DNA damage response-related immune activation signature predicts the response to immune checkpoint inhibitors: from gastrointestinal cancer analysis to pan-cancer validation
Junya Yan*, Shibo Wang*, Jing Zhang*, Qiangqiang Yuan, Xianchun Gao, Nannan Zhang, Yan Pan,Haohao Zhang, Kun Liu, Jun Yu, Linbin Lu, Hui Liu, Xiaoliang Gao, Sheng Zhao, Wenyao Zhang,Abudurousuli Reyila, Yu Qi, Qiujin Zhang, Shundong Cang, Yuanyuan Lu, Yanglin Pan, Yan Kong,Yongzhan Nie
1State Key Laboratory of Holistic Integrative Management of Gastrointestinal Cancers and National Clinical Research Center for Digestive Diseases, Xijing Hospital of Air Force Military Medical University, Xi’an 710032, China; 2Department of Oncology,Henan Provincial People’s Hospital, Zhengzhou University People’s Hospital, Zhengzhou 450003, China; 3Faculty of Life Science, Northwest University, Xi’an 710069, China; 4Unit 73211 of the People’s Liberation Army, Nanjing 211800, China; 5Key Laboratory of Resource Biology and Biotechnology in Western China (Ministry of Education), School of Medicine, Northwest University, Xi’an 710069, China; 6Shaanxi University of Chinese Medicine, Second Clinical Medicine Faculty, Xi’an 712046,China; 7Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Renal Cancer and Melanoma, Peking University Cancer Hospital and Institute, Beijing 100142, China
ABSTRACT Objective: DNA damage response (DDR) deficiency has emerged as a prominent determinant of tumor immunogenicity.This study aimed to construct a DDR-related immune activation (DRIA) signature and evaluate the predictive accuracy of the DRIA signature for response to immune checkpoint inhibitor (ICI) therapy in gastrointestinal (GI) cancer.
KEYWORDS DNA damage response-related immune activation; immune checkpoint inhibitors; biomarker; gastrointestinal cancer; pan-cancer
Introduction
Gastrointestinal (GI) cancer has the highest disease burden among all malignancies with global incidence and mortality rates of 26% and 35%, respectively1.Traditional treatments for GI cancer, including surgical resection, chemotherapy,radiotherapy, and targeted therapy, have not achieved satisfactory outcomes2.In recent years the development of immuno therapy has revolutionized the therapeutic landscape for multiple cancer types, including a subset of GI cancers3.Immune checkpoint inhibitor (ICI) targeting PD-1/PD-L1 signaling have been approved as standard first-line therapy for advanced esophagogastric and colorectal cancer4,5.The objective response rate (ORR) of ICI therapy, however, is only 10%~20% in patients with GI cancer6, highlighting the urgent need to identify optimal biomarkers for precise treatment.
Notably, some biomarkers have been developed to predict the efficacy of ICI therapy in patients with GI cancer, including PD-L1 expression7, tumor mutational burden (TMB)8, microsatellite instability (MSI) status9, and Epstein-Barr virus (EBV)status10.To date, only MSI-high (MSI-H) has been confirmed to be a useful predictive biomarker for GI cancer.Nevertheless,the MSI-H subtype is present in only a small proportion (0%-5%) of colorectal cancers11.PD-L1 expression is one of the most widely studied immunotherapy biomarkers; however, clinical trials have yielded controversial results7,12,13, which limits the predictive value of PD-L1 expression in patients with GI cancer.In addition, TMB, as a biomarker of GI cancer, faces several challenges, such as platform uniformity, intra- tumoral heterogeneity, and lack of consensus on thresholds6,14,15.Overall, the currently available biomarkers do not meet the criteria for clinical application in ICI therapy for GI cancer.
Genomic instability, as one of the hallmarks of various cancers, is mainly caused by the increasing accumulation of damaged DNA and DNA damage response (DDR) deficiency16.Although DDR deficiency drives genomic instability and tumor progression, DDR deficiency also provides potential therapeutic opportunities.The excellent efficacy of olaparib in ovarian cancer patients withBRCA1/2mutations is probably the best example of this interplay17.In addition, data from multiple studies have shown that patients responding to ICI therapy commonly harborBRCA2mutations18.Other investigations also demonstrated that deleterious alterations in genes involved in DDR pathways induce a hyper- mutational phenotype and improve the survival outcomes following ICI therapy19,20.DDR impairment can be induced not only by genomic alterations, but also epigenetic changes in DDR pathways21.Indeed, both mechanisms underlying DDR impairment induce the accumulation of DNA damage and activate the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway to create an inflammatory microenvironment21, thereby defining a distinct subgroup of patients suitable for ICI therapy.
Notably, a recently developed immune-driven 44-gene signature [DNA damage immune response (DDIR) assay22],summarized the common immune active process from the accumulation of DNA damage, which created an inflammatory microenvironment characterized by increased CD8+T cell infiltration and PD-L1 expression23.Furthermore, DDIR positivity has been reported to predict the clinical response to chemotherapy among patients with breast cancer24,25and esophageal adenocarcinoma26.Although the DDIR signature cannot predict an improved response to oxaliplatin chemotherapy in patients with colorectal cancer, a refined 9-gene DDIR signature showed a strong association with MSI and the consensus molecular subtype27.Based on the above analyses, we propose the DDIR signature potential as a classifier in identifying GI cancers that may benefit from ICI therapy.
In the current study we first established a novel DDRrelated immune activation (DRIA) signature by detecting a smaller panel of genes that can be easily translated into an easy-to-use clinical assay.By collecting the clinical and transcriptome profile data of two ICI-treated GI cancer cohorts,we explored and validated the association between the DRIA signature and clinical response to ICI therapy.To broaden the applicable certificate of DRIA, we retrospectively collected one ICI-treated melanoma cohort from our center and three pancancer cohorts from published datasets for extended validation of the DRIA predictive robustness.This study developed and revealed a novel signature (DRIA) consisting of three genes that better predict therapeutic efficacy of ICI therapy in GI cancer and pan-cancer.
Materials and methods
Patients and transcriptome profile data
Fifty-five patients with melanoma who received anti-PD-1 monotherapy between March 2016 and March 2019 were recruited for this study from Peking University Cancer Hospital(PUCH).Formalin-fixed paraffin-embedded (FFPE) biopsy specimens were collected from each patient before undergoing immunotherapy.Whole-transcriptome RNA sequencing was performed on the Illumina NovaSeq 6000 platform (San Diego, California, USA).The details of processing the gene expression data are described in our previous study28.
All the clinical and pathologic data, including age, gender,primary site, metastasis, and clinical response, were extracted from the medical record review.Patient response was determined using the Response Evaluation Criteria in Solid Tumors[RECIST (version 1.1)] criteria as follows: complete response(CR); partial response (PR); stable disease (SD); and progressive disease (PD).The ORR was defined as the proportion of patients with a CR or PR.Progression-free survival (PFS) was defined as the time from the initiation of ICI therapy to the date of disease progression or death from any cause.Patients without disease progression were censored on the date of their last scan.Overall survival (OS) was calculated from the date ICI therapy commenced to the date of death.Patients who did not die were censored on the date of their last scan.In this study, the ORR, PFS, and OS were the primary clinical outcomes.This study was approved by the Institutional Review Board of PUCH (2019KT92) and was in accordance with the Declaration of Helsinki.Written informed consent was obtained from all participants.
External cohort acquisition
The RNA-seq and clinical data from five publicly available cohorts treated with ICI therapy were collected for analysis in this study, as follows: (1) the Kim18 cohort consisted of 45 patients with metastatic gastric cancer who received anti-PD-1 therapy at the Samsung Medical Center10; (2) the Parikh22 cohort included 22 patients with metastatic microsatellite- stable (MSS)colorectal and pancreatic adenocarcinoma who received dual PD-1 and CTLA-4 blockade with radiation therapy in a phase II trial (NCT03104439)29; (3) the Gide19 cohort was comprised of 41 patients with advanced melanoma treated with anti-PD-1 therapy at the University of Sydney30; (4) the IMvigor210 cohort included 298 patients with metastatic urothelial cancer who received anti-PD-L1 therapy at the Memorial Sloan Kettering Cancer Center31; and (5) the Pender21 cohort comprised 56 pan-cancer patients across 17 solid tumor types treated with anti-PD-(L)1 therapy at the University of British Columbia32.Detailed information for the five immunotherapy cohorts is summarized in Table S1.
Definition of the DRIA signature
Two previously reported DDIR signatures in breast and colorectal cancers22,27contain 44 genes and 9 genes, respectively (Table S2).Three overlapping genes between the two DDIR signatures were defined as the DRIA signature, and the DRIA score was calculated as the geometric mean of the three genes.Based on the DRIA scores and immunotherapy responses, receiver operating characteristic (ROC) curve analysis was performed and the cut-off value that generated the maximum Youden index was used to define DRIA status.A DRIA score greater than the threshold was classified as DRIA-high and a DRIA score less than the threshold was classified as DRIA-low.
The Cancer Genome Atlas (TCGA) cohort
We downloaded the transcriptome profiling and survival data of patients with GI cancer (esophageal cancer,n= 161; gastric cancer,n= 375; and colorectal cancer,n= 622) from the TCGA database (http://xena.ucsc.edu/).Some immune-related molecular features, including the PD-L1 score, TMB, MSI status, CD8A score, and IFN-γ score were extracted from cBioPortal (https://www.cbioportal.org/) and the TCGA-GDC website(https://gdc.cancer.gov/about-data/publications/panimmune)to analyze the correlations with the DRIA signature.
Assessment of DDR mutation status
Previous studies defined a DDR gene list (Table S3), including 34 genes involved in 6 major DDR pathways19,20,33.According to the co-mutation counts of these DDR genes, the patients were divided into DDR-deficient (≥ 2 mutations) and DDRproficient groups (< 2 mutations).The mutation information regarding the 34 genes in TCGA-STAD cohort were retrieved from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/).The non- synonymous mutations included TRUNC (Frameshift del, Frameshift ins, nonsense, nonstop,splice region, splice site), INFRAME (Inframe del and Inframe ins), and MISSENSE mutations.
Biomarker analysis
Detailed information regarding clinically known biomarkers for immunotherapy were available in the Kim18 and IMvigor210 cohorts10,31.PD-L1 expression [combined positive score (CPS)],MSI status, TMB, and EBV status data were extracted from the Kim18 cohort.Data from the IMvigor210 cohort with respect to CD8+T cell infiltration, PD-L1 expression in tumor-infiltrating immune cells (ICs) and tumor cells (TCs), TMB, and tumor neoantigen burden (TNB) were also separated.We performed correlation analyses between these biomarkers and the DRIA score, and compared the predictive accuracy to determine the efficacy of immunotherapy.We also included published signatures (Table S4) based on the transcriptome profile data and evaluated the predictive performance based on the Kim18 cohort response to immunotherapy.
Statistical analyses
SPSS (version 23.0) and GraphPad Prism software (version 8) were used for statistical analyses and graphical representations.For normally distributed continuous variables, a t-test was used to compare mean values, whereas the Mann-Whitney U test was used for non-normally distributed continuous variables.Correlations between the DRIA status and categorical measurements, such as tumor response, were assessed using a chi-square test or Fisher’s exact test.Kaplan-Meier estimates of survival outcomes, including PFS and OS, were calculated using a log-rank test.ROC curve analyses were performed to evaluate the predictive accuracy of the DRIA score and other biomarkers with respect to ICI therapy.For all statistical tests, aP< 0.05(two-tailed test) was considered statistically significant.
Results
Immune characteristics and prognosis of DRIA in GI cancer
We identified three overlapping genes (CXCL10,IDO1, andIFI44L) in the two previously reported DDIR signatures(Table S2) and defined the overlapping genes as the DRIA signature.Using these three consensus genes to generate an unweighted cumulative DRIA score, we observed strong positive correlations between the DRIA score and the two original DDIR scores in the TCGA-GI cancer cohort (44-gene: Spearmanr= 0.621, Figure S1A; 9-gene: Spearmanr= 0.963, Figure S1B).Notably, the level ofCXCL10,IDO1, andIFI44Lgene expression was increased in DDR-deficient tumors compared with to DDR-proficient tumors (Figure S2A-C), which indicated the necessity of the three genes for DDIR characteristics.By analyzing the immune-related molecular features of the TCGA-GI cancer cohort, we demonstrated a markedly increased DRIA score in MSI patients compared to MSS patients (Figure S1C).The DRIA score was positively correlated with the TMB (Spearmanr= 0.154, Figure S1D), PD-L1 score (Spearmanr= 0.695, Figure S1E), CD8A score (Spearmanr= 0.737, Figure S1F), and IFN-γ score (Spearmanr= 0.832, Figure S1G).Therefore, patients with different DRIA scores had distinct tumor immune microenvironment characteristics, indicating that the DRIA signature has the potential to predict the response to ICI therapy in patients with GI cancer.
Survival analyses were performed according to DRIA status in the TCGA-GI cancer cohort to investigate the potential prognostic role of DRIA.No significant survival difference was observed between the DRIA-high and -low subsets in patients with esophageal, gastric, and colorectal cancers without ICI therapy (Figure S3A-C), indicating that DRIA was not a prognostic factor.
Exploration of the association between DRIA and response to ICI therapy in gastric cancer
The flow diagram depicting the multi-cohort analysis of the DRIA predictive value for the efficacy of ICI therapy is presented in Figure 1.A total of 517 patients treated with ICI from 6 cohorts, including GI cancer, melanoma, urothelial cancer, and pan-cancer, were included in this study.The clinical baseline characteristics are summarized in Table S5.The Kim18 cohort(n= 45) served as the discovery cohort with which to determine the association between DRIA and the efficacy of ICI therapy in patients with GI cancer.Furthermore, we used the Parikh22 cohort (n= 22) to validate the predictive value of DRIA in patients with GI cancer.Finally, pan-cancer extended validations of the DRIA were performed in the PUCH (n= 55), Gide19(n= 41), IMvigor210 (n= 298), and Pender21 (n= 56) cohorts.The median DRIA score of the patients in the CR/PR group was significantly higher than the patients in the SD/PD group of the Kim18 cohort (Figure 2A).Next, we integrated the DRIA score with the clinical response data to create an ROC curve and quantify the prediction accuracy.The results revealed that the area under the ROC curve (AUC) was 0.838(95% CI, 0.680-0.997; Figure 2B).The patients were divided into a DRIA-high group (n= 11) and a DRIA-low group(n= 34) using a cut-off value that generated the maximum Youden index.The DRIA-high group had a markedly higher ORR than the DRIA-low group (81.8%vs.8.8%; Figure 2C).Moreover, the waterfall plot demonstrated a positive correlation between the DRIA score and a favorable clinical response(Figure 2D).These results supported DRIA as a superior predictor of the clinical benefits of ICI therapy in patients with gastric cancer.
Comparison of DRIA with known clinical biomarkers in gastric cancer
Several emerging predictors have been developed for gastric cancer immunotherapy, including PD-L1 expression,MSI status, EBV status, and TMB.We performed correlation analyses between DRIA status and these biomarkers based on the available data in the Kim18 cohort.Detailed information regarding the DRIA status and other biomarkers in each patient is shown in a heatmap (Figure 3A).When patients were classified into 3 groups according to the PD-L1 CPS, the CPS ≥ 10 group had a significantly increased DRIA score combined with the 1 ≤ CPS < 10 and CPS = 0 groups (Figure 3B).Correlation analysis also showed a remarkably positive correlation between the PD-L1 CPS and the DRIA score (Spearmanr= 0.385; Figure 3C).Additionally, the median DRIA score was significantly higher in EBV-positive patients than EBV-negative patients (Figure 3D).Similarly, MSI-H and TMB-high patients had relatively increased DRIA scores (Figure 3E, F), although the difference did not reach significance.Furthermore, we applied ROC curve analysis to compare the DRIA predictive accuracy and these biomarkers.The DRIA predicting AUC was 0.845, which was higher than PD-L1, TMB, MSI, and EBV(Figure 3G).We further determined whether synergizing DRIA with these biomarkers enhanced the prediction accuracy of the response to ICI therapy.The pairwise combination of DRIA with PD-L1, TMB, MSI, and EBV showed improved patient stratification (Figure S4A).Remarkably, the predicting AUC of the DRIA with PD-L1 combination reached 0.947 (95% CI,0.876-1.000).According to the scoring system constructed by incorporating DRIA and PD-L1, patients were stratified into 3 groups (score = 0, 1, and 2).The proportion of patients who achieved a CR/PR was 90.0%, 15.4%, and 0% for the 2, 1, and 0 score groups, respectively (Figure S4B).
Additionally, the expression signature of the gene series has been used to predict the response to ICI therapy.Therefore, we investigated the association between the DRIA score and these signature scores in the Kim18 cohort.The heatmap showed that these signature scores were dramatically upregulated in the DRIA-high group compared to the DRIA-low group(Figure 3H).We also performed ROC curve analyses.The DRIA predicting AUC was higher than these signatures, indicating that the DRIA signature had a stronger predictive ability for ICI therapy than these previously published immune-related signatures (Figure 3I).
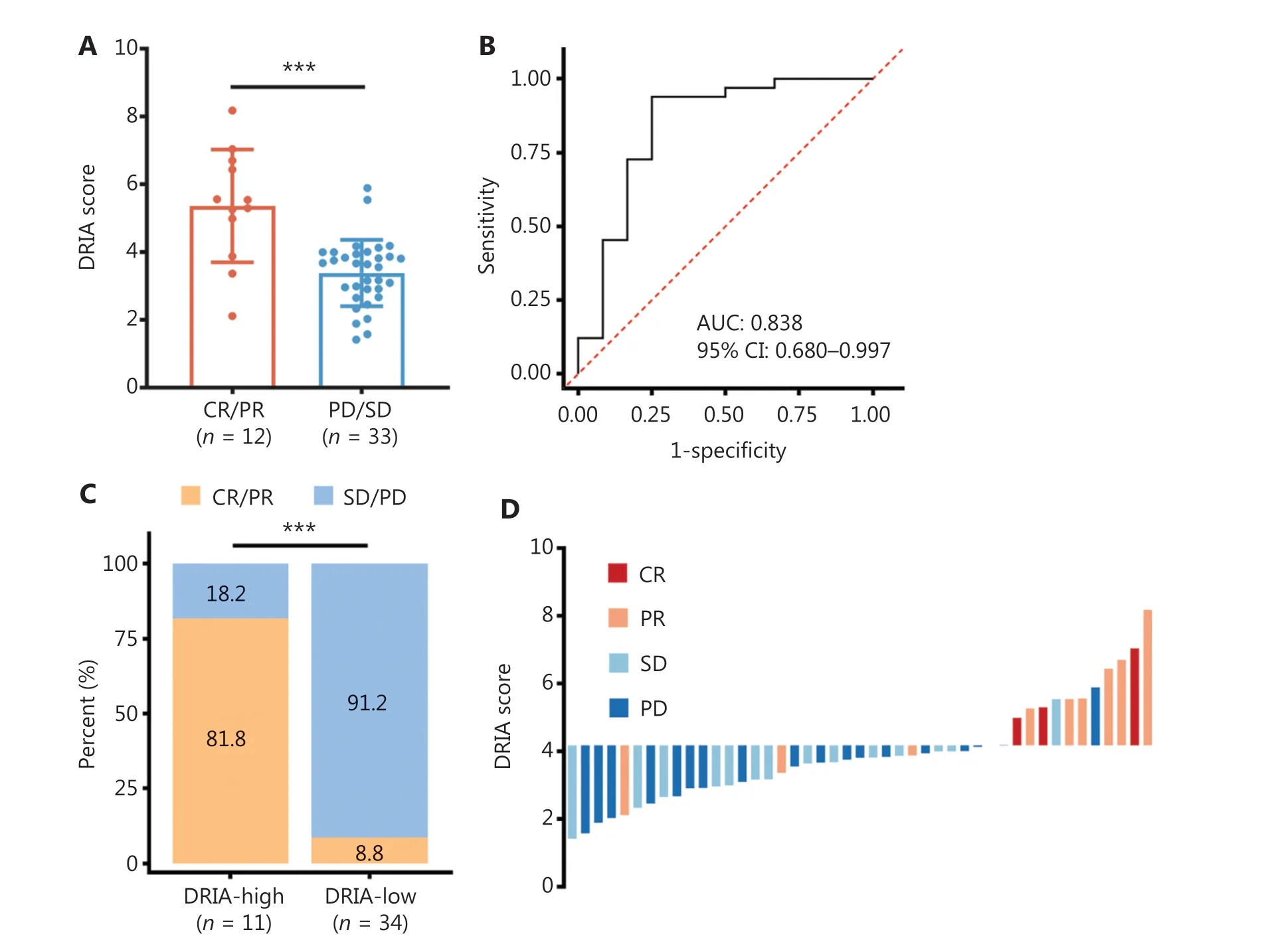
Figure 2 Association between DRIA and the clinical response to ICI therapy in gastric cancer.(A) Distribution of DRIA scores for distinct clinical response groups in the Kim18-gastric cancer cohort.(B) ROC curve of sensitivity versus 1-specificity for the DRIA score predictive value to the clinical response.(C) Stacked bar plot depicting different fractions of clinical response for patients in DRIA-high and -low groups.(D) Waterfall plot of DRIA score for distinct clinical response groups.DRIA, DNA damage response-related immune activation; ICIs, immune checkpoint inhibitors; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; AUC, area under the ROC curve;CI, confidence interval.***P < 0.001.
Validation of the DRIA predictive performance in MSS colorectal cancer and pancreatic adenocarcinoma
The addition of radiation therapy to dual PD-1 and CTLA-4 blockade has demonstrated some efficacy in MSS colorectal cancer and pancreatic adenocarcinoma29.Therefore, we attempted to validate the DRIA predictive value for the clinical benefits of this combination therapy in the Parikh22 cohort.ROC curve analysis indicated that the predictive accuracy was excellent, with an AUC of 0.806 (95% CI, 0.580-1.000;Figure 4A).The ORR in the DRIA-high group was superior to the DRIA-low group (42.9%vs.0%; Figure 4B).Notably,the waterfall plot suggested that all patients responsive to combination therapy were in the DRIA-high group (Figure 4C).As expected, the DRIA-low patients had a worse PFS (median PFS, 1.77vs.2.76 months; Figure 4D) and OS (median OS,6.14vs.11.73 months; Figure 4E) than DRIA-high patients.
Extended validation of the DRIA predictive robustness for ICI therapy efficacy in pancancer cohorts
To confirm the robustness of DRIA in predicting the benefits of ICI therapy, we included four immunotherapy cohorts with multiple types of cancer (melanoma, urothelial cancer, and pan-cancer) for subsequent analyses.Patients in the CR/PR group tended to have higher DRIA scores than the PD/SD group in the PUCH, which is consistent with the two GI cancer (Figure 5A), Gide19 (Figure 5B), Pender19 (Figure 5C),and IMvigor210 (Figure S5A) immunotherapy cohorts.The waterfall plot showed a positive association between a higher DRIA score and a better clinical response (Figure S6A-C).ROC curve analyses revealed that the resulting AUCs were 0.760(95% CI, 0.612-0.907; Figure 5D), 0.845 (95% CI, 0.715-0.974;Figure 5E), and 0.766 (95% CI, 0.596-0.936; Figure 5F), respectively.In addition, patients in the DRIA-high group had a better ORR than the DRIA-low group in the PUCH (40.0%vs.8.0%; Figure 5G), Gide19 (78.9%vs.18.2%; Figure 5H), and Pender21 (50.0%vs.6.8%; Figure 5I) cohorts.Furthermore, we compared the clinical outcomes between the two DRIA groups in the four cohorts.The DRIA-low group had a significantly poorer PFS (Figure 6A-C) and OS (Figures S5B and S7A, B)than the DRIA-high group.In agreement with these results, the multivariate Cox regression analyses showed that the higher DRIA score was an independent prognostic factor for predicting a favorable PFS (Figure 6D-F) and OS (Figures S5C and S7C,D) in patients receiving ICI therapy.
We also validated the correlation between the DRIA and these biomarkers based on the available CD8+T cell infiltration, PD-L1 expression, TMB, and TNB data in the IMvigor210 cohort.When the tumors were divided into three classic immune phenotypes according to CD8+T cells infiltration,we observed that patients with an immune-inflamed phenotype had the highest DRIA scores compared to the other phenotypes (Figure S5D).As predicted, the DRIA-high patients had a relatively higher TMB (Figure S5E) and TNB (Figure S5F).Similarly, a higher DRIA score was positively correlated with higher PD-L1 expression (Figure S5G, H).Above all, the results of the four immunotherapy cohorts fully demonstrated that DRIA precisely predicts the clinical benefit of ICI therapy in pan-cancer.
Comparison of the DRIA predictive performance with two original DDIR signatures
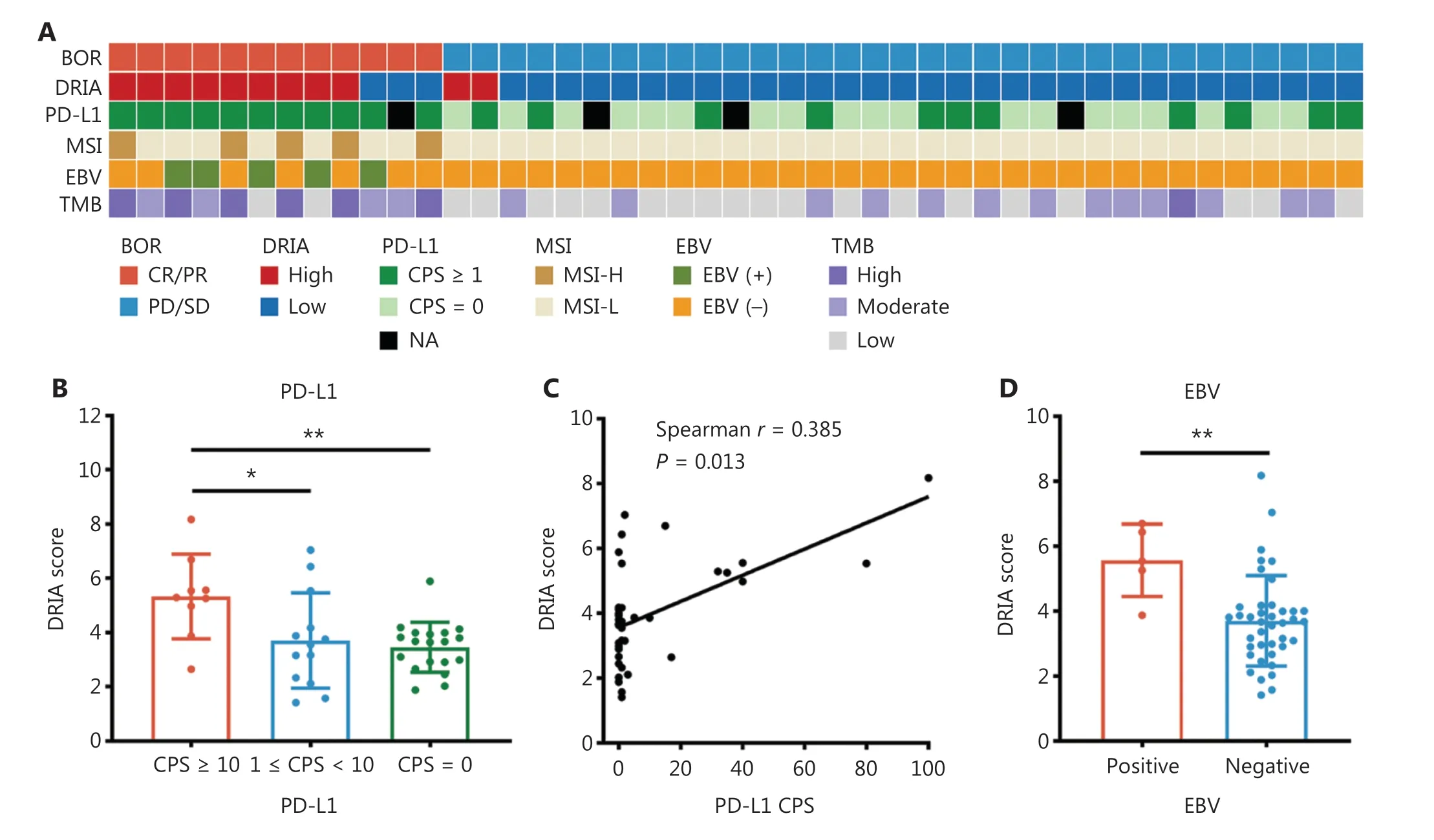
Figure 3 Continued
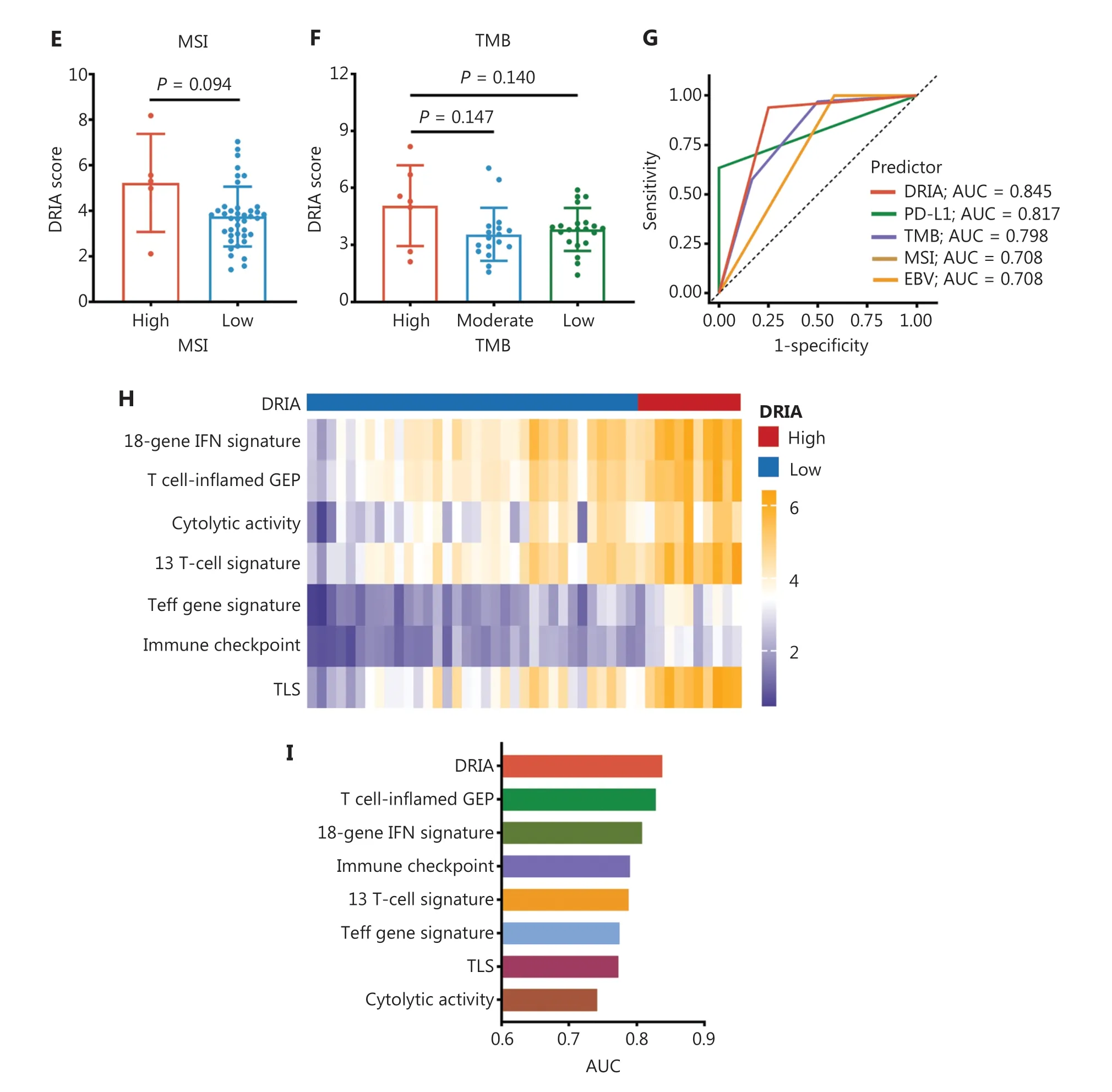
Figure 3 Comparison of the predictive accuracy of DRIA with known clinical biomarkers in gastric cancer.(A) Heatmap of DRIA distributions and known clinical biomarkers for distinct clinical response groups in the Kim18-gastric cancer cohort.(B) Comparison of the DRIA score in three groups according to the level of PD-L1 expression.(C) Correlation of the DRIA score with the level of PD-L1 expression.(D-F) Comparison of the DRIA score in different groups according to EBV (D), MSI (E), and TMB status (F).(G) ROC curves measuring the predictive accuracy of DRIA, PD-L1,TMB, MSI, and EBV for response to ICI therapy.(H) Heatmap showing differently expressed immune-related signatures between the DRIA-high and -low groups.(I) Comparison of the DRIA AUCs and seven published immune-related signatures in predicting the response to ICI therapy.DRIA, DNA damage response-related immune activation; BOR, best of response; MSI, microsatellite instability; EBV, Epstein-Barr virus; TMB,tumor mutational burden; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; CPS, combined positive score;ICIs, immune checkpoint inhibitors; AUC, area under the ROC curve; TLS, tertiary lymphoid structures; NA, not available.*P < 0.05, **P < 0.01.
To facilitate the clinical translation of DRIA, we further conducted ROC analyses to compare the predictive performance of DRIA with two original DDIR signatures as a function of response to ICI therapy in five immunotherapy cohorts.The predicting DRIA AUCs were essentially equivalent to the 9-and 44-gene DDIR signatures (Table S6).Taken together, the DRIA signature conferred superior predictive accuracy for ICI therapy efficacy, which warrants further investigations in prospective clinical trials.
Discussion
In this study we developed and validated a novel immune gene signature, the DRIA, which consists of three genes that robustly predict clinical benefit from ICI therapy in GI cancer and pan-cancer patients.Our results revealed that DRIA positivity effectively identifies patients with improved response rates and survival outcomes in the context of ICI therapy.Indeed, the predictive accuracy of DRIA was superior to the known clinical biomarkers in GI cancer, such as PD-L1 and TMB.
Currently, development of effective biomarkers is required to guide ICI therapy decisions for different types of cancer,including GI cancer.A series of biomarkers, including PD-L1,TMB, MSI, and EBV, have been widely used in the clinical management of patients with GI cancer6.These biomarkers have encountered several issues6,7,11-13, such as prediction instability, intra-tumoral heterogeneity, and fewer predicted benefits to populations, which incites us to explore more markers with universal mechanisms.DDR deficiency has recently emerged to increase the level of neoantigens and tumor immunogenicity, and activate the cGAS-STING pathway to reshape the tumor microenvironment, ultimately leading to the effectiveness of ICI therapy21.In view of the high frequency of DDR deficiency in tumorigenesis and progression, extensive efforts have been made to identify DDRrelated biomarkers for predicting the response to ICI therapy.Several investigations have demonstrated that gene mutations in different DDR pathways result in a durable clinical benefit from ICI therapy in various types of cancer, including melanoma34, lung cancer35, bladder cancer19, and GI cancer20.Previous studies, however, have also shown that even thoughBRCA1/2mutations may confer sensitivity to ICI therapy, this is not held in common for all patients because not all mutations induce DDR deficiency or compensate for by alternate mechanisms36.Conversely,BRCA1/2wide-type tumors confers a DDR-deficient phenotype due to epigenetic silencing ofBRCA1/237,38.Therefore, a transcriptome-based signature which assesses the downstream effects of genomic and epigenetic changes of altered DDR pathways may more accurately predict the sensitivity to ICI therapy.
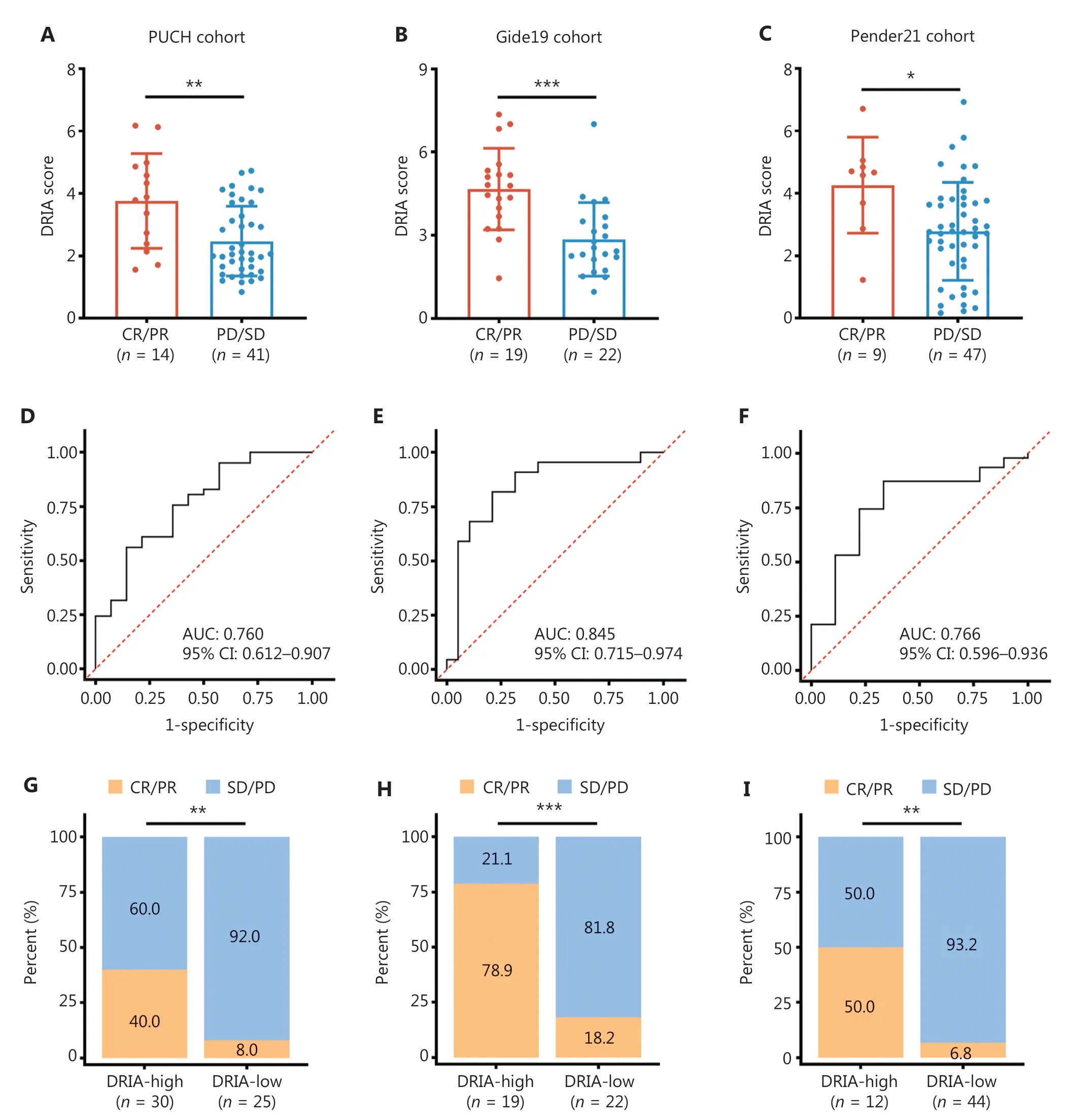
Figure 5 Validation of the predictive robustness of DRIA in ICI-treated pan-cancer cohorts.(A-C) Comparison of the DRIA scores between responders and non-responders in PUCH melanoma (A), Gide19 melanoma (B), and Pender21 pan-cancer cohorts (C).(D-F) ROC curves measuring the predictive value of the DRIA score in PUCH melanoma (D), Gide19 melanoma (E), and Pender21 pan-cancer cohorts (F).(G-I) Proportion of objective response for patients with DRIA-high and -low status in PUCH melanoma (G), Gide19 melanoma (H), and Pender21 pan-cancer cohorts (I).DRIA, DNA damage response-related immune activation; ICIs, immune checkpoint inhibitors; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; AUC, area under the ROC curve; CI, confidence interval.*P < 0.05;**P < 0.01; ***P < 0.001.
The DDIR signature summarizes the cascade of changes in immune activation from the resulting accumulation of DNA damage22,23.Therefore, the DDIR signature could serve as a promising marker to predict the clinical response to ICI therapy.Considering the relatively larger panel of genes in the original DDIR signature that cannot be easily translated into an easy-to-use assay, we used three consensus DDR-related genes (CXCL10,IDO1, andIFI44L) from the 44- and 9-gene DDIR signatures to generate a novel DRIA signature.As predicted, the DRIA signature had a strong correlation with two original DDIR signatures and effectively classified patients into different subsets with distinct tumor immune microenvironment characteristics.Moreover, the level ofCXCL10,IDO1, andIFI44Lgene expression was shown to be increased in DDR-deficient tumors compared to DDR-proficient tumors, which indicated the need for three genes within the DDIR characteristics.In agreement with our results, Parkes et al.23concluded that DDR-deficient tumor cells express significantly higher CXCL10 than DDR-proficient tumor cells.Vidotto et al.39reported that the increased expression of IDO1 driven by DDR genetic deficiency modulates the response to immunotherapy.These data provided a theoretical basis for the DRIA signature to predict the response to ICI therapy.
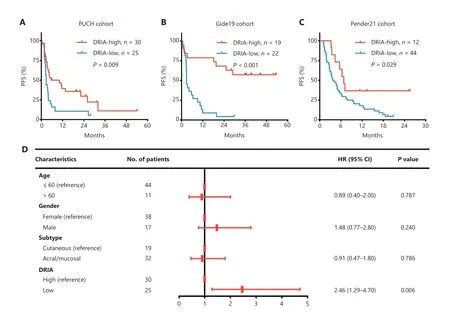
Figure 6 Continued
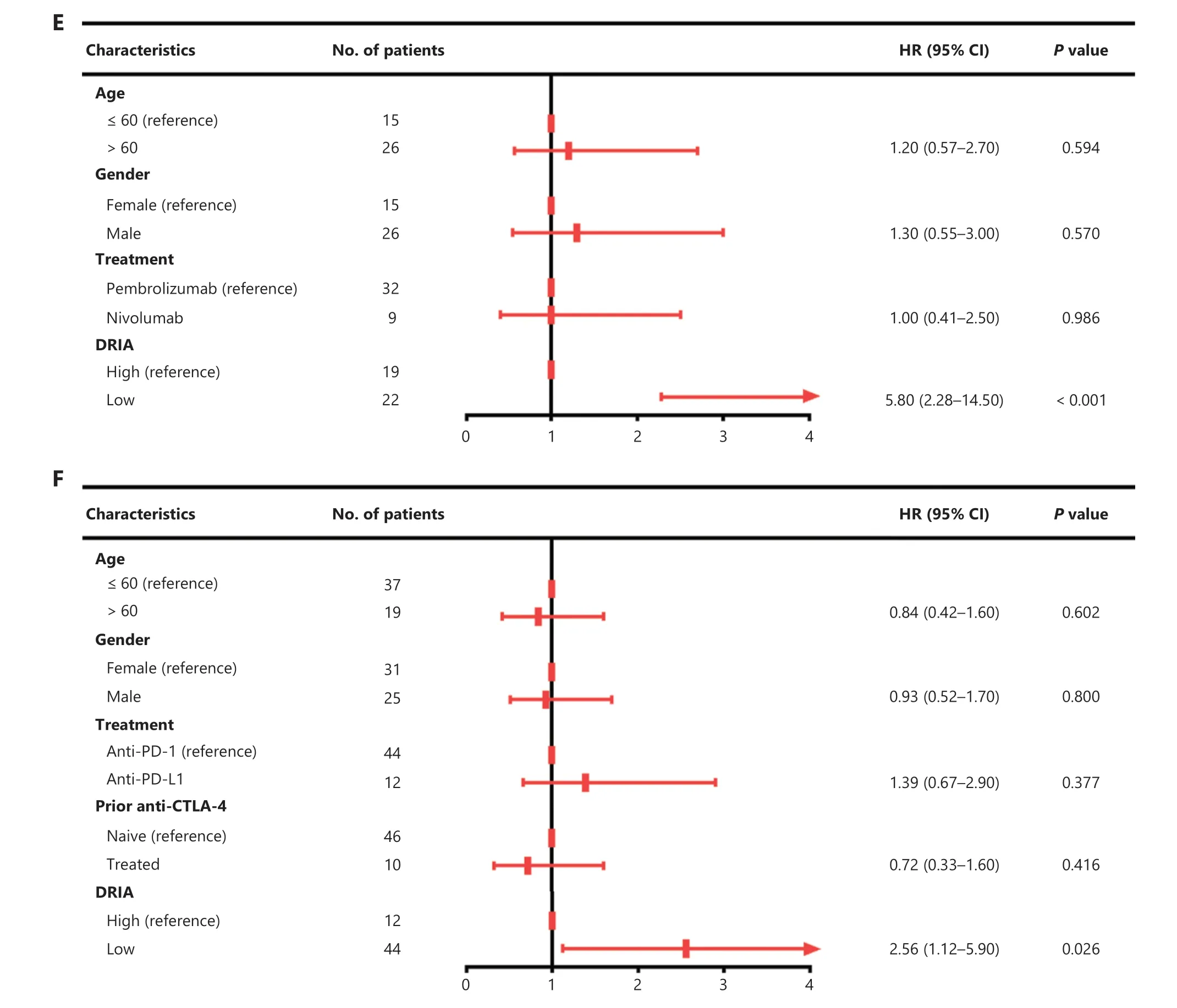
Figure 6 Validation of the prognostic value of DRIA in ICI-treated pan-cancer cohorts.(A-C) Kaplan-Meier plots of PFS segregated by the DRIA score in PUCH melanoma (A), Gide19 melanoma (B), and Pender21 pan-cancer cohorts (C).(D-F) Multivariate Cox regression analyses of DRIA and clinicopathologic factors associated with PFS in PUCH melanoma (D), Gide19 melanoma (E), and Pender21 pan-cancer cohorts (F).DRIA, DNA damage response-related immune activation; ICIs, immune checkpoint inhibitors; PFS, progression-free survival; HR, hazard ratio;CI, confidence interval.
The muti-cohort analysis and validation showed that the DRIA signature is predictive of clinical response and survival benefit following ICI therapy in patients with GI cancer and pan-cancer.The predictive accuracy of DRIA was superior to PD-L1, TMB, EBV, and MSI in patients with GI cancer.We also showed that the DRIA predictive performance was essentially equivalent to the two original DDIR signatures.In fact, these data fully demonstrated that the DRIA signature might serve as a promising predictor for ICI therapy decisions.Further investigations are warranted to develop the detection system of three genes using a PCR array or liquid biopsy and conduct prospective evaluation of the DRIA signature.In addition, we showed that the pairwise combination of DRIA with PD-L1,TMB, MSI, and EBV had improved patient stratification for ICI therapy.These data suggested that the DRIA signature can also be used to complement these current biomarkers to select potential responders for ICI therapy.Similarly, our study included one MSS GI cancer (Parikh22) cohort and successfully predicted three patients who responded to ICI-based combination therapy using the DRIA signature.Other studies have confirmed the presence of DDR pathway aberrations in patients with MSS colorectal cancer40,41.Further investigations are needed to extensively evaluate the predictive value of DRIA for ICI-based combination therapy in additional cohorts.
This retrospective study had several limitations, including unresolved concerns and potential perspectives.First, the DRIA signature was assessed based on endoscopic or needle biopsy specimens with limited tumor clonality.A high level of intra-tumoral heterogeneity is associated with the response to ICI therapy42,43, thereby indicating the limitation of a single biopsy in the development of a predictive biomarker.This limitation can be partially resolved by the pooling of biopsy fragments from multiple sites within the tumor.Second,several factors that may have inevitably produced bias, such as the sequencing platform used, line of treatment, ICI therapeutic regimen used, and the limited types of solid tumors.Moreover, although one real-world pan-cancer cohort was included in this study to validate the predictive value of DRIA,the small sample sizes weakened the accuracy of the results.Of note, however, results were validated in multiple cohorts spanning Asian, European, American, and Australian populations.Considering the limitations and concerns mentioned above,prospective multicenter clinical trials with a larger number of patients with multiple solid tumors are warranted to assess the predictive value of DRIA.Furthermore, comprehensive analyses of the relationship between DRIA and known clinical biomarkers based on a standardized testing system may provide additional information for guiding clinical application of ICI therapy.
Conclusions
In summary, we constructed an individualized DDR-based signature (DRIA) that identified potential patients with improved response rates and survival outcomes in the context of ICI therapy.Our study also demonstrated that this signature has superior predictive power for the clinical benefit of ICI therapy compared to known clinical biomarkers,including PD-L1 TMB, MSI, and EBV.Therefore, DRIA may potentially act as a novel predictive marker for ICI therapy.More in depth corollary studies are warranted in prospective clinical trials.
Grant support
This study was supported by the National Natural Science Foundation of China (Grant Nos.81972761 and 82202837)and the National Key R&D Program of China (Grant Nos.2016YFC1303200 and 2022YFC2505100).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Yongzhan Nie, Yan Kong.Collected the data: Junya Yan, Shibo Wang, Jing Zhang,Qiangqiang Yuan, Xianchun Gao, Nannan Zhang, Haohao Zhang, Kun Liu, Jun Yu, Linbin Lu, Hui Liu, Xiaoliang Gao,Sheng Zhao, Wenyao Zhang, Abudurousuli Reyila, Yu Qi,Qiujin Zhang.
Provided the materials or patients: Yongzhan Nie, Yan Kong,Yan Pan, Shundong Cang, Yuanyuan Lu, Yanglin Pan.
Analyzed and interpreted the data: Yongzhan Nie, Yan Kong,Junya Yan, Shibo Wang, Jing Zhang.Wrote the manuscript: Junya Yan.
Data availability statement
The results shown here are in part based on data generated by The Cancer Genome Atlas project (TCGA, http://cancergenome.nih.gov/) and data available in published articles10,29-32.The raw sequence data from PUCH dataset reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, Beijing Institute of Genomics(China National Center for Bioinformation), Chinese Academy of Sciences, under accession number HRA000524 that are publicly accessible at http://bigd.big.ac.cn/gsa-human.
 Cancer Biology & Medicine2024年3期
Cancer Biology & Medicine2024年3期
- Cancer Biology & Medicine的其它文章
- Erratum to Treatment strategies for patients with HER2-positive gastric cancer
- Bronchoalveolar lavage fluid assessment facilitates precision medicine for lung cancer
- A retrospective analysis of mature T- and NK-cell lymphomas
- Current status of early gastric cancer screening research
- Cervical cancer prevention in China: where are we now,and what’s next?
- Reducing the global cancer burden with gastrointestinal screening: China’s 30 years practice
