OsDA1 positively regulates grain width in rice
Cong Li, Jun Liu, Liy Zhng, To Li, Hongyu Li, Bin Liu, To Zho,*
a Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
b Institute of Nanfan & Seed Industry, Guangdong Academy of Sciences, Guangzhou 510316, Guangdong, China
Keywords:Rice Osda1 Grain size Grain shape Grain length/width ratio TCP
ABSTRACT The size and shape of rice grains influence their yield and commercial value.We investigated the role of OsDA1,a rice homolog of the Arabidopsis DA1 gene,in regulating grain size and shape.OsDA1 was highly expressed in young spikelets and glumes.Its overexpression led to enlarged seeds with increased width and decreased length/width ratio (LWR) and knocking out OsDA1 reduced grain width and increased grain length and LWR.A R310K point mutation in the DA1-like domain is a potential target for breeding for increased grain width and length.OsDA1 interacted with TCP gene-family proteins to regulate grain size and shape.Our findings deepen our understanding of the molecular mechanisms underlying grain size regulation and provide useful information for improving grain yield.
1.Introduction
In cereal crops, grain size is a major factor influencing grain yield and is determined largely by genetic controls during seed development.Elucidation of the molecular mechanisms regulating grain size and discovering novel genes associated with the trait may lead to increased rice yields [1].
In the past two decades, extensive research has revealed that rice grain shape and weight traits are controlled by a highly complex genetic network that involves multiple genes and quantitative trait loci(QTL).So far,approximately 100 QTL controlling grain size(length, width, thickness, and length/width ratio) have been identified and located on 12 chromosomes of rice[2–4].GS3(Grain Size 3)encodes a trans-membrane protein and is a negative regulator of grain size and organ size [5,6].GL3.1 (Grain length 3.1) is another major QTL that has a similar function to GS3.It encodes a PPKL(phosphatase kelch) family-Ser/Thr phosphatase that regulates grain length by mediating cell cycle progression via changes in the phosphorylation status of cell-cycle proteins [7–9].Several QTL associated with grain width have been cloned: GW2 (Grain width 2) [10], GW5 [11,12], GW8[13,14], TGW6 [15,16], GL7/GW7[17,18,19,20], GLW7[21], and GS5 [22].GW2, GW5, and GW7 regulate grain size negatively and GW8, GLW7, and GS5 positively[13,22].Loss of GW2 function conferred increased grain width, as did the mutant allele of GW5[10–12].Upregulation of GW7 expression was associated with the production of slenderer grains [14].GW8 overexpression promoted cell division and increased grain width [13].Higher expression of GS5 increased grain width [22].GLW7,identified in a genome-wide association study[21,increases grain length and yield.
The recently identified ubiquitin-related protein DA1 in Arabidopsis plays a pivotal role in the regulation of seed size through intricate molecular mechanisms [23].DA1 contains two ubiquitin-interacting motifs (UIMs), a single zinc-binding LIM domain, and a DA1-like domain that was first identified in the DA1 family [24,25].The UIM domain and the conserved Cterminal DA1-like domain were required for DA1 function [26].The da1-1 mutant produced larger flowers, siliques, and leaves and larger and heavier seeds than wild-type plants by restricting the period of cell proliferation [24].Mutants of both DA1 and its closest family member DAR1 displayed larger seeds and organs,and upregulation of da1-1 cDNA led to the same effect.Although the mutant is caused by an arginine-to-lysine mutation at position 358 of the DA1-like domain,disruption of DA1 by T-DNA insertion or its overexpression did not cause obvious changes in seed and organ size phenotypes [23,24].Overexpression of DA1R358Kin B.napus increased the biomass and size of the seeds, cotyledons,leaves, flowers, and siliques of transgenic plants [23,24,27–29].Similarly,overexpression of a mutated ZmDA1,the maize homolog of Arabidopsis da1-1,increased grain yield by regulating starch synthesis[30].The latest report[26,29,31]suggests that the ubiquitinactivated peptidase DA1 activates two RING E3 ligases,Big Brother(BB) and DA2, which are subsequently cleaved by the activated peptidase and destabilized.DA1 peptidase activity limits the duration of cell proliferation by cleaving the deubiquitylase UBIQUITINSPECIFIC PROTEASE 15 (UBP15), which promotes cell proliferation in the integuments of ovules and developing seeds, and the transcription factors TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP15)and TCP22,which promote cell proliferation and repress endoreduplication.In a study[32]of the role of HOMOLOG OF DA1 ON RICE CHROMOSOME 3(HDR3)),a homolog of DA1 in Arabidopsis,in regulating grain size in rice,HDR3 interacted with the histone acetyltransferase GW6a to exert joint control of grain size.HDR3 stabilized GW6a,which regulated the expression of genes involved in grain development.
Previously [33], we used a large-scale hybrid transcription factor (HTF) approach to generate rice transgenic populations for investigating the role of various transcription factors in plant growth and development.The HTF approach involved the fusion of coding sequences of TFs with universal transcriptional activation module VP64 (tetrameric repeats of VP16) or repression module 4EAR (tetrameric repeats of EAR).By surveying the phenotypes of the rice HTF transgenic population,we identified a positive regulator of grain size in rice, named OsDA1.OsDA1 is a homologous gene of the HDR3, which is the gene with the highest homology with Arabidopsis DA1 in rice.To investigate the impact of OsDA1 on rice grain width, we employed knockout and overexpression techniques to introduce various mutant versions of OsDA1.Simultaneously, the study identified a TCP gene family that interacts physically with OsDA1 through yeast two-hybrid and firefly luciferase complementation imaging assays.This discovery sheds light on the underlying molecular mechanisms involved in the regulation of rice grain morphology.
2.Materials and methods
2.1.Primers and accession numbers
All PCR primers used in this study are listed in Table S1.Sequences were retrieved from the MSU Rice Genome Annotation Project database (https://rice.plantbiology.msu.edu) [34].The accession numbers are OsDA1 (LOC_Os06g08400.1) and OsACT1(LOC_Os05g36290.1).
2.2.Field planting and trait measurement
The wild-type (WT) rice cultivar Kitakii (Oryza sativa L.subsp.japonica)was used for rice transformation.Rice plants were grown in a field in Beijing (39°54′N, 116°23′E) under natural conditions from May to October.Rice grains were stored at room temperature for one month after harvest before measurement.A WSEEN SC-E(Hangzhou Wanshen Testing Technology Co.,Ltd.)rice appearance quality detector was used to measure grain width, grain length,1000-grain weight,and grain length-to-width ratio (LWR)of filled grains.
2.3.Vector construction and plant transformation
To construct plant transformation vectors, Attb-F and Attb-R primers were used to insert OsDA1 cDNA (1458 bp) into the entry vector pDONR 201 by the Gateway system following the manufacturer’s instructions (Invitrogen Corporation).Then the Gateway system was used to insert OsDA1 cDNA into pGWVP64B for Pubi::DA1-VP64, pGWBEARB for Pubi::DA1-EAR4, pCAMBIA1301-Bar-FLAG for Pubi::DA1-3flag, and pANDA for the DA1 RNAi vector.To construct the PROOsDA1::GUS vector,Gus-F and Gus-R primers were used to amplify a 1897-bp fragment of the OsDA1 promoter(-1897 bp to -1 bp from the ATG) from genomic DNA of the rice cultivar Nipponbare and the amplicon was inserted into PRO::GUS,which had been digested with XmalI and NcoI.To obtain Osda1 mutants, the CRISPR-P resource (https://crispr.hzau.edu.cn) was used to design a 19-bp gRNA-F primer and this was inserted into the pCas9-OsU3-sgRNA vector following XbalI digestion.Several OsDA1 cDNAs with mutated domains were amplified using UIMs,LIM,DA1-Like,and R310K primers.All of the constructs were introduced into Agrobacterium tumefaciens strain EHA105, and transgenic rice plants were generated by A.tumefaciens-mediated transformation[35].The final construct P35S::OsDA1-GFP was transiently expressed in leaves of N.benthamiana plants as previously described [36].
2.4.RNA isolation and qRT-PCR analysis
Total RNA was extracted from rice tissues using TRIzol reagent(Invitrogen).Complementary DNA was synthesized from DNasetreated total RNA(5 μg)with a reverse transcription kit(TRAN Corporation).qRT-PCR (Quantitative Real-Time Polymerase Chain Reaction) was performed in 96-well optical plates using a SYBR Green RT-PCR kit (Takara Corporation) and a Roche Light Cycler 480.Three independent biological replicates were used, and three replicate reactions were used for each sample.All Ct(Cycle threshold) values were normalized to OsACT1.
2.5.Histological analysis
Spikelet hulls were fixed in FAA solution(60% (v/v) ethanol,5%(v/v) glacial acetic acid, and 5% (v/v) formaldehyde), vacuumevacuated for 40 min, dehydrated in an ethanol series (70%, 80%,85%, 90%, 95%, and 100% (v/v) ethanol), destained with 3:1, 1:1,and 1:3 ethanol:xylene mixtures and 100%ethanol,and embedded in paraffin.Tissue sections were cut with a Leica rotary microtome,fixed on glass slides, and stained with 0.05% TBO (Toluidine Blue O).All spikelet hulls were soaked in the ethanol and ethanol:xylene mixtures for 2 h.
2.6.GUS staining (histochemical GUS assay)
Transgenic plant tissues carrying ProOsDA1::GUS were infiltrated with GUS staining solution(100 mmol L-1NaH2PO4buffer,pH 7.0;0.5%Triton X-100;0.5 mg mL-1X-Gluc;and 20%methanol)under vacuum for 10 min and incubated overnight at 37 °C.The stained tissues were then rinsed and photographed.
2.7.Transactivation activity and yeast two-hybrid assays
The full-length OsDA1 cDNA and several derivatives of OsDA1 containing deletions were inserted into a bait pGBKT7 vector that had been digested with EcoRI.OsTCP5 (LOC_Os02g51280),OsPCF2 (LOC_Os08g43160), OsPCF5 (LOC_Os01g11550), OsTCP2(LOC_Os01g55750), OsTCP10 (LOC_Os04g44440), OsTCP11(LOC_Os05g43760), OsREP1 (LOC_Os09g24480), OsTCP20(LOC_Os12g02090), OsPCF7 (LOC_Os01g55100), OsTCP18(LOC_Os09g34950), OsTCP13 (LOC_Os07g04510), OsTCP6(LOC_Os02g51310), OsPCF6 (LOC_Os03g57190), OsTCP21(LOC_Os12g07480), OsTCP15 (LOC_Os08g33530), OsTCP19(LOC_Os06g12230) and OsPCF1 (LOC_Os04g11830) cDNA were inserted into EcoRI and BamHI sites of the prey vector pGADT7.The bait constructs were co-transformed separately with empty pGADT7 vector into yeast strain AH109 to test their transactivation activity.OsTCPs-AD were co-transformed separately with OsDA1-BD into yeast strain AH109 to test their interactions.Cultures containing the fusion protein were plated onto SD/-L-W and SD/-L-W-H-Ade plates and allowed to grow for 48 h before being photographed.The empty vector (BD) was used as a negative control.
2.8.Firefly luciferase complementation imaging assays in N.benthamiana
The CDS sequences of OsPCF1, OsTCP18, and OsPCF3 were individually inserted into the pCambia1300-LUCNvector at the KpnI and SalI sites.The CDS of OsDA1 were individually cloned into the pCambia1300-LUCCvector at the BglII and MluI sites.The LUCNand LUCCplasmids were individually introduced into A.tumefaciens strain EHA105 by electroporation and then infiltrated into N.benthamiana leaves in various combinations.A low-light cooled charge-coupled device camera (Tanon 5200) was used to capture the LUC image with an exposure time of 10 min.
2.9.Subcellular localization
The In-fusion system(Clontech Corporation)was used to insert the OsDA1 full-length cDNA (1458 bp) and its deletion derivatives into a PA7-YFP vector (2× 35S::YFP) that had been digested with BamHI and SmaI.The OsDA1-YFP (2× 35S:: OsDA1-YFP) fusion protein and its deletion derivatives were transiently expressed in rice protoplasts under the control of a 2× 35S promoter.An AHL-RFP fusion protein was used as a nuclear marker.The fluorescence signal was observed with a confocal microscope at 14 h after transformation.Subcellular localization of fusion proteins was examined with a Zeiss LSM 510 confocal microscopy system.Excitation was achieved with an argon laser at 458 nm(ECFP)and 514 nm(EYFP)and with a diode laser at 405 nm(Hoechst 33342, Sigma Corporation).Fluorescence was observed using emission filter settings of 470–500 nm (ECFP), 530–600 nm (ERFP), and 420–480 nm(Hoechst 33342).
3.Results
3.1.OsDA1 positively regulates grain width in rice

Fig.1.Phenotypes of OsDA1V- and OsDA1E-overexpressing plants.(A) Wild-type and transgenic plants grown under natural conditions (NCs) in a Beijing field after grain filling.(B) The grain length phenotypes of WT, OsDA1V-04, OsDA1V-06, OsDA1E-07, and OsDA1E-13 (from left to right).(C) The grain width phenotypes of WT, OsDA1V-04,OsDA1V-06,OsDA1E-07,and OsDA1E-13(from top to bottom).(D–G)Comparisons of grain widths(D),grain lengths(E),grain length/width ratios(F),and 1000-grain weight(G) between WT and transgenic plants.Values are mean ± SD.Student’s t-test was used for comparisons (n = 10 spikelets; **, P < 0.01).
By surveying the phenotypes of the rice HTF transgenic population, we identified a pair of HTFs that displayed opposite grainwidth phenotypes,referred to as OsDA1V and OsDA1E.We obtained multiple independent OsDA1V and OsDA1E lines and selected two transgenic lines, using immunoblotting and qRT-PCR, for further phenotypic identification (Fig.S1A, B).The two OsDA1V lines showed increased grain widths (7.56% and 7.85%) and 1000-grain weights (7.58% and 6.88%) but decreased LWRs (-7.32%and -5.37%), whereas the two OsDA1E transgenic lines showed decreased grain widths (-4.38% and -7.74%) and 1000-grain weight (-2.03% and -5.52%) but increased LWRs (2.44% and 4.39%) (Fig.1D, G; Table S2).Moreover, all the transgenic lines obtained in the study differed in grain length,indicating that grain length was not affected by the HTFs and that the decreased/increased LWR in OsDA1Vs/OsDA1Es was due solely to changes in grain width.
To avoid phenotypic artifacts caused by the additional VP64 and 4EAR motifs, we reconstructed an OsDA1 overexpression vector and an OsDA1 RNAi vector and obtained multiple independent lines.Two OX lines (OX-08 and OX-09) and RNAi lines (RNAi-01 and RNAi-12), which had been confirmed by immunoblotting or qRT-PCR, were selected for further study (Fig.S1C).OsDA1-overexpressing transgenic lines, OX-08 and OX-09, showed increased grain widths(14.24%and 12.79%)and 1000-grain weight(6.10%and 11.79%),with differing degrees of a decreased LWR phenotype (-11.71% and -6.34%).Two RNAi lines, RNAi-01 and RNAi-12, showed grain width (-3.20% and -7.56%), 1000-grain weight(-8.11% and -13.80%), and LWR (4.88% and 2.44%) phenotypes that were opposite to those observed in the overexpressing lines(Fig.S2;Table S2).RNAi lines displayed large variations and differences in grain length,whereas overexpression lines showed negligible changes.The expression of all OsDA1 homologs in RNAi lines was significantly reduced to differing degrees.We speculate that there are functional differences among these homologous genes,resulting in variations in the grain-length phenotype of RNAi strains.
To further confirm the function of the gene OsDA1, we generated two OsDA1 mutants, Osda1-1 and Osda1-2, using the CRISPR/Cas9 system.The results revealed that both mutants showed reduced grain width, by respectively 2.40% and 2.99%.Both mutants showed decreases in 1000-grain weight, by respectively 2.71% and 2.32%.But their grain lengths were increased by 0.89%and 4.90%.These changes increased their LWRs by respectively 3.47% and 3.96% (Fig.2; Table S2).
3.2.OsDA1, a homolog of DA1, was highly expressed in the inflorescence
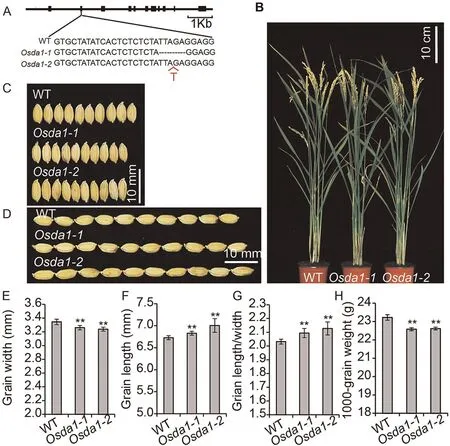
Fig.2.Phenotypes of OsDA1 mutants.(A)The mutant sequences of two homozygous mutants at the T1 generation.(B)Wild type and Osda1 grown under natural conditions in a Beijing field after grain filling.(C) The grain length phenotype of WT, Osda1-1, and Osda1-2 (from top to bottom).(D) The grain length phenotypes of WT, Osda1-1, and Osda1-2(from top to bottom).(E–H)Comparisons of grain widths(E),grain lengths(F),grain length/width ratios(G),and 1000-grain weight(H)between WT and transgenic plants.Values are mean ± SD.Student’s t-test was used for comparisons (n = 10 spikelets; **, P < 0.01).
OsDA1 corresponds to LOC_Os06g08400, which encodes a 486 amino acid protein with a predicted molecular mass of 55.1 kD.OsDA1 contains two UIMs (ubiquitin-interacting motifs, amino acids 46–65 and 77–96) in its respective N-terminal regions, a LIM (amino acids 123–175) in the middle region and a DA1-like(peptidase domain,amino acids 273–481)in the C-terminal region(Fig.3A).The rice genome contains four homologs of the Arabidopsis DA1 gene, with OsDA1 showing the highest degree of similarity to Arabidopsis DA1.In a phylogenetic tree, OsDA1 clustered with homologs in rice (LOC_Os03g42820, LOC_Os12g40490 and HDR3(LOC_Os03g16090)), Arabidopsis (DA1, DAR1 and DAR2) and maize(ZmDA1 and ZmDAR1) (Fig.3B).Amino acid alignment indicated that OsDA1 shares 65.11%identity with DAR1 and 61.05% identity with DA1 (Fig.S3).
To determine the tissue-specific expression pattern of OsDA1,we monitored GUS activity in the various tissues of pOsDA1::GUS transgenic plants.GUS staining indicated that OsDA1 is expressed in germinating seed, lemma, root, stem node, young leaf, and young panicle, but most highly in the inflorescence (Fig.3C).The OsDA1 mRNA expression pattern was also detected by qRT-PCR.OsDA1 was detectable in almost all the tissues and was highest in the inflorescence (Fig.3D).
To draw a comparison between the regulatory roles of DA1 homologs in plant grain length and width, we obtained LOC_Os03g42820, LOC_Os12g40490, and HDR3-overexpressing plants and found that they positively regulate both grain length and width(Figs.S5,S6).Grain shape varied among the three transgenic plants.Overexpression of OsDA1 reduced the LWR of grains,making them more rounded.Grains overexpressing HDR3 showed increased LWR,resulting in a narrower shape.There was no significant variation in the LWR of grains overexpressing LOC_Os03g42820 or LOC_Os12g4049.
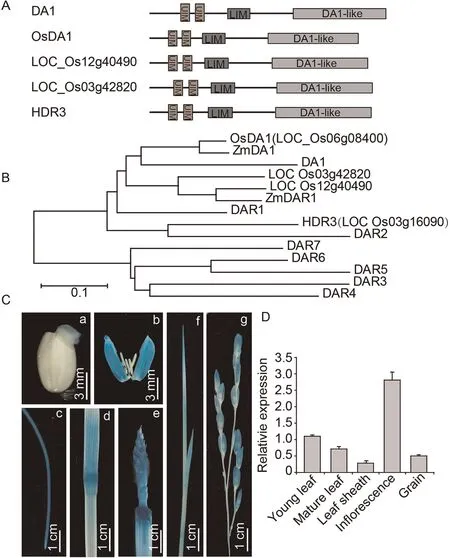
Fig.3.OsDA1 is a homolog of Arabidopsis DA1 and is expressed in almost all rice tissues.(A)Predicted protein structure of OsDA1 proteins.UIM,ubiquitin interacting motif;LIM,a protein structure composed of two contiguous zinc finger domains separated by a two-amino acid residue hydrophobic linker.(B)Phylogenetic tree of OsDA1 proteins in rice and Arabidopsis.(C) OsDA1 promoter expression pattern in OsDA1 promoter-GUS transgenic rice plants.[a–g] indicate germinated seed (2 days), germinating seed,lemma,root,stem node,inflorescence,young leaf,and young panicle.(D)RT-PCR analysis of OsDA1 expression in various organs.Total RNA was extracted from the young leaf,mature leaf, leaf sheath, inflorescence, and young grain of wild type.Actin was used as an internal control.Three replicate experiments were performed.
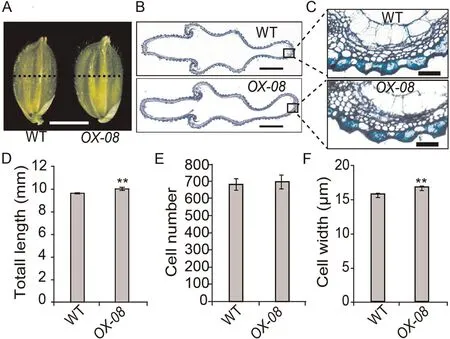
Fig.4.Histological analyses of spikelet hulls before heading in WT and OX-08.(A) Spikelets.Scale bar, 3 mm.(B) Cross-section of spikelet hulls.Scale bar, 100 μm.(C)Magnified view of the spikelet hull cross-sections boxed in B.(D–F)Comparisons of total length(D),total cell number(E),and cell width(F)in the outermost parenchymal cell layers of spikelet hulls of WT and OX-08.Cell width was calculated as total length divided by total cell number.Values are mean ± SD.Student’s t-test was used for comparisons (n = 10 spikelets; **, P < 0.01).

Fig.5.Phenotypes of OsDA1R310K overexpressing plants.(A) Part of an amino acid alignment of the OsDA1 protein and its Arabidopsis homologs.(B) A Wild-type and OsDA1R310K transgenic plants grown under natural conditions in a Beijing field after grain filling.(C) Grain width phenotypes of WT, OsDA1R310K-02 and OsDA1R310K-20 (D)Grain length phenotypes of WT, OsDA1R310K-02 and OsDA1R310K-20.(E–H) Comparisons of grain widths (E), grain lengths (F), grain length/width ratios (G), and 1000-grain weight (H) between WT and transgenic plants.Values are mean ± SD.Student’s t-test was used for comparisons (n = 10 spikelets; **, P < 0.01).
3.3.OsDA1 increased cell size in rice glumes
In Arabidopsis, DA1 controls the size of seeds and organs by restricting cell proliferation[24,28].In rice,in contrast,the number and size of cells in the spikelet hull of a mature grain determine its width.To determine the cause of the increased grain width observed in OsDA1-overexpressing transgenic plants,we compared the cell numbers and cell lengths in spikelet-hull cross sections of OX-08 and WT plants.
The total length of the outermost parenchymal cell layers of spikelet hulls was longer in OX-08 than in WT plants.There was no difference in cell numbers between the two groups (Fig.4).These findings suggest that the increase in the width of spikelet hulls in OX-08 plants is due to an increase in the length of cells on the grain width axis.If so, OsDA1 may be involved in the regulation of cell elongation or expansion in rice, with a role different from that of DA1 in Arabidopsis.
3.4.The DA1-like domain is necessary for the regulation of grain width and length
In previous studies [27,32], da1-1 overexpression increased seed and organ size in Arabidopsis and B.napus,whereas DA1 overexpression did not cause obvious phenotypic changes.The da1-1 mutation occurs at position 358 in the DA1-like domain, which corresponds to position 310 in OsDA1.Unlike in DA1, OsDA1 overexpression resulted in increased grain width in rice.We hypothesized that mutating the 310th position in OsDA1 would exhibit a different phenotype compared to Arabidopsis.To test this hypothesis, we replaced the arginine residue at position 310 with a lysine residue in OsDA1 and overexpressed it in rice.
OsDA1R310Koverexpression lines displayed varying degrees of the larger grain width phenotype, indicating that replacing the arginine residue at position 310 did not affect the function of OsDA1 in grain width regulation (Fig.5; Table S3).The grain lengths of the overexpression lines increased dramatically, suggesting that this amino acid residue is involved in the regulation of grain length.
To test this hypothesis,we constructed ΔUIM,ΔLIM,and ΔDA1-like overexpression vectors and introduced them into rice.Overexpression of ΔLIM led to an increase in grain width,but this increase was less than that in OsDA1R310Kand OsDA1-overexpressing lines.The grain width of ΔUIM and ΔDA1-like-overexpressing lines did not differ from that of wild-type plants,but their grain lengths significantly increased, increasing their LWRs.The increase in grain length in the ΔDA1-like lines was greater than that in ΔUIM lines.The grain length of ΔDA1-like lines was similar to that of the OsDA1R310Klines.This observation suggested that the DA1-like domain participates in regulating two opposing functions:increasing grain width and reducing grain length.The R310 residue appeared to be necessary for the DA1-like domain function of regulation of grain length (Fig.6; Table S3).
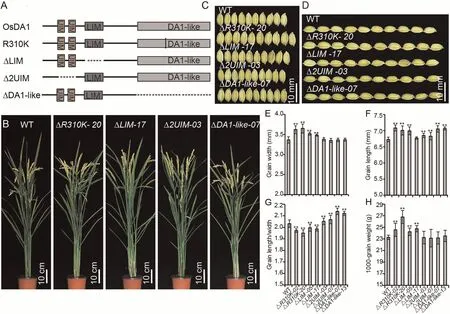
Fig.6.Phenotype analysis of OsDA1’s deletion derivatives.(A) Construction of OsDA1’s deletions.(B) Wild-type and transgenic plants grown under natural conditions in a Beijing field after grain filling.(C) The grain length phenotype in wild-type and transgenic plants.(D) The grain width phenotype of wild-type and transgenic plants.(E–H)Comparisons of grain widths (E), grain lengths (F), grain length/width ratios (G), and 1000-grain weight (H) between WT and transgenic plants.Values are mean ± SD.Student’s t-test was used for comparisons (n = 10 spikelets; **, P < 0.01).

Fig.7.Phenotype analysis of OsTCPs HTF-overexpressing plants.(A–C) Boxplot showing the phenotypic distribution of grain width (A), grain length (B), and grain length/width ratio(C)of OsPCF1V vs.OsPCF1E,OsTCP18V vs.OsTCP18E,OsPCF3V vs.OsPCF3E,and WT.Two-sample t-test P-value shows a significant difference.In boxplots,the bold line represents the median of the data.The full data are presented in Table S4.(D) The grain width phenotype of WT, OsPCF1E, OsTCP18E, OsPCF3E, OsPCF1V, OsTCP18V, andOsPCF3V.(E) The grain length phenotypes of WT, OsPCF1E, OsTCP18E, OsPCF3E, OsPCF1V, OsTCP18V, and OsPCF3V.
3.5.OsDA1 regulated rice grain length via the TCP gene family
Peng et al.[28] reported that DA1 interacts with transcription factors TCP14 and TCP15 to repress endoreduplication by regulating the expression of cell-cycle genes in Arabidopsis.DA1 modulates the stability of TCP14 and TCP15, thereby regulating endoreduplication.We accordingly aimed to investigate whether OsDA1 could interact with TCP homologs in rice.The rice genome contains 22 members of the TCP gene family, of which 17 were cloned to assess their interactions with OsDA1 using the yeast two-hybrid assay (Fig.S7).OsDA1 physically interacted with 14 TCP genes, albeit with slightly weaker interactions in six of them(Fig.S8A).For confirmation, three proteins (OsTCP2, OsTCP13,and OsPCF7) that strongly interacted with OsDA1 in yeast were selected for further validation in vivo by firefly luciferase complementation imaging (LCI) assays.The LCI assay results confirmed the physical interaction between OsDA1 and OsTCPs in N.benthamiana (Fig.S8B).
So when she had rubbed the sleep out of her eyes, and wept till she was weary, she set out on her way, and thus she walked for many and many a long day, until at last she came to a great mountain
In Arabidopsis,TCP14 and TCP15 act as transcriptional activators to increase the expression of RBR and CYCA2;3 while repressing endoreduplication, leading to a reduction in cell and organ size.To determine if similar mechanisms operate in rice, we searched their HTF transgenic library and identified three pairs of TCP hybrid transcription factors: OsPCF1V and OsPCF1E, OsTCP18V and OsTCP18E, and OsPCF3V and OsPCF3E.Each HTF harbored several independent transgenic events.All the transgenic events of TCP fusing with EAR showed an increasing trend in both grain length and width,while the TCP fusing without VP64 showed the opposite trend (Figs.7, S9; Table S4).Fusing with the EAR domain may reverse TCP gene transcription activity, leading to a phenotype similar to that of a mutant, while fusing with the VP64 domain may enhance their roles as activators.The expression levels of RBR and CYCA2;3 were also measured in OsDA1V, OsDA1E, PCF1-V,PCF1-E, PCF3-V and PCF3-E plants (Fig.S10).CYCA2;3 was significantly upregulated in OsDA1E, PCF1V, and PCF3V, while RBR was significantly upregulated only in OsDA1E and PCF1V.These findings provide further evidence of the involvement of osDA1 in regulating grain size via modulation of TCP-family gene expression.
4.Discussion
In Arabidopsis, DA1 encodes a ubiquitin receptor that acts as a negative regulator in the control of organ size [24,25].The da1-1 mutant, which is caused by an arginine-to-lysine substitution at position 358 in the DA1-like domain, produced large organs,including seeds, relative to wild-type plants, whereas neither TDNA insertion into Arabidopsis DA1 or its overexpression in plants led to markedly different seed and organ size phenotypes [23,24].In contrast to DA1,OsDA1 acts as a positive regulator to adjust grain shape in rice.OsDA1 is highly expressed in inflorescences, and the encoded protein was uniformly distributed in the nucleus and cytoplasm and on the plasma membrane.Overexpression of OsDA1 resulted in increased grain width and heavier grain,whereas RNAi and Osda1 mutant plants showed a slight decrease in grain width and 1000-grain weight (Figs.2, S2).
The rice genome encodes four homologs of the Arabidopsis DA1 gene, and OsDA1 has been identified as the most similar to Arabidopsis DA1.Of the identified homologs, the first reported was HDR3, which acts as a positive regulator of grain size.Overexpression of HDR3 resulted in an increase in grain size in both width(5%)and length (10%), whereas knocking out its gene led to a decrease in these parameters (width (-5%) and length (-10%)).We also found that OsDA1 is a positive regulator of rice grain width.Overexpression of the OsDA1 gene leads to increased grain width and weight.However,unlike HDR3,the OsDA1 gene has a reverse effect on grain length regulation, with OsDA1 mutants showing a decrease in grain width and an increase in grain length.This finding suggests that OsDA1 and HDR3 have different functions in regulating grain size,with HDR3 exerting a greater influence on grain length and OsDA1 affecting grain width.Overexpression of LOC_Os03g42820 and LOC_Os12g40490, the other two homologs of DA1, also resulted in increased grain width and length(Figs.S5,S6).However,there were phenotypic differences between these homologs.LOC_Os03g42820 and LOC_Os12g40490 showed equal increases in grain length and width while maintaining LWR.These findings suggest that the homologs of DA1 in rice are differentiated in their function of grain regulation, and OsDA1 may be involved more in the regulation of LWR.
Like DA1 in Arabidopsis, OsDA1 contains three types of conserved domains: two UIMs, a LIM, and a DA1-like (Fig.4A).The UIM domain is a ubiquitin-binding domain, the LIM domain is a DNA-binding domain, and the DA1-like domain is a peptidase domain.The da1-1 mutation,caused by an arginine-to-lysine substitution at position 358 in the DA1-like domain,resulted in larger organs, including seeds, than those of wild-type plants [23–25].Similarly, overexpression of OsDA1R310K, which corresponds to position 358 of the DA1-like domain in Arabidopsis DA1, increased both grain width and length.Overexpression of the truncation mutants of OsDA1 revealed that removing the DA1-like and UIM domains resulted in a loss of grain width regulation while increasing grain length.The effect of removing DA1-like domains was stronger than that of the UIM domain.In contrast, removing the LIM domain retained the regulation of grain width, although the function was weakened.These findings suggest that the DA1-like and UIM domains are necessary for regulating grain width but inhibit the regulation of grain length.Mutations of the R310K amino acid residue retained the positive effect on grain width while losing the inhibitory effect of the DA1-like domain on grain length and caused the increase of both grain width and length,suggesting that the mutation of the R310K amino acid residue is a promising target for improving rice grain weight and shape.
OsDA1 regulated rice grain length via the TCP gene family.In Arabidopsis, DA1 functions in the regulation of cell proliferation and the transition to differentiation by destabilizing regulatory proteins.Recent experimental findings [26] have added to this understanding by showing that DA1 functions as a peptidase,being activated by ubiquitylation caused by two other proteins,DA2 and BB.Once activated, DA1 cleaves DA2 and BB, leading to decreased stability.Beyond its interactions with DA2 and BB, activated DA1 can cleave and destabilize a range of growth-regulatory proteins,including UBP15, the E3 ligase PROTEOLYSIS 1, and the transcription factors TCP15 and TCP22.Gao et al.[28] showed that HDR3 has ubiquitin-binding activity, which promotes the ubiquitylation and stability of GW6a via a UIM-dependent pathway.The stabilizing effect of HDR3 on GW6a subsequently leads to an increase in acetyltransferase activity and expression of downstream target genes.Our study found OsDA1 to interact with 14 of the 17 TCP genes detected, and the functional analysis of three among the interacting TCP genes confirmed their ability to regulate grain size.The increase in grain width observed in OsDA1-overexpressing plants was due to cell enlargement,whereas HDR3 overexpression resulted in cell number increase, implying that OsDA1 and HDR3 regulate grain size via distinct pathways.
CRediT authorship contribution statement
Cong Li:Conceptualization, Investigation, Writing – original draft, Writing – review & editing.Jun Liu:Conceptualization,Investigation, Writing – original draft, Writing – review & editing.Liya Zhang:Conceptualization, Investigation, Writing – original draft,Writing–review&editing.Tao Li:Conceptualization,Investigation,Writing–original draft,Writing–review&editing.Hongyu Li:Supervision,Investigation,Resources.Bin Liu:Supervision,Investigation, Resources.Tao Zhao:Conceptualization, Supervision,Funding acquisition,Project administration,Writing–review& editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported in part by the National Transgenic Science and Technology Program (2016ZX08010-002), National Natural Science Foundation of China(157101834),and Agricultural Science and Technology Innovation Program of CAAS.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.10.012.
- The Crop Journal的其它文章
- Corrigendum to ‘‘GmTOC1b negatively regulates resistance to Soybean mosaic virus”.[Crop J.11 (2023) 1762–1773]
- Decoding the inconsistency of six cropland maps in China
- Wetting alternating with partial drying during grain filling increases lysine biosynthesis in inferior rice grain
- A polygalacturonase gene OsPG1 modulates water homeostasis in rice
- The ABA synthesis enzyme allele OsNCED2T promotes dryland adaptation in upland rice
- Trehalose: A sugar molecule involved in temperature stress management in plants

