et al.(16) examined 184 glioma patients and found that EGFRvIII status was not related to OS or PFS in patients who received adjuvantradio-chemotherapy. Viana-Pereira et al. (18) analyzed 55 patients with glioma, including 27 primary glioblastomas (GBM), 24 anaplastic oligodendrogliomas (AO) and 4 anaplastic oligoastrocytomas(AOA), and found no association between EGFRvIII and patient survival. However, some other studies hold contradictory opinions. Shinojima et al. (19) studied 87 GBM patients and found that in patients with EGFR amplification, EGFRvIII overexpression was a significant and independent predictor of a shorter OS. Layfield et al.(40) also found that EGFR amplification with EGFRvIII overexpression was a strong indicator of poor survival in 32 GBM patients. In our patient cohort, we found that high EGFRvIII expression was associated with a significantly worse PFS (mean 35.1 vs. 47.8 months, P=0.001) and OS(mean 43.8 vs. 54.5 months, P=0.002) than low EGFRvIII expression by Kaplan-Meier survival analyses (Figure 2A, B,Table 4). However, the Cox proportional hazards regression analysis for PFS and OS showed that EGFRvIIIexpression was not an independent prognostic factor (Table 5, 6). Univariate analysis revealed that patient age ≥50 years(P=0.001), KPS score ≤80 (P=0.037), a higher tumor grade(P<0.001), IDH wild type (P<0.001), Ki-67 labeling index>5% (P=0.020) and EGFRvIII nuclear translocation(P<0.001) were correlated with higher EGFRvIII expression, and all of these factors predicted a shorter survival time (Table 2, 4). In previous studies, EGFR amplification and EGFRvIII mutation have been strongly correlated with increasing age (20) and a higher tumor grade (18), both of which are factors leading to a poorer prognosis.
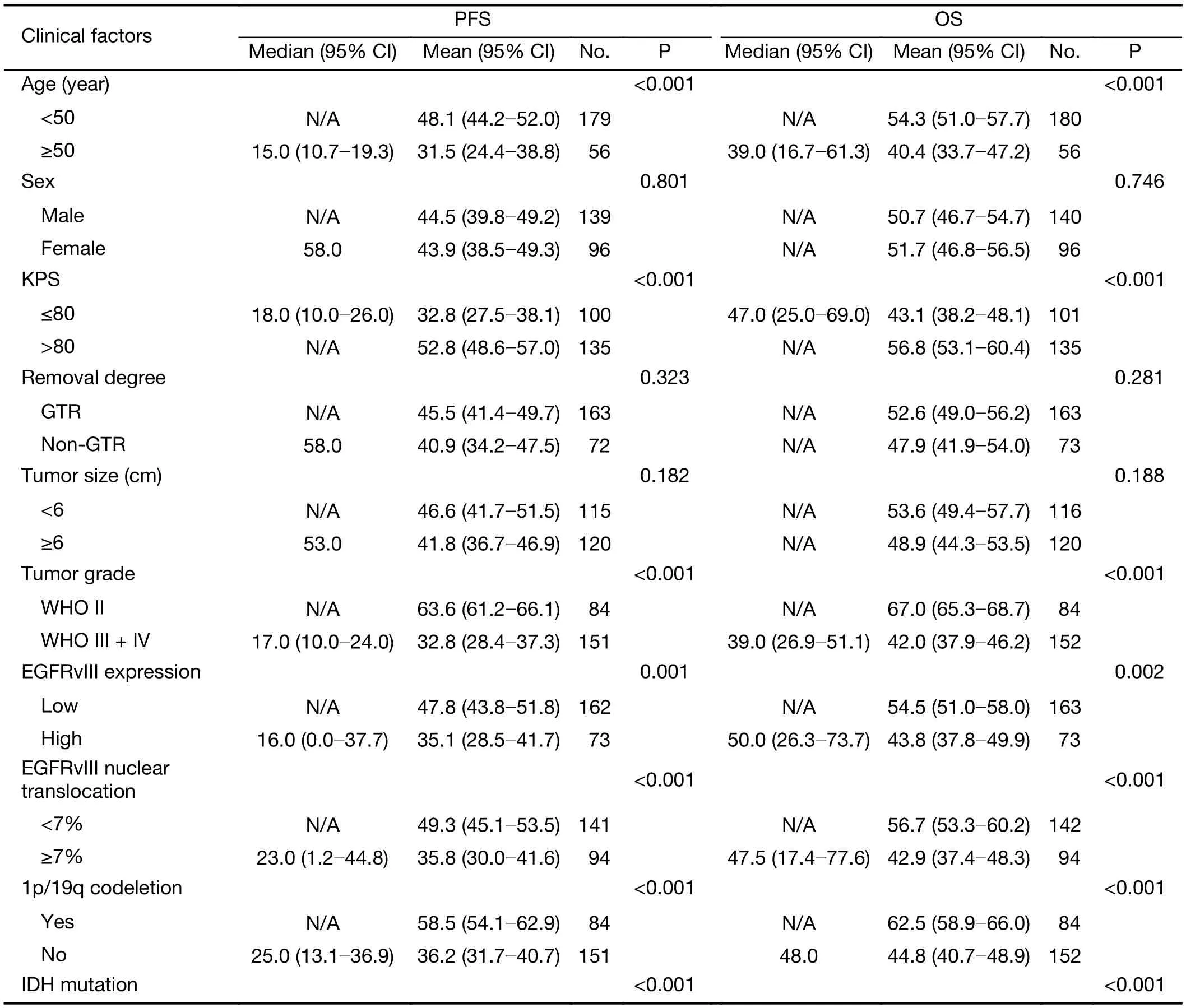
Table 4 Clinical prognostic factors associated with PFS and OS analyzed by Kaplan-Meier survival analyses
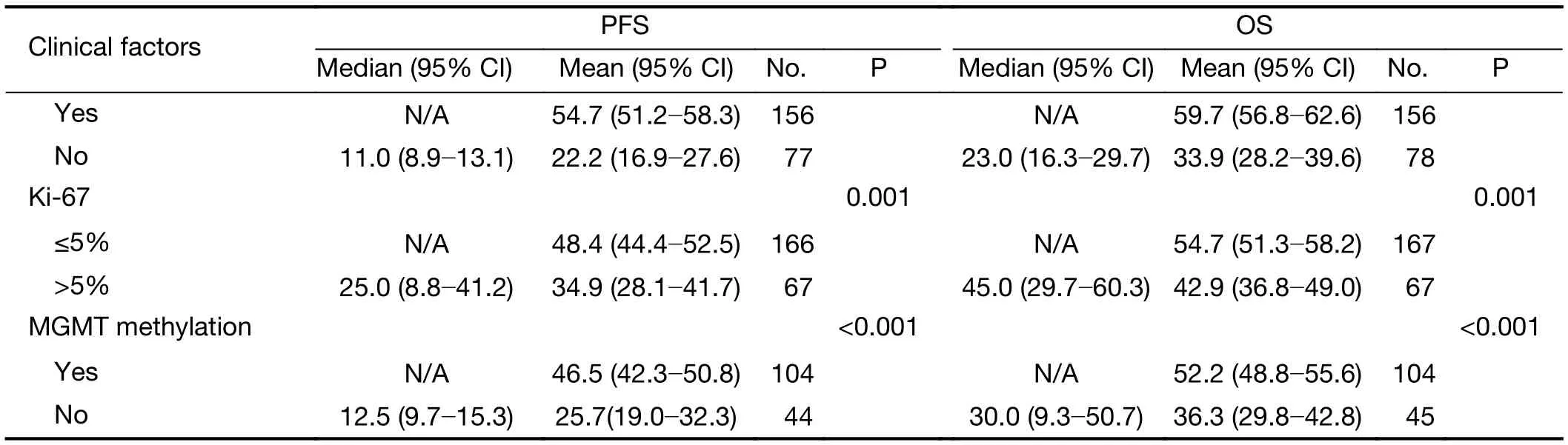
Table 4 (continued)
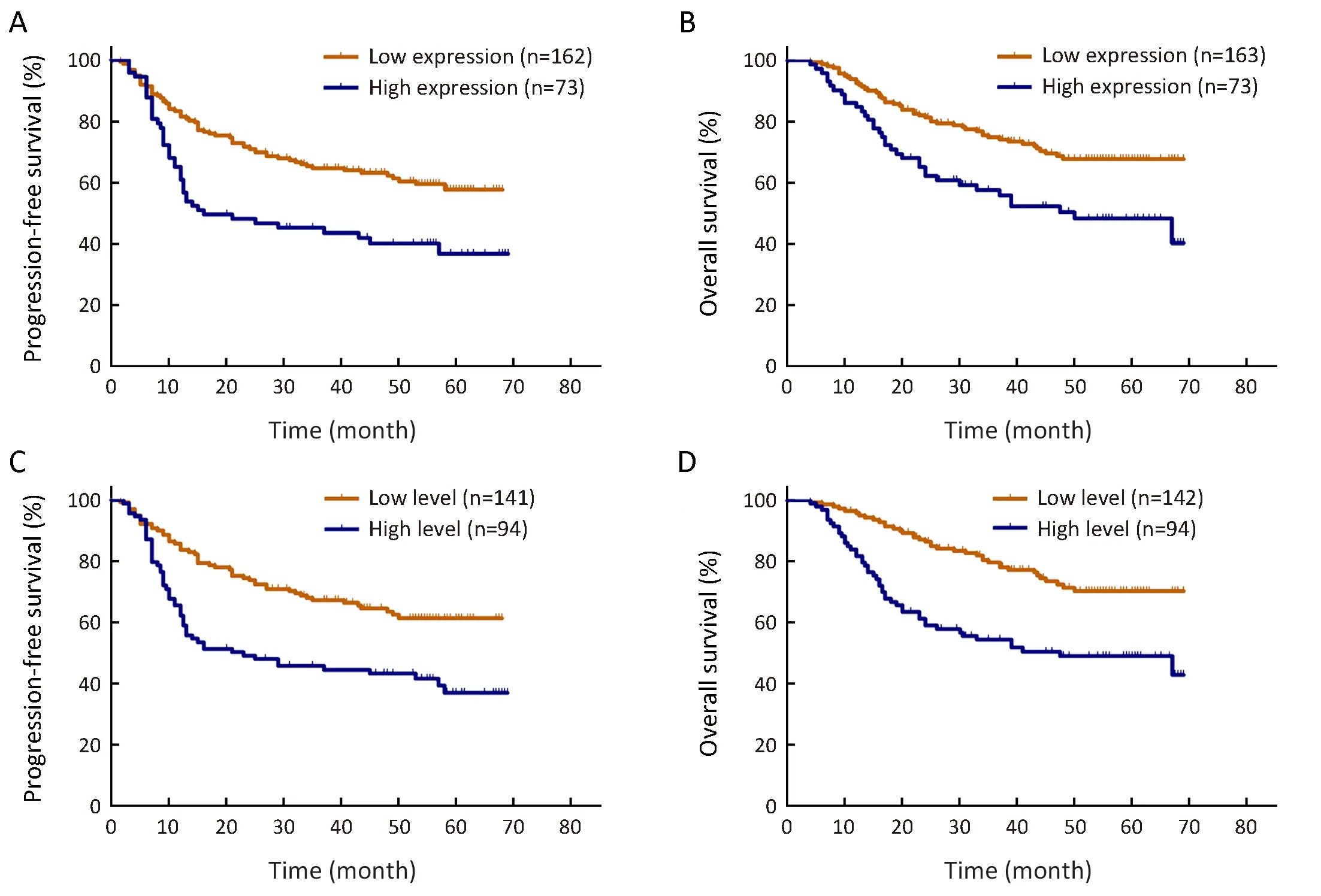
Figure 2 Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) rates in our patient cohort. High expression of epidermal growth factor receptor variant III (EGFRvIII) predicted a shorter survival for PFS (P=0.001) (A) and for OS (P=0.002) (B), and a high level of EGFRvIII nuclear translocation also predicted a shorter survival in glioma for PFS (P<0.001) (C) and for OS (P<0.001) (D).
We found that EGFRvIII nuclear translocation is an independent factor indicating a poor prognosis in patients with glioma (Table 5, 6). In our patient cohort, patients with a high level of EGFRvIII nuclear translocation had asignificantly worse PFS (mean 35.8 vs. 49.3 months,P<0.001) and OS (mean 42.9 vs. 56.7 months, P<0.001)than those with a low level of EGFRvIII nuclear translocation by Kaplan-Meier survival analyses (Figure 2C,D, Table 4). In the Cox proportional hazards regression analysis, patients with a high level of EGFRvIII nuclear translocation had both a significantly shorter PFS (HR:1.661, 95% CI: 1.116-2.471, P=0.012) and OS (HR: 1.920,95% CI: 1.228-3.003, P=0.004) than those with a low level of EGFRvIII nuclear translocation (Table 5, 6). Univariate analysis revealed that patient age ≥50 years (P=0.030), KPS score ≤80 (P=0.019), a higher tumor grade (P=0.045), IDH wild type (P=0.038) and higher EGFRvIII expression(P<0.001) were correlated with a high level of EGFRvIII nuclear translocation, and all of these factors indicated a poor prognosis (Table 3, 4).
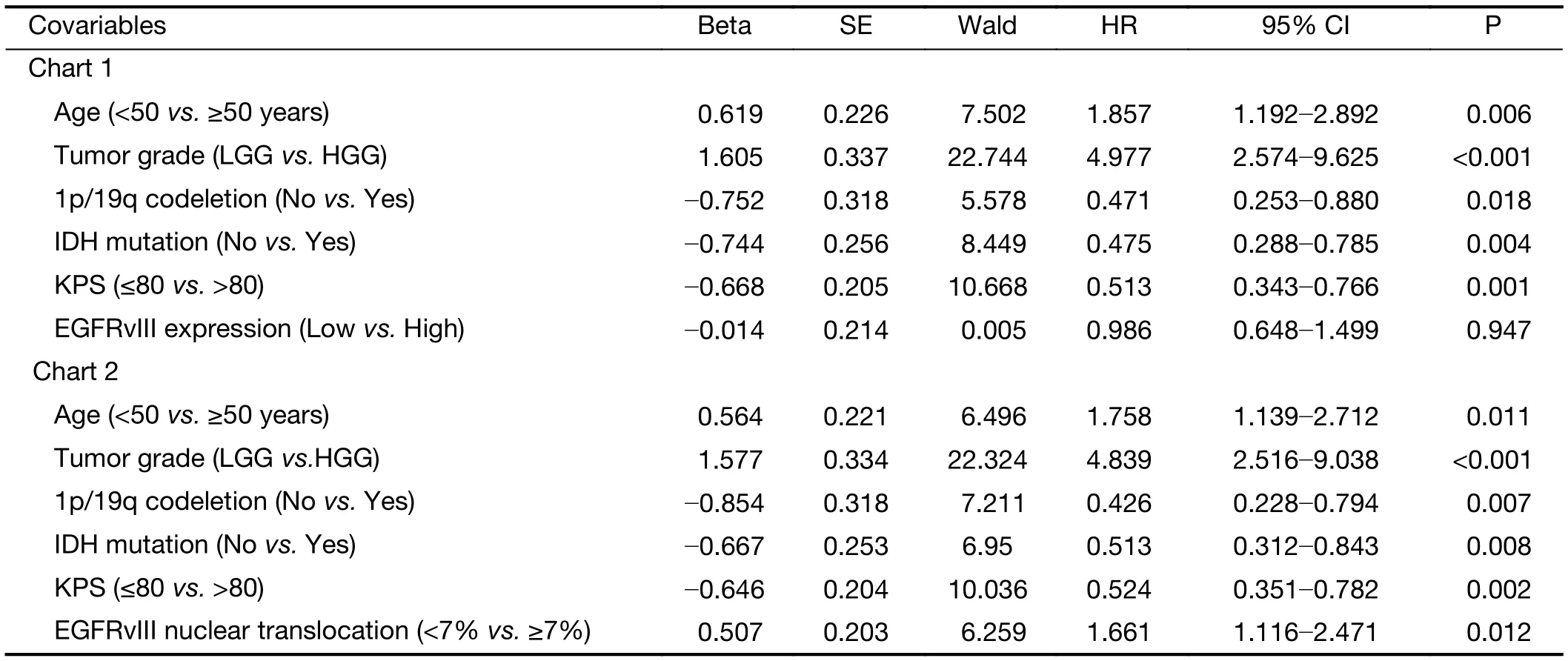
Table 5 Multivariate Cox regression of risk factors associated with PFS
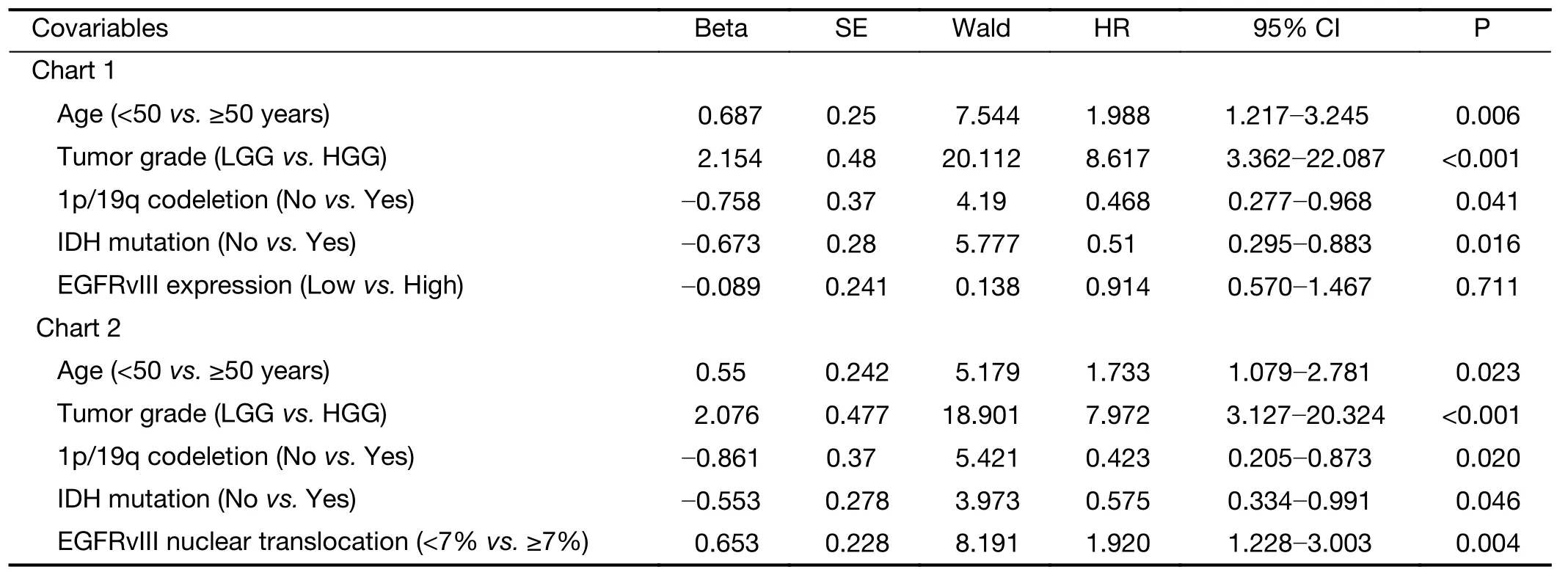
Table 6 Multivariate Cox regression of risk factors associated with OS
To date, to the best of our knowledge, no studies have confirmed the specific mechanism of the nuclear translocation of EGFRvIII; however, the pathway of EGFR nuclear translocation has been previously reported. The mechanism of EGFR nuclear translocation reported by previous research can be summarized into two categories.First, the transmembrane protein EGFR is a liganddependent receptor molecule; after binding to its ligand molecules, the extracellular signal is transmitted into cells.The ligand molecules then induce endocytosis of receptor molecules, and cell surface EGFR is translocated into the inner cytoplasm (41). Subsequently, one form of the EGFR nuclear transposition pathway is, after endocytosis mediated by clathrin (42), the sorting of EGFR to the early endosome followed by the binding of nuclear importin-β,alone or together with importin-α, to the nuclear localization signal of EGFR to eventually guide EGFR into the nucleus (43). Another possible form of EGFR nuclear translocation involves the reverse transport pathways mediated by the Golgi apparatus and endoplasmic reticulum (44). After endocytosis, EGFR vesicles fuse with the early endosome, loop through the Golgi apparatus and endoplasmic reticulum mediated by a variety of other proteins, and then, with the help of importin-β or/and importin-α, EGFR traverses the nuclear pore complexes and finally translocates into the nuclear matrix or inner nuclear membrane (45). EGFR may play an important role in regulating downstream signaling pathways when it enters the nucleus through the nuclear translocation pathway, including the PLC-γPKC pathway, Ras-MAP kinase pathway, PI3K/AKT pathway and JAK2-STAT3/5 pathway (7), which are known to be critical for the maintenance of GBM cancer stem cells (46,47) and closely related to the self-renewal, proliferation, invasion,migration, apoptosis and tumor-related angiogenesis of tumor cells (6).
In recent years, the nuclear translocation phenomenon of EGFR has been found in various malignant tumors such as glioma (13), non-small lung cell carcinoma (14), breast cancer (48) and ovarian cancer (49), and it is closely related to tumor progression and migration. Lo HW et al. (49)analyzed 130 patients with breast carcinomas and found that 37.7% of the patients showed positive immunostaining for nuclear EGFR and that nuclear EGFR expression was significantly associated with high levels of Ki-67 and cyclin D1, both of which were indicators for cell proliferation and poor prognoses. In addition, they also analyzed EGFR expression in 37 patients with ovarian cancer and found that 24.3% of the patients had moderate or high levels of nuclear EGFR, which was associated with a shorter survival time. The phenomenon of EGFR nuclear translocation has also been found in GBM patients, leading to a poorer prognosis and more severe progression of the tumor (13).In our patient cohort, a high level of EGFRvIII nuclear translocation was detected in 95 of 240 (39.6%) gliomas,with WHO grade II, III and IV demonstrating 26/84(31.0%), 20/84 (23.8%) and 49/72 (68.1%) cases,respectively (Table 1). It was also significantly correlated with the tumor grade (P=0.045) and a poor prognosis(Table 3, Figure 2C, D). Our findings reveal for the first time the clinical significance of EGFRvIII nuclear translocation, which may replace EGFRvIII expression as a novel clinical prognostic indicator.
The greatest advantage of this study is that we demonstrated and compared the clinical prognostic significance of EGFRvIII overexpression and EGFRvIII nuclear translocation in Chinese glioma patients for the first time. However, this study has some limitations. First,the sample size in our patient cohort was not very large,and thus more samples are needed for further analysis.Second, this was a retrospective analysis including samples from 2011 to 2015, and prognostic biomarkers such as ATRX, TERT and H3K27M were not available for all samples and were not analyzed in the present study. Thus,the study was vulnerable to potential biases from other unobserved biomarkers. Third, this study did not confirm the specific mechanism of the nuclear translocation of EGFRvIII, necessitating further research.
Conclusions
We found that a high proportion of glioma specimens in our cohort exhibited EGFRvIII overexpression and EGFRvIII nuclear translocation. We also demonstrated the prognostic significance of EGFRvIII overexpression and EGFRvIII nuclear translocation in glioma. Although EGFRvIII overexpression is not an independent prognostic indicator, the high level of EGFRvIII nuclear translocation independently predicted a shorter survival and poor prognosis in the present study. Therefore, our results highlight the importance of further studies to confirm the specific mechanism of EGFRvIII nuclear translocation and suggest the potential of specific therapies targeting EGFRvIII in patients with glioma.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81771309).
Footnote
Conflicts of Interest: The authors have no conflicts of interests to declare.
 Chinese Journal of Cancer Research2019年1期
Chinese Journal of Cancer Research2019年1期
- Chinese Journal of Cancer Research的其它文章
- STIM1 expression is associated with osteosarcoma cell survival
- Risk-stratification model to select conversion surgery for advanced gastric cancer patients
- Metastatic patterns and surgical methods for lymph nodes No. 5 and No. 6 in proximal gastric cancer
- Prognostic value of 18F-fluorodeoxyglucose positron emission tomography using Deauville criteria in diffuse large B cell lymphoma treated with autologous hematopoietic stem cell transplantation
- Clinical features and outcomes of diffuse large B-cell lymphoma based on nodal or extranodal primary sites of origin: Analysis of 1,085 WHO classified cases in a single institution in China
- Incidence and mortality of thyroid cancer in China, 2008-2012
