Application of the back-error propagation artificial neural network (BPANN) on genetic variants in the PPAR-γ and RXR-α gene and risk of metabolic syndrome in a Chinese Han population
Xu Zho, Kng Xu, Hui Shi, Jinluo Cheng, Jinhu M, Ynqin Go, Qin Li, Xinhu Ye, Ying Lu, Xiofng Yu, Jun Du, Wenong Du, Qing Ye, Ling Zhou,
aDepartment of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu 211166, China;
bDepartment of Endocrinology, the Affiliated Changzhou Second Hospital, Nanjing Medical University, Changzhou, Jiangsu 213003, China;
cDepartment of Endocrinology, the First Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu 210029, China;
dDepartment of Endocrinology, the Third Affiliated Hospital, Nanjing Medical University, Yizheng, Jiangsu 211400, China.
Application of the back-error propagation artificial neural network (BPANN) on genetic variants in the PPAR-γ and RXR-α gene and risk of metabolic syndrome in a Chinese Han population
Xu Zhaoa,△, Kang Xua,△, Hui Shia, Jinluo Chengb, Jianhua Mac, Yanqin Gaod, Qian Lic, Xinhua Yeb, Ying Lua, Xiaofang Yua, Juan Dua, Wencong Dua, Qing Yea, Ling Zhoua,
aDepartment of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu 211166, China;
bDepartment of Endocrinology, the Affiliated Changzhou Second Hospital, Nanjing Medical University, Changzhou, Jiangsu 213003, China;
cDepartment of Endocrinology, the First Affiliated Hospital, Nanjing Medical University, Nanjing, Jiangsu 210029, China;
dDepartment of Endocrinology, the Third Affiliated Hospital, Nanjing Medical University, Yizheng, Jiangsu 211400, China.
This study was aimed to explore the associations between the combined effects of several polymorphisms in the PPAR-γand RXR-αgene and environmental factors with the risk of metabolic syndrome by back-error propagation artificial neural network (BPANN). We established the model based on data gathered from metabolic syndrome patients (n = 1012) and normal controls (n = 1069) by BPANN. Mean impact value (MIV) for each input variable was calculated and the sequence of factors was sorted according to their absolute MIVs. Generalized multifactor dimensionality reduction (GMDR) confirmed a joint effect of PPAR-γand RXR-αbased on the results from BPANN. By BPANN analysis, the sequences according to the importance of metabolic syndrome risk factors were in the order of body mass index (BMI), serum adiponectin, rs4240711, gender, rs4842194, family history of type 2 diabetes, rs2920502, physical activity, alcohol drinking, rs3856806, family history of hypertension, rs1045570, rs6537944, age, rs17817276, family history of hyperlipidemia, smoking, rs1801282 and rs3132291. However, no polymorphism was statistically significant in multiple logistic regression analysis. After controlling for environmental factors, A1, A2, B1and B2(rs4240711, rs4842194, rs2920502 and rs3856806) models were the best models (cross-validation consistency 10/10, P = 0.0107) with the GMDR method. In conclusion, the interaction of the PPAR-γand RXR-αgene could play a role in susceptibility to metabolic syndrome. A more realistic model is obtained by using BPANN to screen out determinants of diseases of multiple etiologies like metabolic syndrome.
back-error propagation artificial neural network (BPANN), metabolic syndrome, peroxisome proliferators activated receptor-γ(PPAR) gene, retinoid X receptor-α(RXR-α) gene, adiponectin
INTRODUCTION
Metabolic syndrome is characterized by the simultaneous presence of interrelated metabolic risk factors, such as visceral obesity, high blood pressure and carbohydrate and lipid metabolism abnormalities[1], and it leads to an increased risk for type 2 diabetes mellitus and cardiovascular diseases[2,3]. Although the criteria used to diagnose the metabolic syndrome may be different, several studies have demonstrated that the prevalence of metabolic syndrome has increased dramatically in the past two decades[4-6]. Metabolic syndrome is subjected to multifactorial influences including both environmental and genetic factors. Current researches focus more on the cause of one or certain metabolic syndrome components, but rarely pay attention to the etiology of metabolic syndrome.
Reportedly, adiponectin is an adipose tissue-derived cytokine with anti-inflammatory, anti-atherogenic and cardioprotective properties. Increasing evidence suggested that adiponectin, linked to central obesity and insulin resistance, is a key contributor to the development of metabolic syndrome[7]. The ADIPOQ gene encoding adiponectin is located on chromosome 3q27 and this chromosome region had been mapped as a susceptibility locus for metabolic syndrome and coronary heart disease by genome-wide scans[8]. There are specific functional peroxisome proliferator response elements (PPREs) in the promoter region of adiponectin. PPAR-γbinds to PPREs as a heterodimer with members of the retinoid X receptor (RXR) nuclear receptor subfamily[9], and increases adiponectin promoter activity in cells from humans[10]. The expression of adiponectin in the adipose tissue is maintained and induced by direct binding of endogenous or exogenous PPAR-γ/RXR heterodimer to the PPRE in the adiponectin promoter[10]. Therefore, the PPAR-γand RXR-αgenes in the adiponectin pathway are candidates for obesity and metabolic syndrome.
The back-error propagation artificial neural network (BPANN) is a computer-based algorithm that is trained to recognize and categorize complex patterns. BPANN as a new approach to etiology research avoids the limitations of case control study and logistic regression without the distribution form and independence of variables. Our group has previously found the association of single nucleotide polymorphisms (SNPs) in thePPAR-γand RXR-αgenes with metabolic syndrome or type 2 diabetes risks in different populations[11,12]. In the present study, we further explored the application characteristics of BPANN in studying the combined effects of genetic variants in the PPAR-γand RXR-αgene and metabolic syndrome risks in a Chinese Han population. We compared the results of BPANN with those of traditional logistic regression analysis to better understand and use artificial neural network in the study of diseases of multiple etiologies.
SUBJECTS AND METHODS
Subjects
A total of 2,081 Chinese patients, including 1,012 metabolic syndrome and 1,069 non-metabolic syndrome controls, were recruited for metabolic studies with their informed consent. The protocol was approved by the Research Ethics Committee of Nanjing Medical University. All participants were genetically unrelated ethic Han Chinese. The cases were consecutively recruited from the inpatient or outpatient departments of three affiliated hospitals of Nanjing Medical University (the Second Affiliated Changzhou Hospital, the Third Affiliated Hospital and the First Affiliated Hospital) between March 2008 and August 2010, without any restriction on age and sex (430 males and 582 females with a mean age of 55.35±10.62 years). Age (±5 years) and sex-matched non-metabolic syndrome controls who underwent routine annual health examinations within the same geographical area and the period (478 males and 591 females with a mean age of 55.78±13.10 years) were also recruited for the study. In order to collect demographic data and information in environmental exposure history, each participant was interviewed face-toface using a standard questionnaire. After the interview, 5 mL venous blood sample was collected from each participant. The level of physical activity was defined as walking or riding≥ 15 minutes/day and/or lifting or carrying heavy objects at work daily and/or doing sports or physical exercise > 2 hours/week. Tobacco smokers were defined as patients who smoked at least one cigarette per day for over 1 year. Alcohol drinkers were defined as those who had the sum of milliliters of alcohol per week from wine, beer, cider or spirits.
The new (2009) criteria of metabolic syndrome was based on a joint interim statement of the International Diabetes Federation (IDF); National Heart, Lung and Blood Institute (NHLBI); American Heart Association (AHA); World Heart Federation; International Atherosclerosis Society and International Association, which define metabolic syndrome as the presence of three or more of the following features: triglycerides (TG)≥ 150 mg/dL (1.7 mmol/L) (or drug treatment for elevated triglycerides), high-density lipoprotein cholesterol (HDL-C) < 40 mg/dL (1.0 mmol/L) in mates and < 50 mg/dL (1.3 mmol/L) in females (or drug treatment for reduced HDL-C), fasting plasma glucose(FPG)≥ 100 mg/dL (or drug treatment for elevated glucose), systolic blood pressure (SBP)≥130 and/or diastolic blood pressure (DBP)≥85 mmHg (or antihypertensive drug treatment in a patient with a history of hypertension), and waist circumstances (WC)≥85 cm in men and 80 cm in women (current recommended waist circumstance thresholds for abdominal obesity for people in China)[13].
Measurements
Weight and height were measured by trained personnel, and body mass index (BMI, in kg/m2) was calculated. Blood pressure was measured on the right arm, with the patient in a sitting position and after a minimum 10-min rest, using a standard mercury sphygmomanometer. After an overnight fast, venous blood samples were drawn and promptly centrifuged and the plasma was stored at -20°C. Serum adiponectin, FPG and TC, HDL-C, LDL-C and TG were analyzed using human adiponectin ELISA kit (RapidBio Co., Calabasas, CA, USA), glucose oxidase method and enzymatic colorimetric method (Au5400; Olympus, Japan), respectively. DNA was extracted from blood samples by using a phenol-chloroform technique. All measurements were conducted by the manufacturers' protocols.
SNP selection and genotyping
In the present study, we first used NCBI dbSNPs database (http://www.ncbi.nlm.nih.gov/), the public HapMap SNP database (http://www.hapmap.org/) and previously reported literatures to identify potentially functional SNPs of the PPAR-γand RXR-αgene. Three potentially functional SNPs of the PPAR-γgene (rs2920502, rs3856806 and rs1801282) and four of the RXR-αgene (rs1045570, rs3132291, rs4240711 and rs4842194) with minor allele frequency (MAF)≥0.05 in the Chinese Han population were identified, including four at the coding region and three at the 3' untranslated region. Then, we used public HapMap SNP database to identify PPAR-γand RXR-αgene tagging SNPs by using tagger with greedy algorithm. Finally, two tagging SNPs (rs17817276 and rs38566806) of the PPAR-γ gene and two of the RXR-α gene (rs1045570 and rs6537944) were selected based on pair-wise tagging (r2≥0.80, MAF ≥0.05) by using genotype data from unrelated HapMap CHB individuals. Genomic sequences were obtained from the HapMap database. Primer version 5.0 and polymerase chain reaction (PCR) primer-introduced restriction analysis were used to design the nine primer sets (Table 1).
Genetic analyses were performed on genomic DNA extracted from leucocytes of venous blood. We used the TaqMan allelic discrimination assay to genotype the polymorphisms on the platform of 7900HT Realtime PCR System (Applied Biosystems, Foster City, CA). The information on assay conditions and the primers and probes are available upon request. Two negative controls were included in each 384-well reaction plate and individual genotype identification were determined by SDS software 2.0 (ABI). Moreo-ver, to confirm genotyping results, 10% of samples were randomly selected to repeat the procedure and the results were 100% concordant.
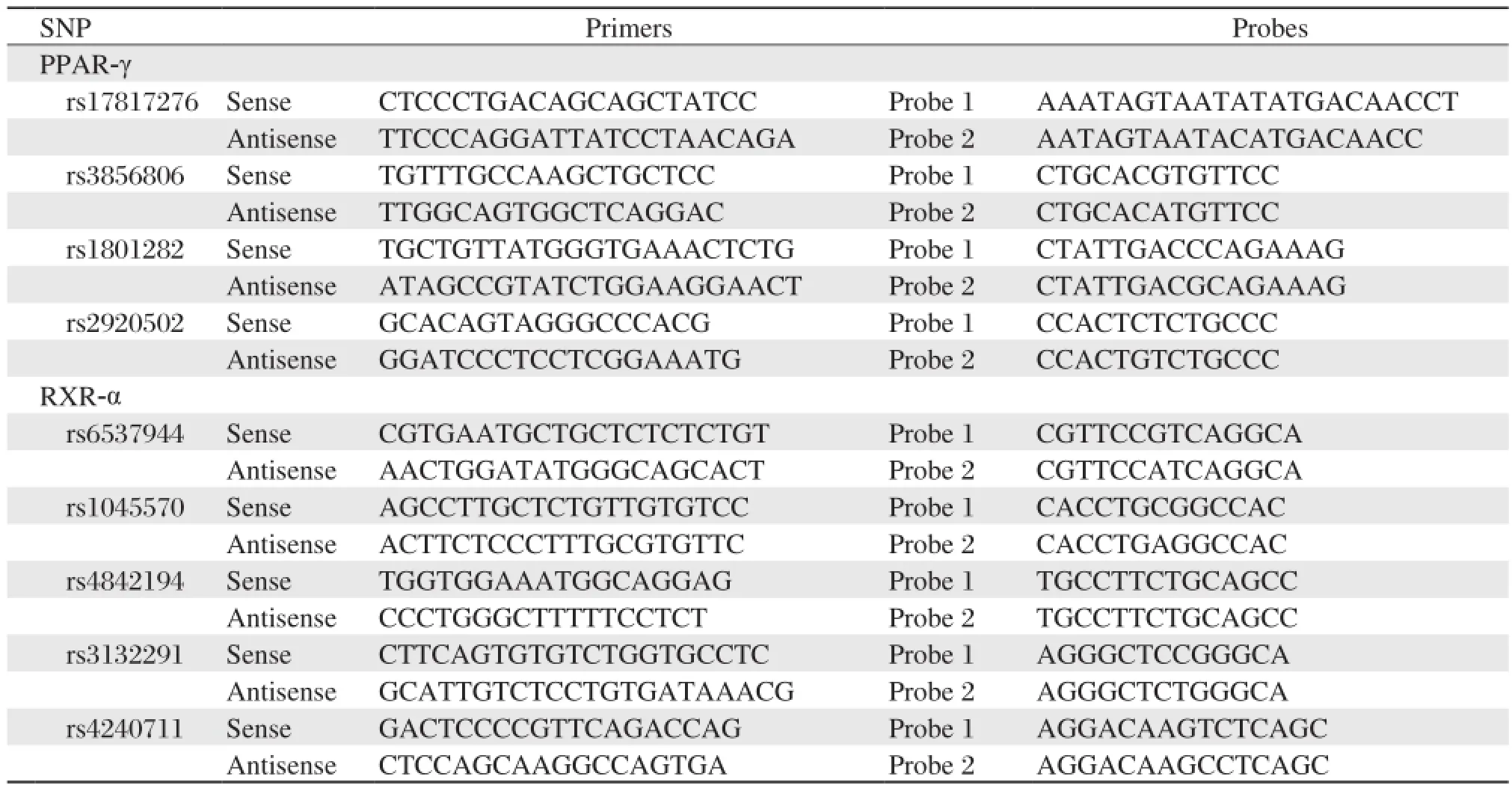
Table 1 Primers and annealing temperature used for RET sequencing
Statistical analysis
Hardy-Weinberg equilibrium was assessed within controls using the goodness-of-fit χ2test. The distribution of the general characteristics between metabolic syndrome and non-metabolic syndrome controls was compared by using two-sided Chi-square test and/ or Student's t test. The significance level was set at P <0.05. Both univariate and multiple logistic regression analyses were performed to estimate crude and adjusted standardized partial regression coefficient (β), odds ratios (ORs) and 95% confidence intervals (CIs) for the association between genotypes and metabolic syndrome risks. All statistical analyses were performed by SPSS software (SPSS version 13.0, Chicago, IL, USA). Generalized multifactor dimensionality reduction (GMDR) software (version 1.0.1) (www. healthsystem.virginia.edu/internet/Addiction-Genomics/) was applied for detecting gene-gene and geneenvironment interactions[14]. BPANN prediction model was established and each input neurons' mean impact value (MIV) was calculated using MATLAB7.0 software (MathWorks, Natick, MA)[15].
RESULTS
Univariate logistic regression analysis of associations between risk factors and metabolic syndrome
The clinical characteristics of the 2081 subjects are shown in Table 2. The genotype distributions of all the SNPs satisfied Hardy-Weinberg equilibrium (all P > 0.05 in controls). The associations between the 19 variables, including four of the PPAR-γgene, five of the RXR-αgene, serum adiponectin concentration and other environmental factors, was estimated by binary logistic regression analysis. Table 3 shows the sequences of the impact of various factors on metabolic syndrome risk. The top ten were family history of hyperlipidemia or type 2 diabetes, physical activity, family history of hypertension, BMI, alcohol drinking, gender, rs6537944, rs1801282 and rs4842194.
Multiple logistic regression analysis of associations between risk factors and metabolic syndrome
Multiple logistic regression analysis was performed to determine the association between the 19 variables and metabolic syndrome with the stepwise regressive method (the removal probability was 0.1). Table 4 illustrates nine factors in the best model and only six factors were statistically significant, including family history of type 2 diabetes or hyperlipidemia, physical activity, gender, alcohol drinking, BMI, rs4240711, rs2920502 and serum adiponectin.
BPANN multiple analysis of associations between risk factors and metabolic syndrome
We used 19 factors as input variables and metabolic syndrome diagnosis as output variables to establish model with all available samples. The transfer function was logsig function. Learning rate and training error were 0.1 and 0.01, respectively. Training steps were set to a maximum of 1000 steps. After the completion of training, the MIV was obtained. Table 5summarizes all related factors, consisting of BMI, serum adiponectin, rs4240711, gender, rs4842194, family history of diabetes, rs2920502, physical activity, alcohol drinking, rs3856806, family history hypertension, rs1045570, rs6537944, age, rs17817276, family history of hyperlipidemia, smoking, rs1801282 and rs3132291 in sequence of the absolute value of MIV.
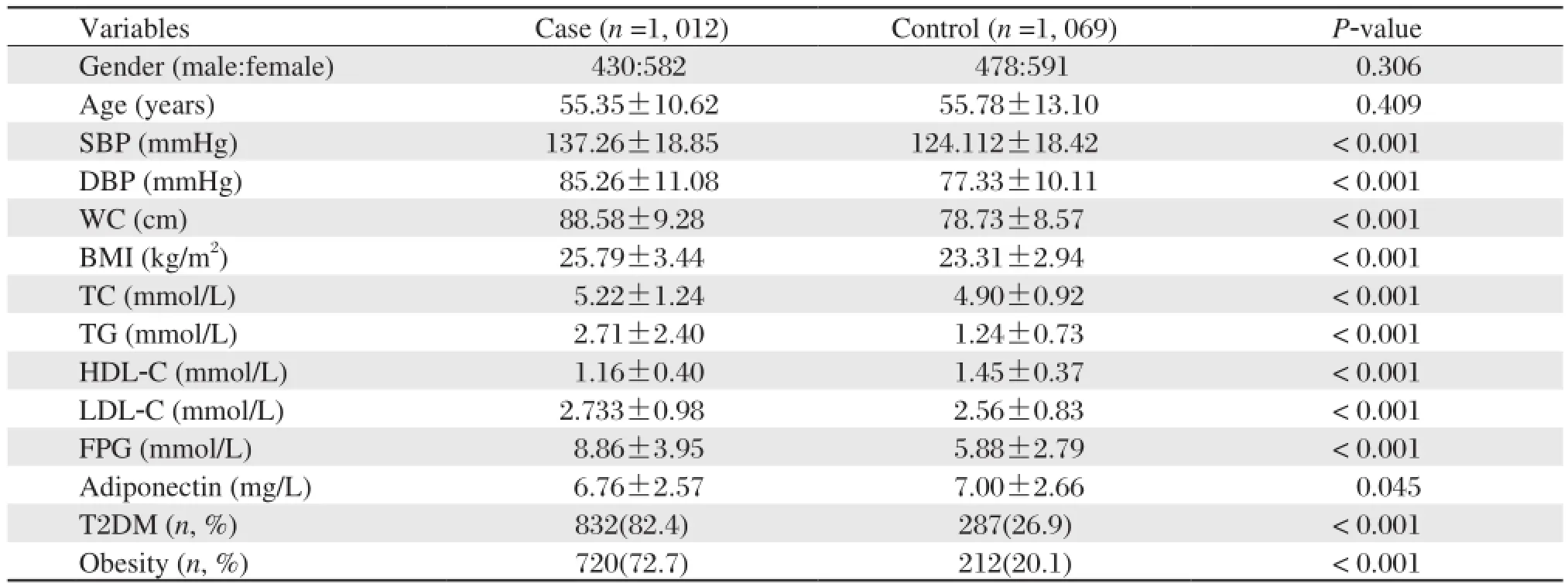
Table 2 Basic characteristics of the case and control groups
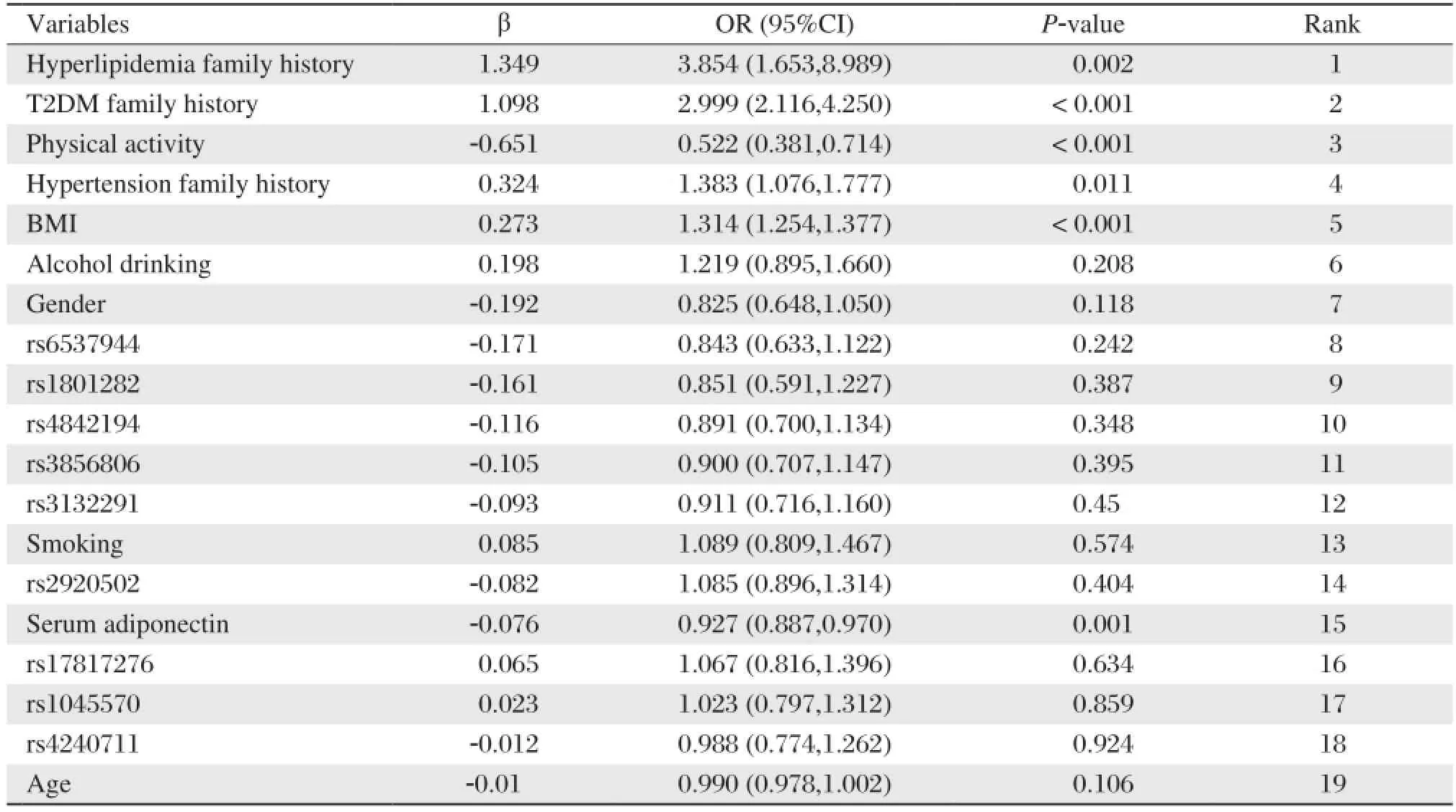
Table 3 Univariate logistic regression analysis results
GMDR for the combined effect of the PPAR-γand RXR-αgene
The SNPs rs4240711, rs4842194, rs2920502 and rs3856806, which were obtained in the top ten in BPANN multiple analysis, were named as A1, A2, B1and B2in turn. GMDR that evaluated the combined effect of the four SNPs detected A1, A2, B1and B2(rs4240711, rs4842194, rs2920502 and rs3856806) model as the best model (Cross-validation consistency 10/10, P = 0.0447) (Table 6). After controlling for age, gender, smoking, alcohol drinking and physical activity, the results showed that A1, A2, B1and B2model still were the best model from GMDR (Crossvalidation consistency 10/10, P = 0.0107) (Table 7). However, we failed to obtain a significant model when all the nineteen SNPs were added into the analysis (Cross-validation consistency 10/10, P = 0.1719) (data not shown).
DISCUSSION
Metabolic syndrome, like virtually all human diseases, results from the interactions between geneticand environmental factors. Environmental factors such as obesity, low levels of physical activity and inappropriate dietary habits are strong determinants of metabolic syndrome[16,17]. Traditional statistical methods due to their own limitation do not show the real relationship between gene and environmental elements with the risk of metabolic syndrome. Binary logistic regression analysis, for example, was used to analyze the association between the 19 variables and metabolic syndrome. Table 3 showed that the locant, according to standardized partial regression coefficient (β) of each variable, of top six environmental factors was hyperlipidemia family history, family history of type 2 diabetes, physical activity, smoking, family history of hypertension and BMI. Relative to environmental factors, the contribution of gene polymorphisms to the susceptibility and causation of diseases was hidden and weaker. Binary logistic regression limited to analyzing the combined action of elements did not provide the evidence for gene-environment interaction.
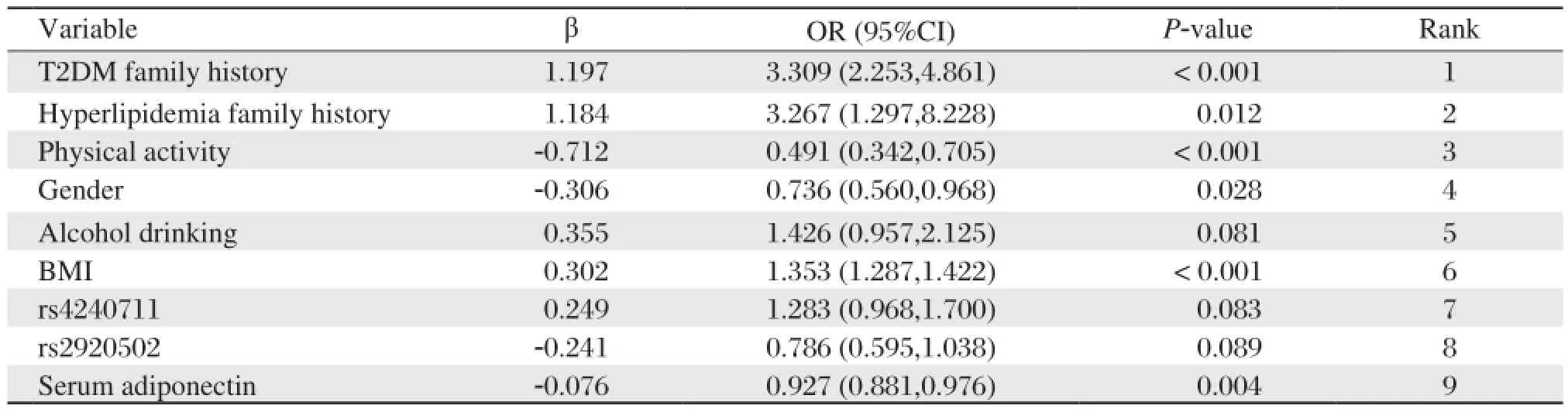
Table 4 Multiple logistic regression analysis results
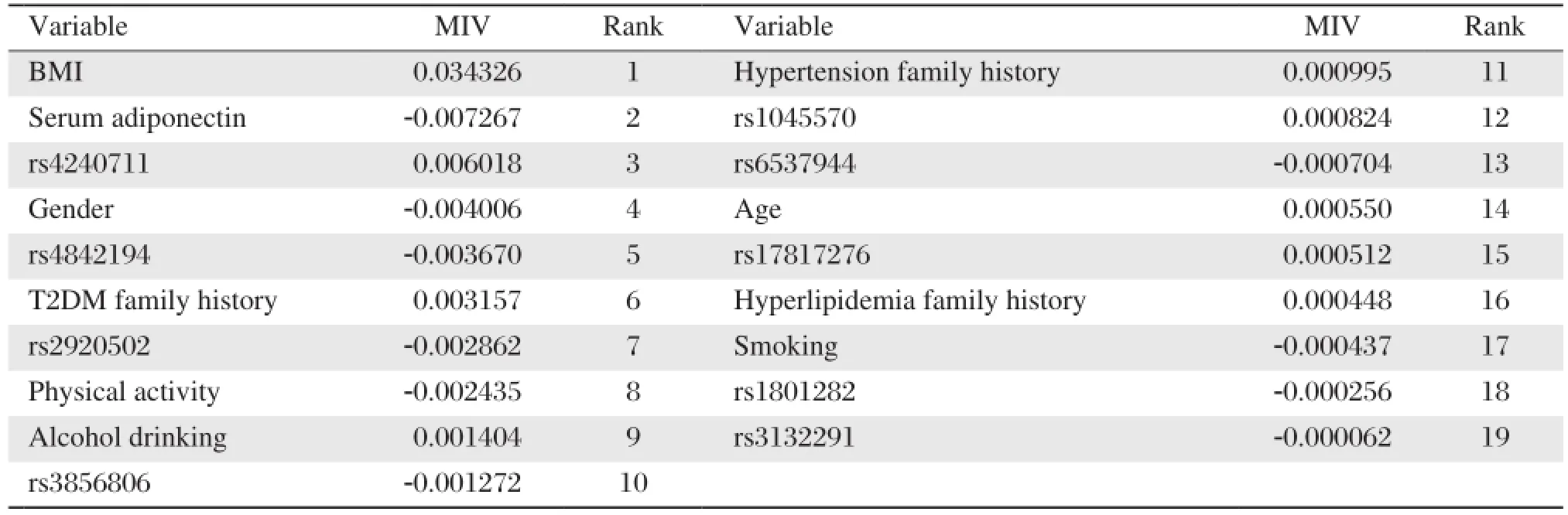
Table 5 Input variables and sorting of mean influence values (MIV)
By using the maximum likelihood method, only six variables were statistically significant, including family history of type 2 diabetes and hyperlipidemia, physical activity, gender, BMI and serum adiponectin. The result of multiple logistic regression analysis did not detect the role that PPAR-γgene and RXR-αgene polymorphisms played in the development of metabolic syndrome, but BPANN analysis had proved the association[12]. Adiponectin has an obvious protective effect on metabolic syndrome and its cardiovascular complications[18], but the contribution of adiponectin was weakened in the model, because of the complex interaction between variables.
Artificial neural networks are a non-linear pattern recognition algorithm consisting of a group of processing units that simulate the function of neurons in human brain and reveal relationships among the input data that cannot always be recognized by conventional models[19]. In recent years, artificial neural networks have been increasingly used in complex medical decision-making, such as the diagnosis of various diseases, investigating the predictive values of disease risk factors and analyzing the complex relationships between gene-environment[20,21]. BPANN has not only been the core of feedforward neural networks, but also embodied the essence of artificial neural networks[15]. The units in a BPANN are highly interconnected by weighted links, very similar to neural synapses. In the constant learning process in which the former feedback of error was used to modify corresponding weights and threshold value, BPANN adjusted the weights of links between neurons in order to associate input data with correct output (such as disease diagnosis). BPANN could fully show all the relationship between factors in a simulation. Therefore, we further used BPANN to identify the real association between PPAR-γand RXR-αgene polymorphisms with susceptibility to metabolic syndrome in southernHan Chinese and the key risk factors for metabolic syndrome. Compared to logistic regression analysis, BPANN showed a better understanding of using artificial neural network in epidemiology. Compared with the results of univariate and multiple logistic regression analyses, the seating arrangements of BMI and serum adiponection concentration were obviously raised in BPANN analysis. The sequence of serum adiponection concentration was raised to the second factor only after BMI from the last sequence in multiple logistic regression analysis. Central obesity is considered a pivotal component in metabolic syndrome. Even in subjects without obesity, a higher BMI tends to correlate with a higher number of positive metabolic syndrome components[22]. It has been reported that adiponectin concentration was an important predictor for the risk of metabolic syndrome[23]. Our result further validated that adiponectin had a great influence on the pathogenesis of metabolic syndrome.
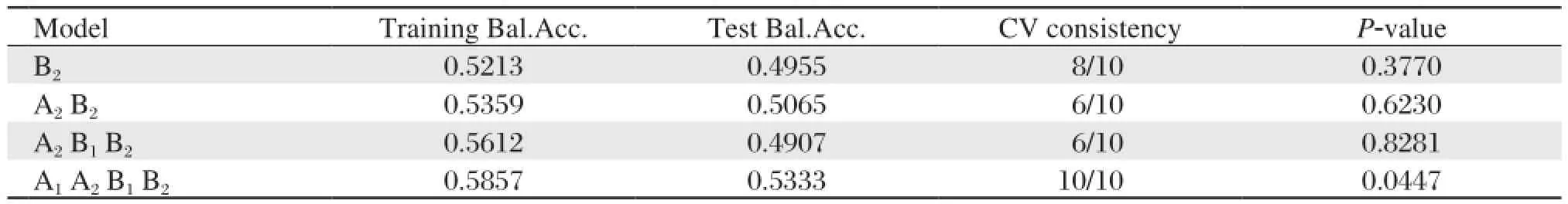
Table 6 The GMDR models for PPAR-γand RXR-αgene interaction on metabolic syndrome
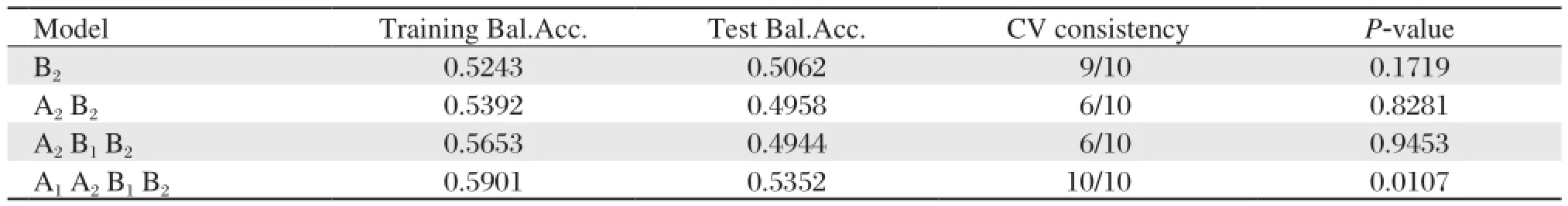
Table 7 The GMDR models for PPAR-γand RXR-αgene-environment interaction on metabolic syndrome
PPAR-γrs2920502, rs3856806 and RXR-αrs4240711, rs4842194 had no association with metabolic syndrome in univariate and multiple logistic regression analyses, but the seating arrangements of the four gene polymorphisms were in front in BPANN analysis, which implied an association between the four genetic variants in PPAR-γand RXR-αand metabolic syndrome. Currently, the studies investigating the associations between rs2920502, rs4240711 and rs4842194 polymorphisms and the risk of metabolic syndrome have been reported rarely. In a previous study[12], we also found that those carrying rs2920502CG and CG/GG genotype had a significantly increased risk of metabolic syndrome and rs4240711GG and AG/GG, rs4842194 CC and CT/ CC genotypes were all associated with prominent protective effects for metabolic syndrome. Up to now, numerous studies have focused on the association between C1431T variant (rs3856806) of PPAR-γand the risk of metabolic syndrome, type 2 diabetes and obesity in several populations[24-26], but the conclusions are conflicting. Li et al.[24]reported that polymorphism C1431T of exon 6 of PPAR-γwas associated with metabolic syndrome risks in a Chinese population study of 423 cases with metabolic syndrome and families without metabolic syndrome. However, in another study of 792 Han Chinese in Beijing, Dongxia et al.[27]failed to show the association, but a significant association between rs3856806 and insulin resistance. In a population-based study of 1910 subjects by our group, divided according to the IDF (2005) criteria, we also did not find differences of rs3856806 among metabolic syndrome patients and non-metabolic syndrome controls by logistic regression analysis[12]. In Meirhaeghe's French population study[17], the C1431T variant was only connected with the other three polymorphisms, including P3-681C > G, P2-689C > T, Pro12Ala, and the GTGC haplotype was associated with metabolic syndrome. It may be that the individual C1431T's role in the pathogenesis of metabolic syndrome is not obvious and the combined effect with other polymorphisms exists, but the traditional statistical methods do not show.
No polymorphisms in the two genes had been found to associate with metabolic syndrome in univariate and multiple logistic regression analyses, but the seating arrangements of the four gene polymorphisms were in front in BPANN analysis. The finding of BPANN analysis and logistic regression analysis suggested that the interactions between four SNPs may be associated with the risk of metabolic syndrome. We used the GMDR method to indentify the combined effect of the four SNPs (rs2920502, rs3856806, rs4240711 and rs4842194). The GMDR is an extended version of the multifactor dimensionality reduction approach, which allows high-dimensional interactions of multiple factors to be simultaneously retrieved. The GMDR method is a non-parametric and genetic model-free approach that efficiently identifies higher-order interactions between genes and/or geneenvironmental factors with both dichotomous and continuous phenotypes in various population-based study designs. The main idea of GMDR is to reduce multi-dimensional genotypes into one-dimensional binary attributes by pooling genotypes of multiple SNPs using a well-defined classifier[14]. Furthermore, GMDR can reduce the complexity substantially and permit adjustment for discrete and quantitative covariates[14]. GMDR has been performed to successfully identifythe combinations of SNPs that significantly influenced complex diseases[28]. The model of A1, A2, B1 and B2 (rs4240711, rs4842194, rs2920502 and rs3856806) were the best model (Cross-validation consistency 10/10, P = 0.0447). It showed that four SNPs loci interact with one another and suggested interactions between the two genes. After controlling age, gender, smoking, alcohol drinking and physical activity, the results showed that A1, A2, B1and B2(rs4240711, rs4842194, rs2920502 and rs3856806) model were still the best model as the highest Test Balanced Accuracy (0.5901) (Cross-validation consistency 10/10, P = 0.0107). After adjusting age, gender, smoking, alcohol drinking and physical activity, the combined effect of PPAR-γgene and RXR-αgene still existed. PPAR-γ acts as a nuclear receptor-transcription factor by forming a heterodimer with RXR. PPAR-γ/RXR heterodimer directly binds to the functional PPAR-responsive element (PPRE) and increases human adiponectin promoter activity in cells. The PPAR-γgene and RXR-αgene are related to the adiponectinsignal transduction pathway. The two genes may play a significant role in insulin resistance, which is a key feature of metabolic syndrome and type 2 diabetes by affecting adiponectin secretion levels.
BPANN is designed to detect patterns in input data, which may match output data even if the nature of such patterns is not known a priori; thus, all richer relations between factors can be simulated and provided at a time than the ordinary model[29]. Simultaneously, BPANN not requiring the distribution form and independence of variables, also can handle the problem of collinearity better. Compared with the traditional analysis methods, BPANN may be better suited to predict outcomes when the relationships between the variables are complex, multidimensional and nonlinear in complex biological systems[30]. Neural network models have the ability to detect all possible interactions between predictive variables and provide a model consistent with practical situations. Even so, BPANN analysis was a "black box" and had limited ability to explicitly identify possible causal relationships[31]. The model builder of logistic regression was able to select variables which were most strongly predictive of an outcome based on the magnitude of the standardized partial regression coefficients (β) and the associated odds ratios.
In this article, we put four SNPs, which was selected by BPANN analysis, into model analyses by GMDR software. We obtained an optimal model with statistical significance. The result suggested an interaction between the RXR-αgene and RXR-αgene. However, we failed to obtain an optimal model when all the 19 SNPs were included in the analysis without screening. Therefore, we conclude that BPANN can be used to select influence factors commonly, especially for early screening of genetic factors. We can further analyze gene interactions by using the results of screening, according to the ranking list of MIV as a relative stable reference.
[1] Sygnowska E, Piwonska A, Waskiewicz A, Broda G. Socioeconomic factors and the risk of metabolic syndrome in the adult Polish population: the WOBASZ study. Kardiol Pol 2012: 70: 718-27.
[2] Laaksonen DE, Lakka HM, Niskanen LK, Kaplan GA, Salonen JT, Lakka TA. Metabolic syndrome and development of diabetes mellitus: application and validation of recently suggested definitions of the metabolic syndrome in a prospective cohort study. Am J Epidemiol 2002; 156: 1070-7.
[3] Onat A, Ceyhan K, Basar O, Erer B, Toprak S, Sansoy V. Metabolic syndrome: major impact on coronary risk in a population with low cholesterol levels--a prospective and cross-sectional evaluation. Atherosclerosis 2002: 165: 285-92.
[4] Ford E, Giles W, Mokdad A. Mokdad, Increasing prevalence of the metabolic syndrome among u.s. Adults Diabetes Care 2004; 27: 2444-9.
[5] Nestel P, Lyu R, Low LP, Sheu WH, Nitiyanant W, Saito I, et al. Metabolic syndrome: recent prevalence in East and Southeast Asian populations. Asia Pac J Clin Nutr 2007; 16: 362-7.
[6] Athyros VG, Bouloukos VI, Pehlivanidis AN, Papageorgiou AA, Dionysopoulou SG, Symeonidis AN, et al. The prevalence of the metabolic syndrome in Greece: the Metabolic syndrome-Greece Multicentre Study. Diabetes Obes Metab 2005; 7: 397-405.
[7] Okamoto Y, Kihara S, Funahashi T, Matsuzawa Y, Libby P. Adiponectin: a key adipocytokine in metabolic syndrome. Clin Sci (Lond) 2006; 110: 267-78.
[8] Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, et al. A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 2001; 10: 2751-65.
[9] Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem 1995; 270: 12953-6.
[10] Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, et al. Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes 2003; 52: 1655-63.
[11] Lu Y, Ye X, Cao Y, Li Q, Yu X, Cheng J, et al. Genetic variants in peroxisome proliferator-activated receptorgamma and retinoid X receptor-alpha gene and type2 diabetes risk, a case-control study of a Chinese Han population. Diabetes Technol Ther 2011; 13: 157-64.
[12] Shi H, Yu X, Li Q, Ye X, Gao Y, Ma J, et al. Association between PPAR-γ and RXR-α gene polymorphism and metabolic syndrome risk: a case-control study of a Chinese Han population. Arch Med Res 2012; 43: 233-42.
[13] Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009; 120: 1640-5.
[14] Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet 2007; 80: 1125-37.
[15] Mukherjee A, Deshpande JM. Application of artificial neural networks in structural design expert s- ystems. Comput Struct 1995; 54: 367-75.
[16] Bragt MC, Popeijus HE. Peroxisome proliferator-activated receptors and the metabolic syndrome. Physiol Behav 2008; 94: 187-97.
[17] Meirhaeghe A, Cottel D, Amouyel P, Dallongeville J. Dallongeville, Association between peroxisome proliferator-activated receptor gamma haplotypes and the metabolic syndrome in French men and women. Diabetes 2005; 54: 3043-8.
[18] Guo W, Che Z, Xu A, Zhou Z. Angiopoietin-like protein 3 and adiponectin levels in patients with metabolic syndrome. Zhong Nan Da Xue Xue Bao Yi Xue Ban (China) 2010; 35: 203-8.
[19] Bartosch-Harlid A, Andersson R. Diabetes mellitus in pancreatic cancer and the need for diagnosis of asymptomatic disease. Pancreatology 2010; 10: 423-8.
[20] Coppedè F, Grossi E, Migheli F, Migliore L. Polymorphisms in folate-metabolizing genes, chromosome damage, and risk of Down syndrome in Italian women: identification of key factors using artificial neural networks. BMC Med. Genomics 2010; 3: 42.
[21] Shi H, Lu Y, Du J, Du W, Ye X, Yu X, et al. Application of Back Propagation Artificial Neural Network on Genetic Variants in Adiponectin ADIPOQ, Peroxisome Proliferator-Activated Receptor-gamma, and Retinoid X Receptor-alpha Genes and Type 2 Diabetes Risk in a Chinese Han Population. Diabetes Technol Ther 2011; 14: 293-300.
[22] Lee IT, Chiu Y, Hwu C, He C, Chiang F, Lin Y, et al. Central obesity is important but not essential component of the metabolic syndrome for predicting diabetes mellitus in a hypertensive family-based cohort. Results from the Stanford Asia-Pacific Program for Hypertension and Insulin Resistance (SAPPHIRe) Taiwan follow-up study. Cardiovasc Diabetol 2012; 11: 43.
[23] Zhuo Q, Wang ZQ, Fu P, Piao JH, Tian Y, Xu J, et al. Association between adiponectin and metabolic syndrome in older adults from major cities of China. Biomed. Environ Sci 2010; 23: 53-61.
[24] Yang LL, Hua Q, Liu RK, Yang Z. Association between two common polymorphisms of PPARgamma gene and metabolic syndrome families in a Chinese population. Arch. Med Res 2009; 40: 89-96.
[25] Jaziri R, Lobbens S, Aubert R, Pean F, Lahmidi S, Vaxillaire M, et al. The PPARG Pro12Ala polymorphism is associated with a decreased risk of developing hyperglycemia over 6 years and combines with the effect of the APM1 G-11391A single nucleotide polymorphism: the Data From an Epidemiological Study on the Insulin Resistance Syndrome (DESIR) study. Diabetes 2006; 55: 1157-62.
[26] Prakash J, Srivastava N, Awasthi S, Agarwal C, Natu S, Rajpal N, et al. Association of PPAR-gamma gene polymorphisms with obesity and obesity-associated phenotypes in north indian population. Am J Hum Biol 2012; 24: 454-9.
[27] Dongxia L, Qi H, Lisong L, Jincheng G. Association of peroxisome proliferator-activated receptorgamma gene Pro12Ala and C161T polymorphisms with metabolic syndrome. Circ J 2008; 72: 551-7.
[28] Suyama Y, Matsuda C, Isomura M, Hamano T, Karino K, Yamasaki M, et al. Effects of six functional SNPs on the urinary 8-isoprostane level in a general Japanese population; Shimane COHRE Study. Dis Markers 2011; 30: 291-8.
[29] Curtis D. Comparison of artificial neural network analysis with other multimarker methods for detecting genetic association. BMC Genet 2007; 8: 49.
[30] Eftekhar B, Mohammad K, Ardebili HE, Ghodsi M, Ketabchi E. Comparison of artificial neural network and logistic regression models for prediction of mortality in head trauma based on initial clinical data. BMC Med Inform Decis Mak 2005; 5: 3.
[31] Jack V. T. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol 1996; 49: 1225-31.
Received 03 October 2012, Revised 06 November 2012, Accepted 16 November 2012, Epub 20 March 2013
The study was supported by the National Natural Science Foundation of China Grant No. 30771858, Jiangsu Provincial Natural Science Foundation Grant No. BK2007229 and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
△These authors contributed equally to this study.
The authors reported no conflict of interests.
10.7555/JBR.27.20120061
 THE JOURNAL OF BIOMEDICAL RESEARCH2014年2期
THE JOURNAL OF BIOMEDICAL RESEARCH2014年2期
- THE JOURNAL OF BIOMEDICAL RESEARCH的其它文章
- Rapid progression of nonculprit coronary lesions six weeks after successful primary PCI in culprit artery:a case report
- Naringin ameliorates acetic acid induced colitis through modulation of endogenous oxido-nitrosative balance and DNA damage in rats
- IRE1α is essential for Xenopus pancreas development
- Clopidogrel improves aspirin response after off-pump coronary artery bypass surgery
- Diagnostic and prognostic value of minor elevated cardiac troponin levels for percutaneous coronary intervention-related myocardial injury:a prospective,single-center and double-blind study
- A clinical perspective on mucoadhesive buccal drug delivery systems
