A single-cell landscape of triptolide-associated testicular toxicity in mice
Wei Zhng , Siyu Xi , Jinhun Ou , Min Co , Gungqing Cheng , Zhijie Li ,Jigng Wng ,,**, Chunin Yng ,*
a Department of Nephrology,Shenzhen Key Laboratory of Kidney Diseases,Shenzhen Clinical Research Centre for Geriatrics,Shenzhen People's Hospital(The Second Clinical Medical College, Jinan University, The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, Guangdong,518020, China
b State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs,Artemisinin Research Center,Institute of Chinese Materia Medica,China Academy of Chinese Medical Sciences, Beijing,10700, China
Keywords:
Single-cell sequence
Transcriptomics
Triptolide
Reproduction toxicity
Testis
A B S T R A C T
Triptolide is a key active component of the widely used traditional Chinese herb medicine Tripterygium wilfordii Hook.F.Although triptolide exerts multiple biological activities and shows promising efficacy in treating inflammatory-related diseases,its well-known safety issues,especially reproductive toxicity has aroused concerns. However, a comprehensive dissection of triptolide-associated testicular toxicity at single cell resolution is still lacking. Here, we observed testicular toxicity after 14 days of triptolide exposure, and then constructed a single-cell transcriptome map of 59,127 cells in mouse testes upon triptolide-treatment. We identified triptolide-associated shared and cell-type specific differentially expressed genes,enriched pathways,and ligand-receptor pairs in different cell types of mouse testes.In addition to the loss of germ cells, our results revealed increased macrophages and the inflammatory response in triptolide-treated mouse testes, suggesting a critical role of inflammation in triptolideinduced testicular injury. We also found increased reactive oxygen species (ROS) signaling and downregulated pathways associated with spermatid development in somatic cells, especially Leydig and Sertoli cells, in triptolide-treated mice, indicating that dysregulation of these signaling pathways may contribute to triptolide-induced testicular toxicity. Overall, our high-resolution single-cell landscape offers comprehensive information regarding triptolide-associated gene expression profiles in major cell types of mouse testes at single cell resolution, providing an invaluable resource for understanding the underlying mechanism of triptolide-associated testicular injury and additional discoveries of therapeutic targets of triptolide-induced male reproductive toxicity.
1. Introduction
Increasing infertility and subsequent declining newborn babies are worldwide issues. It has been reported that infertility affects more than 10% of reproductive-aged couples, among which males account for 20%-30% [1,2]. Increasing evidence shows that the declined sperm quality of men is increased in the last two decades,especially in industrialized countries and areas [2]. Aside from genetic factors, most of the male infertility in humans is due to exposure to exogenous drugs and chemicals such as chemotherapies and endocrine disruptor chemicals [3,4]. Notably, several components in herbal medicines such as aristolochic acid I are also associated with male infertility [5].
Tripterygium wilfordii Hook. F. has been one of the most used herbal medicines for centuries in China and other Asian countries,including Japan and Korea. In China, it is currently marketed as Tripterygium wilfordii multiglycoside tablets and is widely used for treating autoimmune and inflammatory-related diseases including rheumatoid arthritis and systemic lupus erythematosus [6]. As a main active ingredient of Tripterygium wilfordii Hook. F., and Tripterygium wilfordii multiglycoside tablet [7], a diterpene triptolide has numerous biological functions including anti-inflammation,inflammation modulation,and anti-cancer effects [8]. For instance,triptolide is a promising drug candidate for treating colon cancer[9]and reverses epithelial-mesenchymal transition in glioma cells[10].Although it has notable efficacy in treating inflammatory-related disease, its side effects, especially testicular toxicity, hinders its clinical use [8,11]. Although several mechanisms such as the modulation of oxidative stress-related signaling were proposed for triptolide-induced testicular injury[12],its molecular mechanisms,especially at the single-cell level are unclear.
Mammalian spermatogenesis is a conserved, complex, and dynamic process of cell proliferation from the differentiation of spermatogonia into mature spermatocytes involving mitosis,meiosis, and spermiogenesis events. Previous studies regarding triptolide-associated testicular injury have mainly used bulk RNAseq to characterize the expression of genes in the whole testis.The changes in different cell types in response to triptolide in mouse testes, a complex organ, may occur by varied mechanisms.Thus,a holistic study of how triptolide affects the cell-type-specific response in the testis is helpful for better understanding its mechanisms. Advanced single-cell RNA sequencing (ScRNA-seq)comprehensively examines the transcriptional information of individual cells by using next-generation sequencing technologies,allowing for dissection of the cellular function of individual cells at an unprecedented high resolution. Recently, scRNA-seq has been used by multiple studies including our previous work to dissect testes pathology[13-15],which allows the simultaneous detection of gene expression changes in different cell types of the entire testis, faithfully representing the changes in gene expression of single cells in different physiological and pathological conditions.
Herein, male mice were used as a model to investigate triptolide-induced male reproductive toxicity,and deep ScRNA-seq was conducted in testis tissues, aiming to fully dissect the male reproductive toxicity of triptolide exposure at the single-cell level.Our findings are expected to provide novel information for understanding triptolide-induced testicular injury.
2. Methods
2.1. Animals and triptolide treatment
C57BL/6 mice (aged at 6-8 weeks) were maintained under a light-dark cycle of 12 h with water and food. For drug treatment,mice were given triptolide(dissolved in corn oil)via intraperitoneal injection for two weeks at a concentration of 200 μg/kg bw/d[16].This study was performed in strict accordance with the Guidelines of Shenzhen People's Hospital' Animal Care and Use Committee(Approval number:AUP-220801-YCB-0499-01).
2.2. Hematoxylin and eosin(H&E)and Masson's trichrome staining
After fixation,testicular samples from Ctrl and triptolide-treated mice were fixed with paraformaldehyde, followed by embedded with paraffin, and then serially sliced into 4 μm sections, followed by H&E staining or Masson's trichrome staining according to previous standard protocols[15,17-19].Images were acquired by using a microscope.
2.3. Single-cell suspension preparation
The preparation of single-cell suspension was followed by our previous protocols [15,20]. Briefly, testes tissues from the Ctrl and triptolide-treated mice were digested with collagenase IA and deoxyribonuclease I (Thermo Fisher Scientific Inc., Waltham, MA,USA),and then by trypsinization.Next,cells suspension was filtered by using a cell strainer (40 μm), washed, centrifuged, and finally resuspended in magnetic-activated cell sorting buffer (Miltenyi Biotec, Bergisch Gladbach, North Rhine-Westphalia, Germany) for ScRNA-Seq analysis.
2.4. Construction of single-cell libraries and sequencing
In brief, single cells suspensions and beads were mixed with each other by adjusting co-encapsulation occupancy to 0.05. After collection of individual droplets, messenger RNA was reverse transcribed into complementary DNA,followed by amplification of cDNA. Finally, a 3?gene expression library was prepared with the Chromium Next GEM Single Cell 3’ Kit v3.1 (10× Genomics, Pleasanton, CA, USA). Sequencing was conducted by using a NovaSeq 6,000(Illumina,San Diego,CA,USA)by Novogene(Tianjin,China).
2.5. ScRNA-seq data quality control analysis
Raw sequencing reads of mice testes were aligned to the mouse genome reference (GENCODE, mm10) and calculated to single-cell matrix using Cell Ranger (version 6.1.1) with the default parameters. In total, we captured 75,061 cells without any filtration. To filter low quality cells and doublets, genes fewer than 300 or mitochondrial ratio more than 20% in each cell were removed by using single-cell matrix of each sample [21]. Doublets were discarded using DoubletFinder (version 2.0.2) with the default parameters [22].59,127 cells were finally identified after filtration in the dataset.After that the data was normalized,dimensionality was reduced, and cell cluster was performed using Seurat. Then, the clusters with relatively low gene numbers and absence of specific marker genes were also removed.
2.6. Integration and cell type annotation
For each cell, the gene counts matrix of each sample was normalized with the “SCTransform” function implemented in Seurat. To integrate each sample for correction of batch effect, top 2000 variable genes were selected using ‘FindVariableFeatures’ by Seurat,which directly modeled the mean-variance relationship.We found top 2,000 variable genes have a strong enrichment of features to compute the representations. So top 2,000 variable genes were chosen and projected into a low-dimensional subspace by using canonical correlation analysis among different samples.Next,features and anchors for downstream data integration were chosen based on function of “Find Integration Anchors” and “Integrate Data” in Seurat. Following data integration and scaling, “RunPCA” function of principal component analysis(PCA)was conducted,and we identified 40 significant components that have a strong enrichment of low P-value features to compute the representations using the function ‘JackStrawPlot’ implemented in Seurat. At last, “Find-Neighbors” and “FindClusters” function implemented in Seurat was used for dataset cluster. “RunUMAP” function implemented in Seurat was used for dimensionality reduction. Cell types were allocated to each cluster using the abundance of known marker genes as described previously[23].
2.7. Differentially expressed genes (DEGs) analysis
DEGs in each cell types of Ctrl and triptolide-treated sample were analyzed by using “FindMarkers” implemented in Seurat using Wilcoxon Rank Sum test.Genes with adjusted P values < 0.05,|LogFC| >0.25 were regarded to be triptolide-associated DEGs.
2.8. Gene ontology (GO) enrichment analysis
GO enrichment analysis was performed with R packages clusterProfiler [24]. Representative pathways were selected based on the cutoff of adjusted P values < 0.05 and displayed by using ggplot2 R package(version 3.3.2).
2.9. Transcription factor regulatory network analysi s
Briefly, single-cell regulatory network inference and clustering(SCENIC) (version 1.1.2.2) software pipeline with default parameters was used for transcriptional regulatory network analysis [25].The referenced transcription factors (TFs) were obtained from R packages RcisTarget (version 1.6.0) with mm10 [26]. To infer and construct the triptolide-related transcriptional regulatory network,sets of associated DEGs that co-expressed with transcription factors were identified using R packages GENIE3 in R3.6.Next,to filter false positives and indirect targets, RcisTarget was used to identify putative direct-binding targets by cis-regulatory motif analyses and only TFs with significant motif enrichment of the correct upstream regulator were retained. At last, TFs with significant motif enrichment and target genes with high-confidence annotation were used for subsequent downstream data analysis. The transcriptional regulatory network was visualized by using cytoscape (version 3.8.1) software[27].
2.10. Gene set score analysis
Publicly available gene sets were obtained from Gene Ontology Resources and Molecular Signatures Database (MSigDB) [28]. The function “AddModuleScore” implemented in Seurat was used for gene sets scoring for each input cells. R package via the Wilcoxon test was used to calculate the significant difference between Ctrl and triptolide-treated mice.
2.11. Cell-cell communication inference
CellChat (version 1.1.3) [29] was used to dissect intercellular communications via ligand-receptor interactions networks by combining a curated ligand-receptor database based on gene expression data, social network analysis, manifold learning, and pattern recognition. Communication probability was modeled between cell types and representative ligand-receptor pairs were chosen with the cutoff of P value < 0.01.
2.12. Real-time polymerase chain reaction (qPCR) and western blotting analysis
qPCR and western blotting were conducted according to previous protocols [30-32], and primers for PCR are listed in Table S1.
2.13. Statistical analyses
Data analysis were calculated with GraphPad 8 Software. Significance differences between different samples was revealed with Student's t-test, or Wilcoxon Rank Sum test. P values for bioinformatic analyses were shown in indicated figures.
2.14. Data availability
The sequencing data reported here was deposited in the OMIX,China National Center for Bioinformation/Beijing Institute of Genomics(https://ngdc.cncb.ac.cn/omix:accession no.OMIX002591)with public available.
3. Results
3.1. Triptolide-induces testicular toxicity
To examine the potential testicular toxicity of triptolide, mice were exposure to triptolide for 2 weeks at a concentration of 0.2 mg/kg bw/d by intraperitoneal injection according to a previous study [16]. Triptolide treatment reduced testis size and the testicular index (Figs.1A and B, and S1A), suggesting that triptolide induces testicular injury. Further analysis showed that triptolide treatment significantly reduced sperm content, and increased abnormal morphology of sperms(Figs.1C and D).Triptolide-treated mice also showed reduced levels of luteinizing hormone (LH)(Fig.1E), a critical hormone for spermatogenesis. Histological images further showed that triptolide treatment disrupted normal testicular morphology, such as disordered seminiferous tubules and reduced seminiferous tubule diameters (Fig.1F). Masson's trichrome staining further revealed that triptolide-treated mice showed impairment of spermatogenesis and accumulation of collagen around seminiferous tubules within testes(Fig.1G).These results suggest that triptolide induces testicular toxicity, which is consistent with previous findings [11,16].
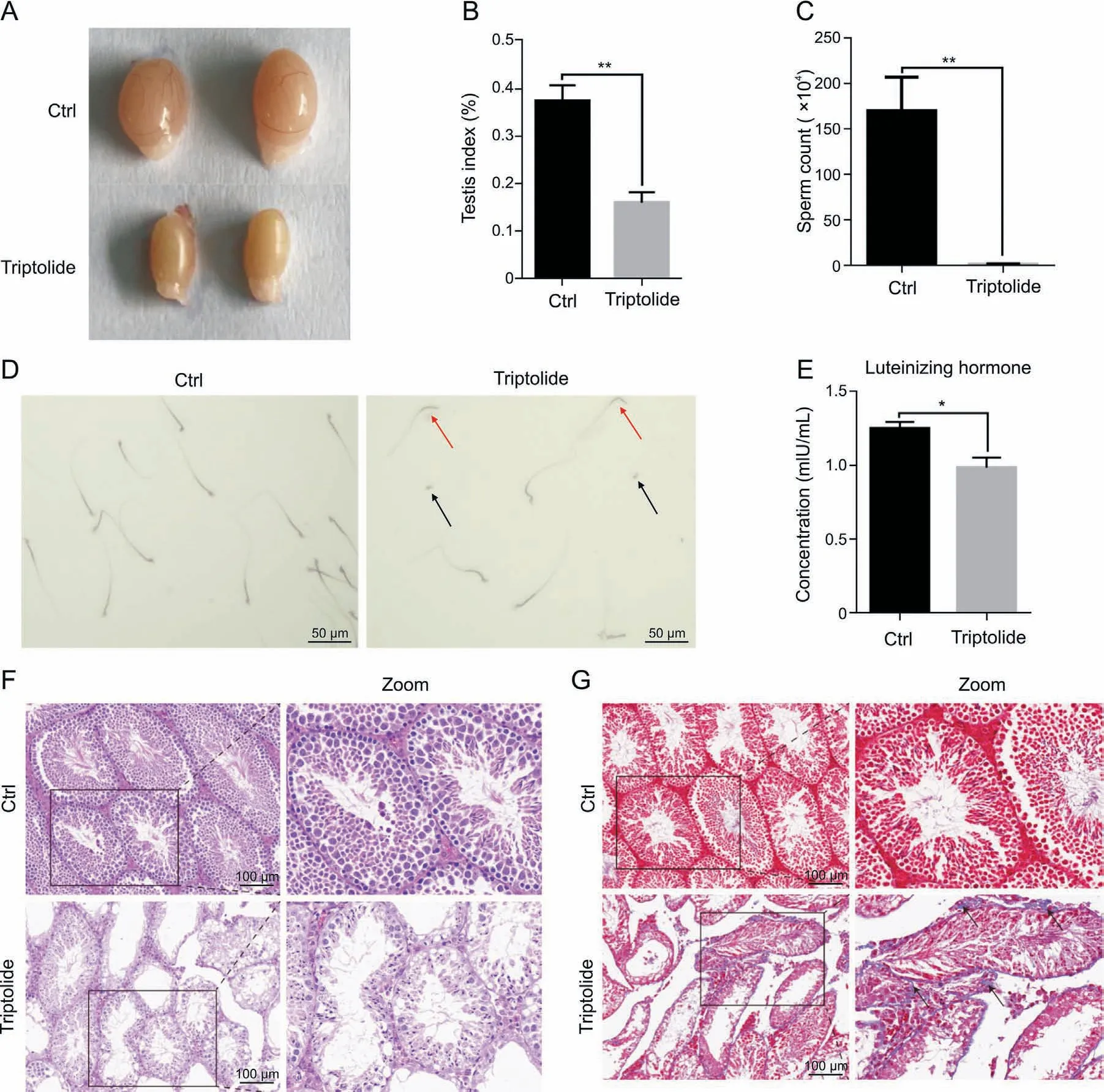
Fig.1. Triptolide induces testicular injury in mice. (A) Representative images showing triptolide-treated mice with reduced testes size. (B) Bar plot showing reduced testes index(testes weight/body weight)in triptolide-treated mice.(C)Bar plot showing triptolide-reduced sperm contents in testes.(D)Representative images showing triptolide-treated mice with abnormal sperm morphology. Arrows indicate abnormal sperms. (E) Bar plot showing triptolide-treated mice with lower luteinizing hormone contents. (F) Representative hematoxylin and eosin (H&E) images showing abnormal testicular morphology in triptolide-treated mice. (G) Representative Masson's trichrome staining results showing the accumulation of extracellular matrix in interstitial testicular tissues after triptolide treatment. Arrows indicate positive cells. Ctrl: control. *P < 0.05, **P < 0.001, compared with vehicle control.
3.2. ScRNA-seq reveals different testicular cell types and triptolideassociated changes in cell composition
To better understand triptolide-induced testicular injury,ScRNA-seq was performed in testicular tissues of mice upon triptolide treatment. After quality control, a pool of 59,127 qualified cells in vehicle and triptolide-treated mice (three mice per group)were used for further analysis and visualized using the uniform manifold approximation and projection (UMAP) algorithm according to the expression levels of canonical marker genes in each cell type according to a previous report [23] (Figs. 2A and B). We identified 9 cell types that represent major cell types within the testes,which include spermatogonia(SPG),spermatocytes(Scytes),round spermatids (STids), elongating spermatids (STids), macrophage, Sertoli, Telocytes, innate lymph (a type II innate lymphoid cell population [33]) and Leydig cells. Detailed marker genes in each cell type were also listed(Fig.2C).According to the expression of cell-type specific top genes, GO enrichment analysis showed their physiological functions (Fig. 2D). Finally, we analyzed and compared relative cell numbers in control and triptolide-treated testis samples. As shown in Figs. 2E and S1B, triptolide treatment dramatically reduced round STids, elongating STids, and Scytes,three major cell types that are critical for forming mature sperm.Consistent with this result,triptolide-treated mice showed reduced sperm contents (Fig.1D). Collectively, we established a single-cell transcriptome map in the testis upon triptolide treatment.
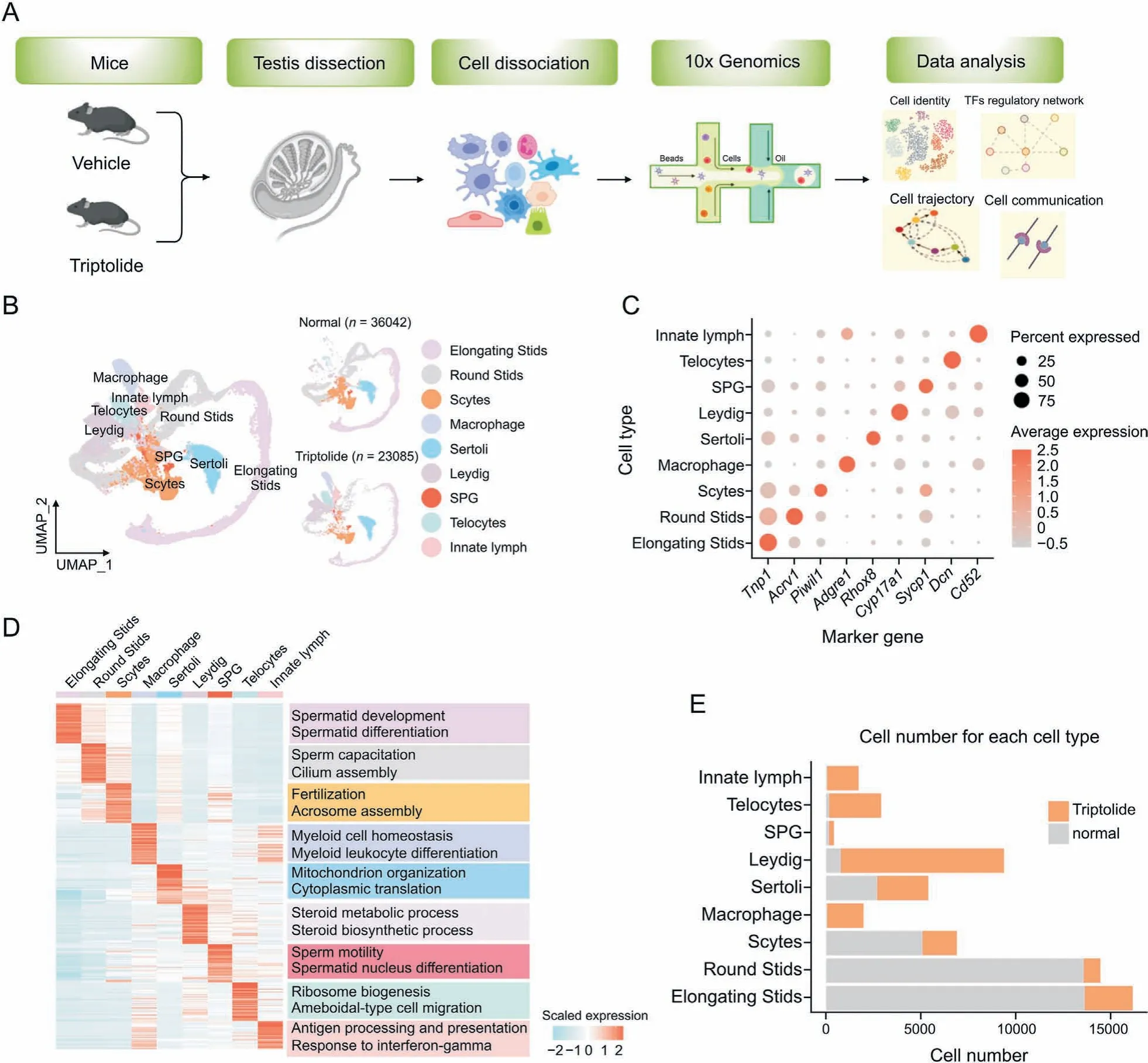
Fig.2. Construction of a single-cell transcriptome map in response to triptolide in mice.(A)Scheme model showing the overall experiment design in the current study.(B)Uniform manifold approximation and projection(UMAP)plot showing the nine cell types of mouse testis in control(Ctrl)(36,042 cells)and triptolide-treated mice(23,085 cells).Cell types are annotated with indicated colors. (C) Dot plot indicating the levels of representative marker genes in indicated cell type. (D) Heatmap indicating gene expression signatures in each cell type. Enriched biological processes in indicated cell types are shown on the right panel. (E) Histogram map indicating the number of indicated cell types present in the normal and triptolide-treated mice used for data analysis.TFs:transcription factors;SPG:spermatogonia;Scytes:spermatocytes;round Stids:round spermatids;elongating Stids:elongating spermatids.
3.3. Triptolide-associated transcriptional changes in testicular germ cells
Based on the above single-cell results, we next identified DEGs in each germ cell type of the testes in response to triptolide treatment. Our results showed that most DEGs upon triptolide treatment in different cell types were highly heterogeneous, and there were also multiple shared DEGs in different germ cell types(Fig. 3A). Among the 4 germ cell types, round STids showed the greatest numbers of DEGs upon triptolide treatment (Fig. 3A). GO enrichment analysis revealed that the top commonly upregulated pathways in different germ cells upon triptolide treatment included ATP metabolic process, glucocorticoid metabolic process,steroid metabolic process,cell death in response to oxidative stress,and cellular response to oxidative stress (Fig. 3B), suggesting that triptolide treatment disrupted steroid metabolism and induced germ cell apoptosis. Interestingly, GO analysis revealed that the shared top-downregulated biological processes in germ cells upon triptolide treatment were associated with spermatid development,spermatid differentiation, sperm motility, germ cell development,cilium organization, and acrosome assembly (Fig. 3C). All these pathways are critical for spermatogenesis. These results may suggest that triptolide not only induces germ cell death but also inhibits spermatogenesis.
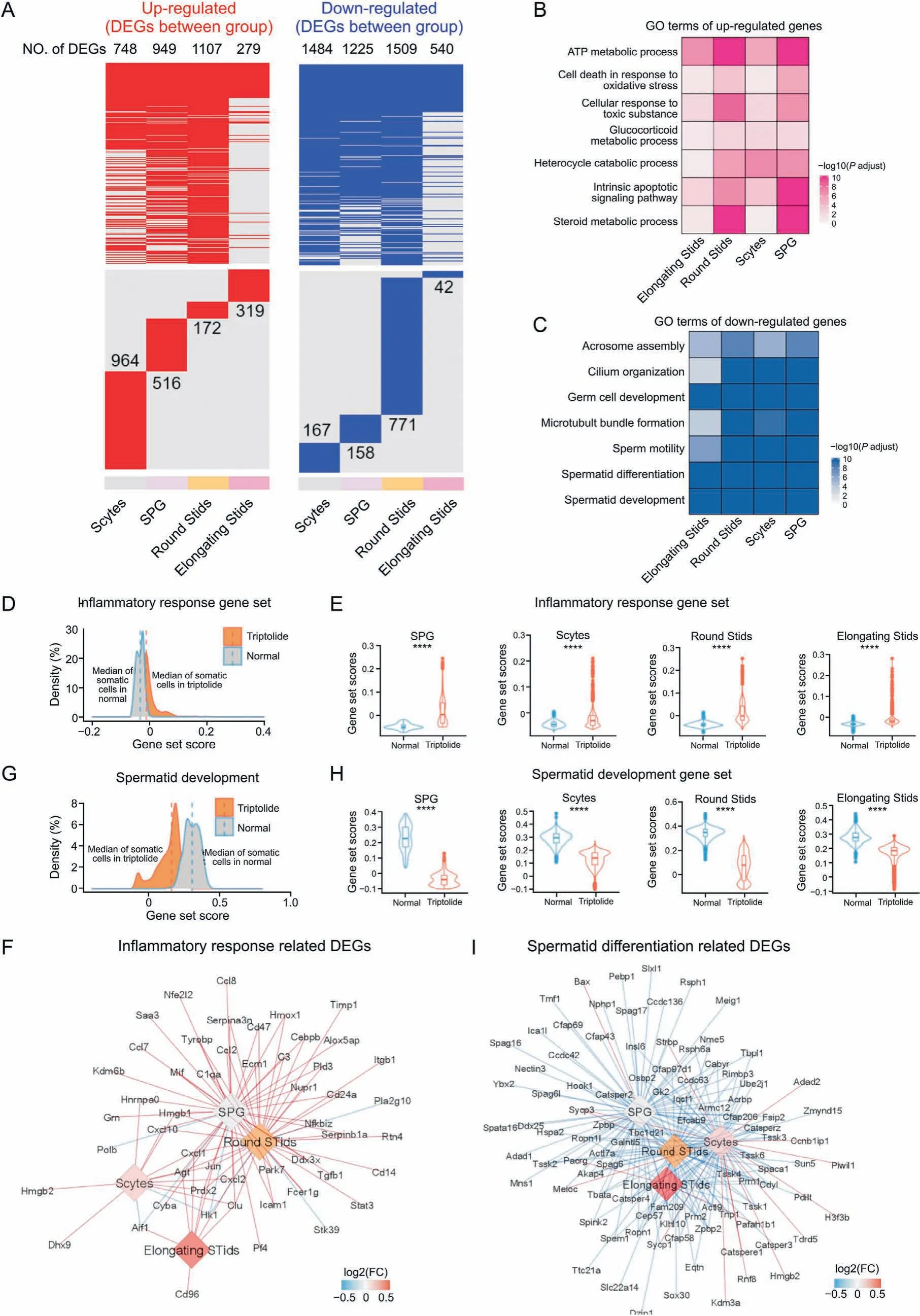
Fig. 3. Triptolide-associated changes in germ cells in mice testes. (A) Heatmaps revealing the distribution of triptolide-associated up-regulated (left, red) and down-regulated differentially expressed genes (DEGs) (right, blue) in indicated cell types of mice testes. The numbers of DEGs in each cell types are listed. The upper panel shows the shared DEGs in different cell types, the lower panel indicates the cell-type specific DEGs.(B,C)Representative shared gene ontology(GO) terms for up-regulated (B)and down-regulated(C)DEGs in indicated cell types.The color keys from white to red(B),or from white to blue(C),represent the range of P-adjust value.(D)Density plot indicating elevated gene set scores of pathways associated with inflammatory response in germ cells. (E) Gene set score results showing biological processes related to inflammatory response in SPG, Scytes,round Stids, and elongating Stids in normal and triptolide group. ****P < 0.0001 (two-sided Wilcoxon rank-sum tests). (F) Network plot indicating the DEGs associated with inflammatory response in indicated cell types. The color keys from blue to red, show the range of fold change (FC) value. (G) Density plot indicating elevated gene set scores of pathways for spermatid development in sperm cells. (H) Gene set score analysis of pathways for spermatid development in indicated cell types between normal and triptolide group. ****P<0.0001(two-sided Wilcoxon rank-sum tests).(I)Network plot indicating DEGs that relevant to spermatid differentiation in indicated cell types. The color keys from blue to red, indicate the range of fold change (FC) value. SPG: spermatogonia; Scytes: spermatocytes; round Stids: round spermatids; elongating Stids: elongating spermatids.
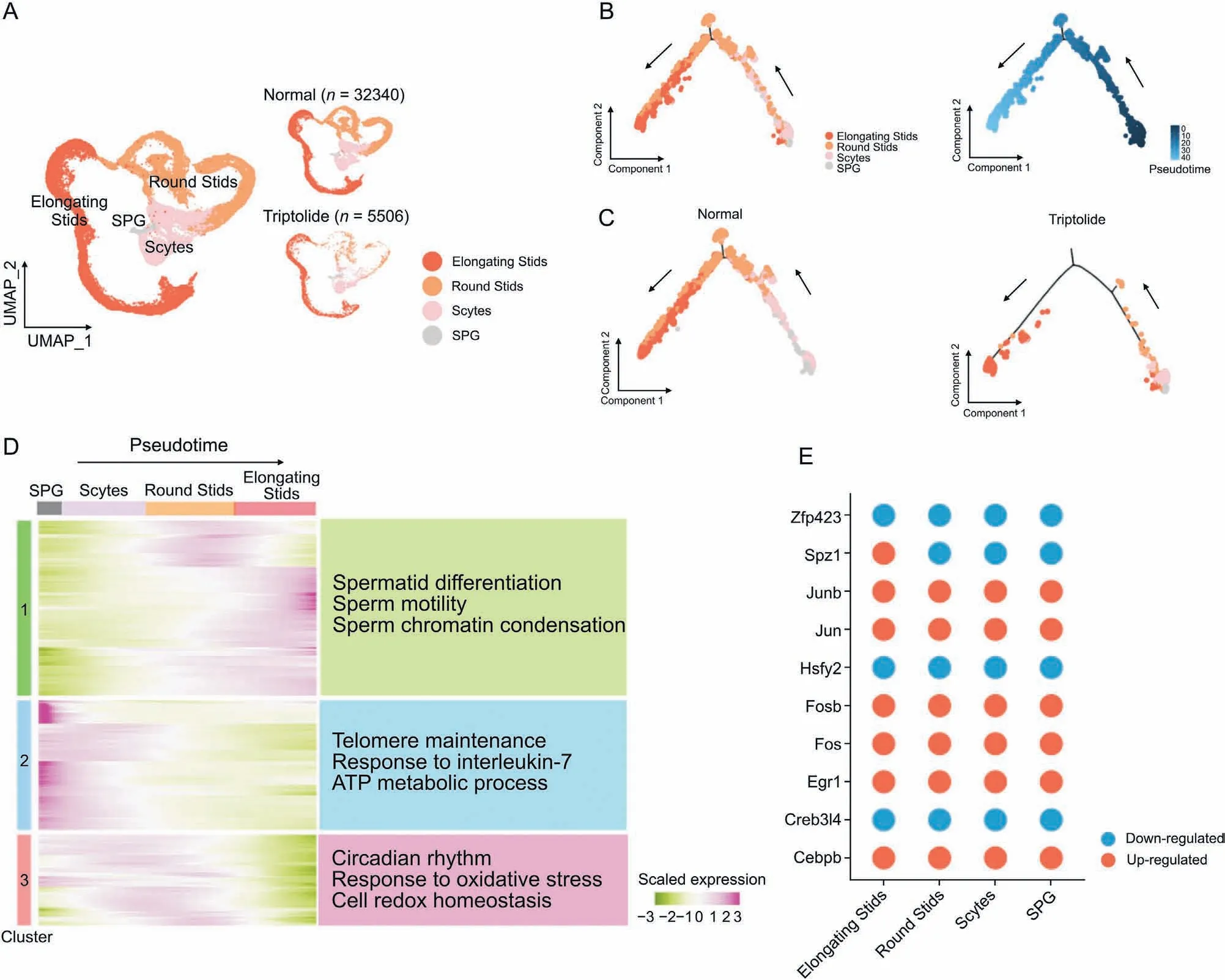
Fig.4. Triptolide-associated changes along the trajectory of germ cell differentiation.(A)Uniform manifold approximation and projection(UMAP)plot indicating the four sperm cell types of mouse testis sequentially and stepwisely developed.(B)UMAP(left)and pseudotime trajectories(right)analysis of indicated germ cell types.Cells were ordered from the beginning(darkest blue color)to the end(lightest blue)as shown with the arrow.(C)Pseudotime trajectories analysis of indicated four germ cell types in normal(left)and triptolide(right) treatment mice. (D) Heatmap showing the gene expression profiles of germ cells along the pseudotime trajectories. Representative enriched biological processes along the pseudotime trajectories are shown on the right. (E) Dot plots showing shared up-regulated (red) and down-regulated (blue) transcription factors (TFs) along the pseudotime trajectories under triptolide treatment conditions shared in different germ cells. SPG: spermatogonia; Scytes: spermatocytes; round Stids: round spermatids; elongating Stids:elongating spermatids.
To further characterize DEGs that are associated with triptolideassociated germ cell injury,we then analyzed the gene set scores of genes for the inflammatory response pathway.As shown in Figs.3D and E, triptolide treatment increased the overall gene set scores associated with the inflammatory response pathway in germ cells and in four germ cell types.Triptolide treatment increased multiple inflammation-related genes, especially in SPG and round STids including Cxcl10,Cxcl1,Ccl7,Ccl8 and Ccl2(Fig.3F),suggesting that a critical role of the inflammatory microenvironment may account for triptolide-induced male infertility. Moreover, triptolide treatment decreased the overall gene set scores of spermatid development signaling in germ cells,and in all four germ cell types(Figs.3G and H). Multiple key genes such as Cfap43, Cfap69, Cfap206, and Cfap58, which are critical for spermatid differentiation, were also decreased (Fig. 3I). Finally, we observed that triptolide treatment increased the overall gene set scores of reactive oxygen species(ROS)in germ cells,and in four germ cell subtypes(Figs.S1C and D).A variety of ROS-associated genes were also dramatically changed after triptolide treatment. For instance, Gpx4, a key gene for inhibiting ROS and ferroptosis, was decreased in triptolide-treated mice (Fig. S1E), suggesting that increased ROS signaling in germ cells may play a critical role in triptolide-induced testicular injury.Collectively, these results indicate that an increase in the inflammatory response, activation of ROS signaling, and an decrease in spermatid development may contribute to germ cell injury upon triptolide treatment.
3.4. Triptolide-associated gene-expression profiles of differentiation trajectories of spermatogenesis
To dissect the germline changes in response to triptolide, we focused on the analysis of four germ cell types that are critical for spermatogenesis.UMAP plot analysis showed that the four types of germ cells developed stepwisely and sequentially from SPG to elongating Stids (Figs. 4A and B). We then showed that the distribution of each germ cells in vehicle control and triptolide treated samples was distinct (Fig. 4A), further confirming that triptolide treatment inhibits spermatogenesis. Next, we established the lineage trajectory of four germ cells along the pseudotime and observed that triptolide-treated mice showed a reduction in the distribution of all four germ cells in the trajectory (Figs.4B and C).Concurrently, branched expression analysis modeling (BEAM)revealed a gene expression signature with respect to pseudotime and revealed three clusters(Clusters 1 to 3)associated with cell fate transition in response to triptolide-treatment (Fig. 4D). Cluster 1 showed downregulated genes along the trajectory,especially at the early stage of spermatogenesis.GO analysis showed that triptolideassociated downregulated genes in Cluster 1 were enriched in pathways for spermatid differentiation,sperm motility,and sperm chromatin condensation (Fig. 4D), suggesting that triptolide inhibits the transition of SPG to Scytes.In contrast to Cluster 1,Cluster 2 and Cluster 3 showed triptolide-upregulated genes,especially at stages of SPG to Scytes, and round Stids, which were enriched in pathways of response to interleukin-7,response to oxidative stress,and cell redox homeostasis (Fig. 4D). These results indicate thattriptolide treatment inhibits spermatid development, which is probably mediated by increased oxidative stress and inflammation,and reduces spermatid differentiation, sperm motility and sperm chromatin condensation. We also identified key transcription factors along the trajectory in different germ cells upon triptolide treatment, and found that upregulated TFs such as Junb and Jun were associated with inflammatory response, and downregulated TFs such as Zfp34 were associated with spermatid development regulation (Fig. 4E), further highlighting the critical roles of increased inflammatory response and spermatid differentiation in triptolide-induced impairment of spermatogenesis.
3.5. Triptolide-associated transcriptional changes in somatic cells of mouse testes
Apart from germ cells, triptolide treatment also induced heterogeneous changes in the expression of genes in different somatic cells including macrophages, Sertoli cells, Leydig cells, innate lymph cells, and Telocytes (Fig. 5A). GO analysis indicated that commonly downregulated genes in different somatic cells of triptolide-treated mice were enriched in several critical pathways for spermatogenesis such as acrosomal assembly,binding of sperm to the zona pellucida, cilium movement, germ cell development,and sperm chromatin condensation(Fig.5B).This finding indicates that triptolide treatment may compromise the function of somatic cells in spermatogenesis. Moreover, GO analysis revealed that the commonly top-upregulated DEGs in the testes of somatic cells of triptolide-treated mice were enriched in several stress-related biological processes such as response to toxic substrate, cellular response to oxidative stress, and regulation of inflammatory response (Fig. 5C). In addition, the upregulated genes after triptolide treatment were also enriched in collagen metabolic process,and fibroblast proliferation, suggesting that triptolide treatment may also promote the formation of a fibrosis-like status in somatic cells, especially in Leydig, Telocytes, and Sertoli cells of the testes(Fig. 5C). Interestingly, gene set score analysis revealed that genes associated with fibrosis were increased in somatic cells of triptolide-treated mice (Figs. S2A and B), and multiple fibrosisrelated genes such as Acta2, Vim, Col8a1, and Timp1 were increased in Leydig and Sertoli cells (Fig. S2C). This result is consistent with Masson's trichrome staining results showing the accumulation of extracellular matrix in interstitial testicular tissues after triptolide treatment in mice (Fig.1G).
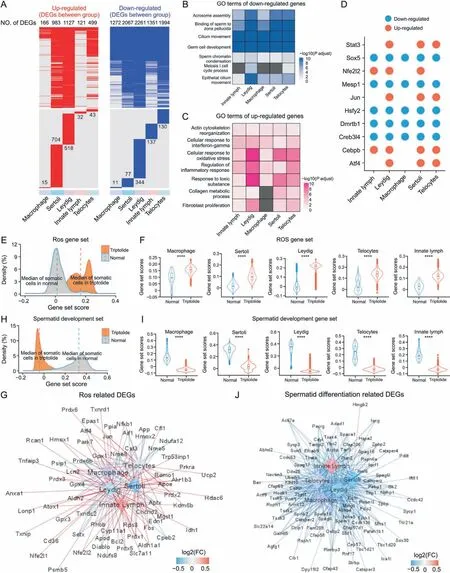
Fig. 5. Triptolide-associated changes in somatic cells in mice testes. (A) Heatmaps revealing the distribution of up-regulated (left, red) and down-regulated (right, blue) genes in indicated cell type after triptolide treatment. Genes that no significantly changed are shown in grey color. The upper reveals the shared differentially expressed genes (DEGs) by different cell types,the lower panel showing cell-type specific DEGs in indicated cell type.The numbers of DEGs are listed on the plots.(B,C)Representative shared gene ontology(GO)terms of down-regulated(B)and up-regulated(C)DEGs in indicated cell types.The color keys from white to blue(B),or from white to red(C)show the range of P-adjust value.(D)Dot plots revealing the shared up-regulated(red)and down-regulated(blue)transcription factors(TFs)in indicated cell types after triptolide treatment.(E)Density plot showing increased gene set scores of pathways related to reactive oxygen species (ROS) in somatic cells after triptolide treatment. (F) Gene set score analysis for pathways of ROS in macrophage,Sertoli,Leydig,Telocytes and innate lymph between normal and triptolide group. ****P<0.0001(two-sided Wilcoxon rank-sum tests).(G)Network plot revealing DEGs associated with ROS signal pathway in indicated somatic cell types after triptolide treatment.The color keys from blue to red indicate the range of fold change(FC)value.(H)Density plot demonstrating increased gene set scores of pathways for spermatid development in somatic cells after triptolide treatment. (I) Gene set score analysis of pathways for spermatid development in macrophage,Sertoli, Leydig,Telocytes and innate lymph between normal and triptolide group. ****P < 0.0001 (two-sided Wilcoxon rank-sum tests).(J)Network plot showing DEGs associated with spermatid differentiation signal pathway in indicated cell types after triptolide treatment.The color keys from blue to red indicate the range of FC value.
SCENIC analysis[31]was then used to reveal key shared or celltype-specific differentially expressed TFs in somatic cells that were involved in triptolide-induced testicular injury (Figs. 5D and S2D). We observed that triptolide treatment upregulated TFs such as Jun and Stat3, which were intimately associated with the inflammatory response. Triptolide treatment downregulated TFs such as Sox5, which are involved in spermatogenesis [34]. These results further highlight the crucial roles of inflammatory response and inhibition of spermatogenesis in triptolide-induced testicular injury.
We further showed that the gene set scores of ROS signaling in somatic cells were upregulated after triptolide treatment (Figs. 5E and F). Gene network analysis showed that multiple ROS-related genes such as Prdx1, Prdx2, Prdx6, Prdx4, and Txnrd1 were significantly increased after triptolide treatment(Figs.5E-G),suggesting that triptolide treatment may increase ROS signaling. In contrast,the gene set scores of spermatid differentiation were decreased after treatment, and a variety of genes associated with spermatid differentiation such as Sycp1,Sycp3,Cep57,Sox30,Zpbp2,and Cfap58 were significantly decreased (Figs. 5H-J). Moreover, the gene set scores of inflammatory responses were also increased in triptolidetreated somatic cells, and numerous inflammatory genes were upregulated (Fig. S3). To verify the scRNA-seq results, our qPCR results confirmed that triptolide inhibited the expression of genes required for spermatid development(Prm1,Spaca1,Catasperz),and antioxidant(Gpx4,Prdx6b,Txndc2)(Figs.S4A and B).In TM3 mouse Leydig cells, as shown in Figs. S4C and D, triptolide reduced cell viability,and induced apoptosis.These results highlight the critical role of several dysregulated critical signaling pathways, such as inhibition of germ cell development and increased ROS and inflammation in somatic cells,in triptolide-induced male infertility.
3.6. Characterization of triptolide-associated changes in Sertoli cells
Sertoli cells are called ‘nurse cells’ and are within the seminiferous tubules of testes, which promote the formation of spermatozoa via direct interacting with germ cells and modulation of the environmental milieu.The function of follicle-stimulating hormone(FSH)and testosterone in testes for spermatogenesis is through the action of Sertoli cells.To better characterize the roles of triptolideinduced testicular injury, we focused on analyzing Sertoli cells.According to the expression of the top cell-type specific genes,Sertoli cells were divided into three subtypes: Sdc4+Sertoli cells,Cypt4+Sertoli cells and Chchd10+Sertoli cells(Figs.6A and B).GO analysis showed that Cypt4+Sertoli cells play a key role in regulating cilium organization,microtubule-based movement,and ADP metabolic process (Fig. 6C). Sdc4+Sertoli cells mainly function to regulate hormone-related signaling pathway, hormone secretion,and hormone transport (Fig. 6C). Chchd10+Sertoli cells regulate protein polymerization(Fig.6C).Next,DEGs in different subtypes of Sertoli cells were identified (Fig. 6D). GO enrichment showed that common downregulated genes in different Sertoli cells were associated with cilium organization, and spermatid development(Figs. 6E-G). In contrast, GO enrichment showed that common upregulated genes were related to oxidative stress,RNA processing,or ATP metabolic process (Figs. 6E-G). Furthermore, key TFs that regulate the expression of DEGs in Sdc4+Sertoli cells were also identified (Fig. 6H). For instance, we showed that Creb3l4, a CREB/ATF family of TFs that is critical for spermatogenesis, was responsible for regulating multiple genes in Sdc4+Sertoli cells after triptolide treatment. These results indicate the dysregulation of the function of Sertoli cells, such as inhibition of spermatid differentiation and increased ATP metabolism in triptolide-induced testicular injury.
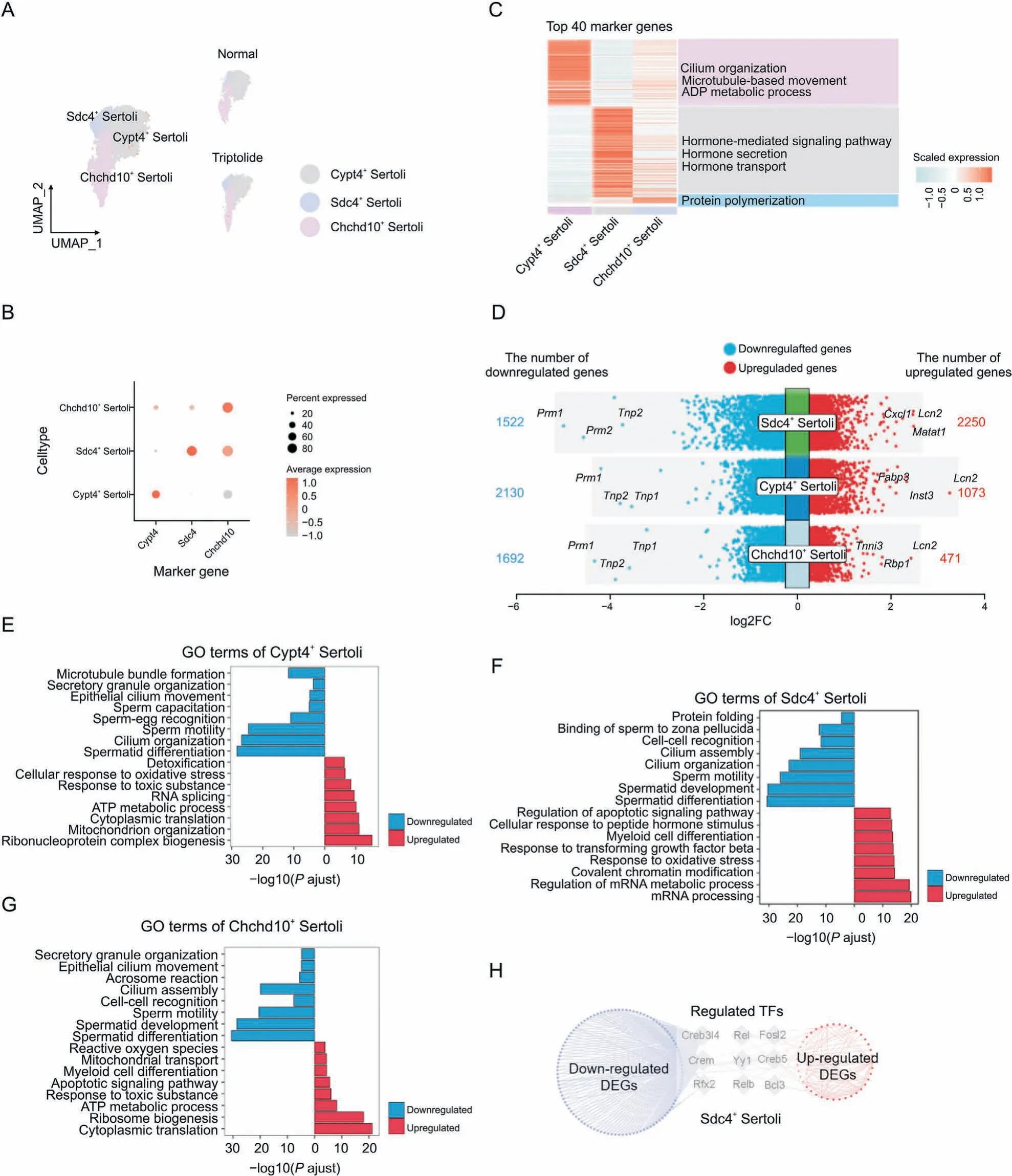
Fig. 6. Triptolide-associated changes in Sertoli cells. (A) Uniform manifold approximation and projection (UMAP) plot indicating the three Sertoli subtypes of mouse testes in normal and triptolide-treated mice. (B) Dot plot revealing the representative genes for indicated Sertoli subtypes. The size of dot indicates the percent expressed in each Sertoli subtypes.The color keys from grey to red indicate the range of scaled gene counts.(C)Heatmap showing the top 40 marker genes of each Sertoli subtypes,along with their enriched gene ontology (GO) terms (listed on the right). (D) Dot plot showing the number and logarithmic fold change (FC) (logFC) of regulated genes (dots) across three Sertoli subtypes.Genes in colored dots are significantly(FDR<0.05 and FC>25%)up-regulated or down-regulated after triptolide treatment,as determined by differentially expressed genes(DEGs)analysis (see details in Methods). (E-G) Bar plot showing enriched biological processes of the three Sertoli subtypes, Cypt4+ Sertoli cells (E), Sdc4+ Sertoli cells (F), and Chchd10+ Sertoli cells (G). Red: upregulation; blue: downregulation. (H) Network plot indicating the differentially expressed transcription factors (TFs) and their corresponding triptolide-associated DEGs (red dots show up-regulated genes and blue dots show down-regulated genes) in Sdc4+ Sertoli cells.
3.7. Triptolide-associated changes in testicular macrophage
Testicular macrophages normally exert immunosuppressive functions and play a crucial role in regulating testes homeostasis[35]. However, excessive activation of macrophages also has a detrimental effect on spermatogenesis. First, immunostaining was used to characterize triptolide-associated lymphocytes and macrophage infiltration, and we found that triptolide increased CD45 (lymphocytes marker) and F4/80 (macrophage marker)-positive cells (Figs. S5A and B). To characterize the function of macrophages in testes in response to triptolide treatment, we identified three subtypes of macrophages, named Lcn2+macrophages, Rcan1+macrophages, and Cd74+macrophages (Fig. 7A).We showed the distributions of these three subtypes of macrophages by using UMAP analysis in vehicle control and triptolidetreated mice, and found that triptolide-treatment significantly increased these three subtypes of macrophages (Fig. 7B). GO enrichment of the top marker genes showed that Lcn2+macrophages play a critical role in regulating oxidative stress response,and cholesterol homeostasis (Fig. 7C). GO enrichments analysis showed that Cd74+macrophages mainly function to antigen processing and presentation,regulation of lymphocyte differentiation,and monocyte chemotaxis processes (Fig. 7C). GO enrichment analysis showed that Rcan1+macrophages mainly function to regulate myeloid cell differentiation, cellular response to interferon-gamma, and cytokine biosynthesis processes (Fig. 7C).We further characterized triptolide-associated DEGs in different macrophage subtypes and found the top commonly downregulated genes in these three macrophage subtypes, including Prm1, a key gene for fertilization [36] (Fig. 7D). In contrast, the top commonly upregulated genes in different macrophage subtypes after triptolide treatment include Lcn2 and Cccl8, two genes that regulate the inflammatory response[37](Fig.7D).GO enrichment revealed that downregulated genes after triptolide treatment in macrophages were associated with sperm capacitation,binding of sperm to zona pellucida, epithelial cilium movement, and sperm chromatin condensation (Fig. 7E). GO enrichment revealed that upregulated genes after triptolide treatment in macrophages were associated with T cell-mediated cytotoxicity, iron ion homeostasis, and macrophage activation (Fig. 7E). These results suggest that apart from regulating the inflammatory response, macrophages also downregulate multiple genes associated with spermatogenesis after triptolide treatment.
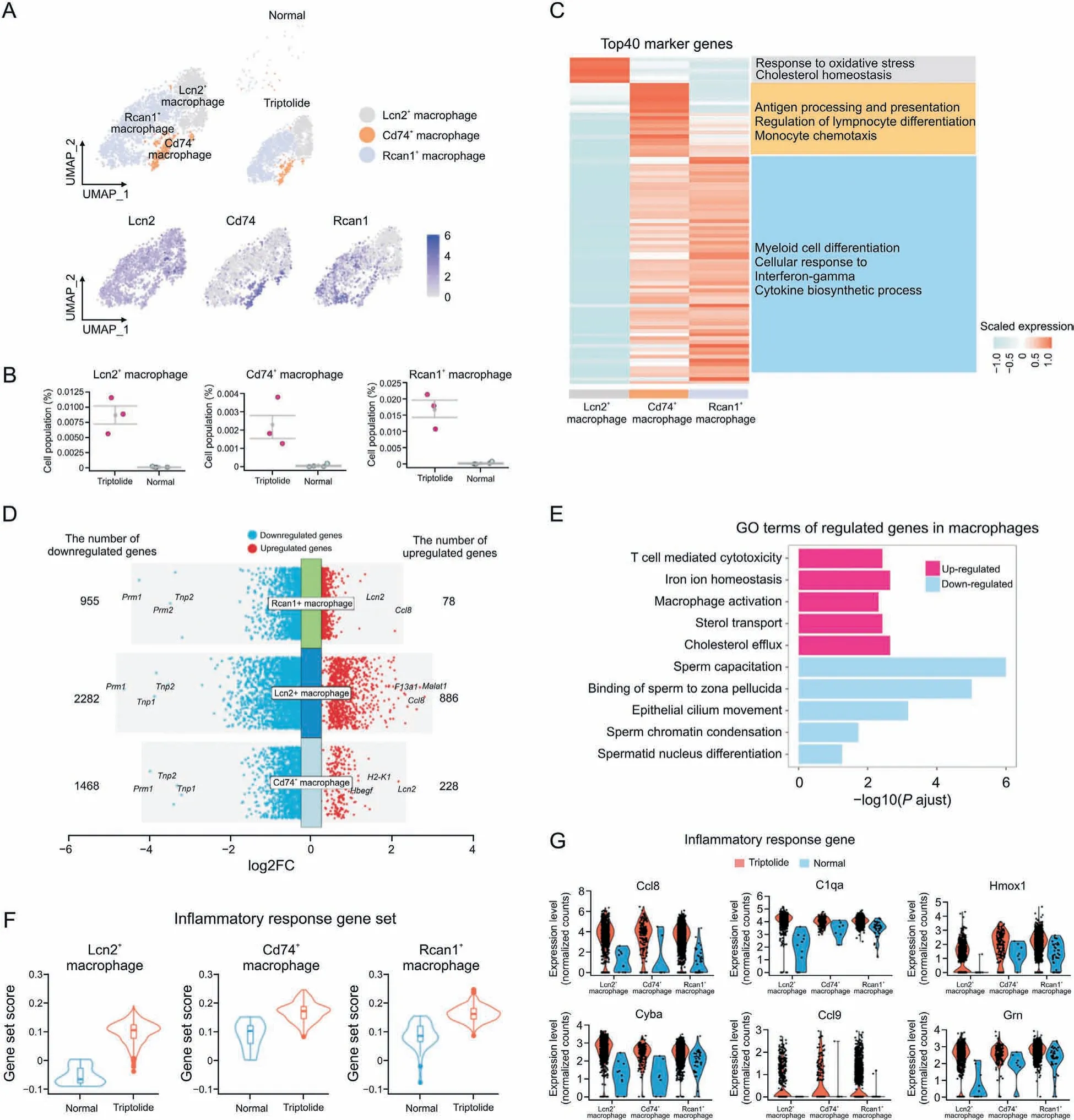
Fig. 7. Triptolide-associated molecular characteristics in macrophages. (A) Uniform manifold approximation and projection (UMAP) plot showing the distribution of three macrophage subtypes in normal and triptolide-treated mice. (B) Dot plot showing the cell percentage of each macrophage subtypes in normal and triptolide-treated mice.(C)Heatmap revealing the top 40 marker genes of indicated macrophage subtypes,along with their enriched gene ontology(GO) terms(listed on the right).(D)Dot plot showing the number and logarithmic fold change(FC)(logFC)of regulated genes(dots)across three macrophage subtypes.Genes in colored dots are significantly(FDR<0.05 and fold change>25%)up-regulated or down-regulated under triptolide conditions, as determined by differentially expressed genes(DEGs)analysis (see details in Methods). (E) Bar plot showing enriched GO terms in macrophages after triptolide treatment. (F) Gene set score analysis for inflammatory response pathway in three macrophage subtypes after triptolide treatment. ****P<0.0001(two-sided Wilcoxon rank-sum tests).(G)Violin plots with dots showing differentially inflammatory response related genes in three macrophage subtypes between normal and triptolide group.
Further analysis showed that these kinds of macrophages showed elevated gene set scores associated with inflammatory response (Fig. 7F). Multiple pro-inflammatory genes such as Ccl8,C1qa, Hmox1, Cyba, Ccl9, and Grn were dramatically increased in triptolide-treated mice (Fig. 7G), suggesting that triptolide treatment increases inflammatory response. Importantly, chemokines associated with macrophage migration such as Ccl8 and Ccl9 were increased in triptolide-treated macrophages (Fig. S5C). Overall,these results showed that testicular macrophage cells not only promote inflammatory response, but also inhibit sperm formation in response to triptolide.
3.8. Triptolide-associated changes in SPG
To understand the changes of SPG in response to triptolide,SPG was further divided into undifferentiated spermatogonia stem cells(SSCs) and differentiated SSCs based on the expression of their marker genes(Figs.S6A and B).A lineage trajectory of undifferentiated SSCs and differentiated SSCs along the pseudotime was established and we observed that triptolide-treated mice showed a reduction of differentiated SSCs in the trajectory (Fig. S6C), suggesting that triptolide may disrupt the transition of undifferentiated SSCs to differentiated SSCs. Meanwhile, BEAM showed gene expression signature with respect to pseudotime and revealed three clusters (Clusters 1 to 3) associated with cell fate transition upon triptolide treatment (Fig. S6D). Cluster 2 showed downregulated genes along the trajectory, especially in undifferentiated SSCs. GO analysis showed that these genes were enrichment in pathways of DNA repair, stem cell population maintenance, and wnt singling pathways(Fig.S6D),indicating that triptolide may inhibit the transition of undifferentiated SSCs to differentiated SSCs via modulating these pathways.These results highlight a critical role of dysregulation of several critical pathways in SPGs in triptolide-associated testicular injury.
3.9. Triptolide-associated changes in ligand-receptor pairs
Based on our ScRNA-seq results, we next established a comprehensive intercellular network of potential ligand-receptor interactions in response to triptolide treatment according to a previously reported database[38].Triptolide-treated mice showed the overall increased numbers of interactions, especially the interaction of several macrophage subtypes with Leydig cells(Figs.8A and B).Furthermore,differential interaction strengths between macrophages and Sertoli cells were increased(Fig.8C).In contrast,the interaction of several germ cell types including elongating STids, and round Stids with other cell types including Sertoli cells and Leydig cells were reduced (Fig. 8C). These results highlight a critical role of testicular microenvironment and the changes in interaction of different cell types within the testis in contributing to triptolide-induced testicular injury.
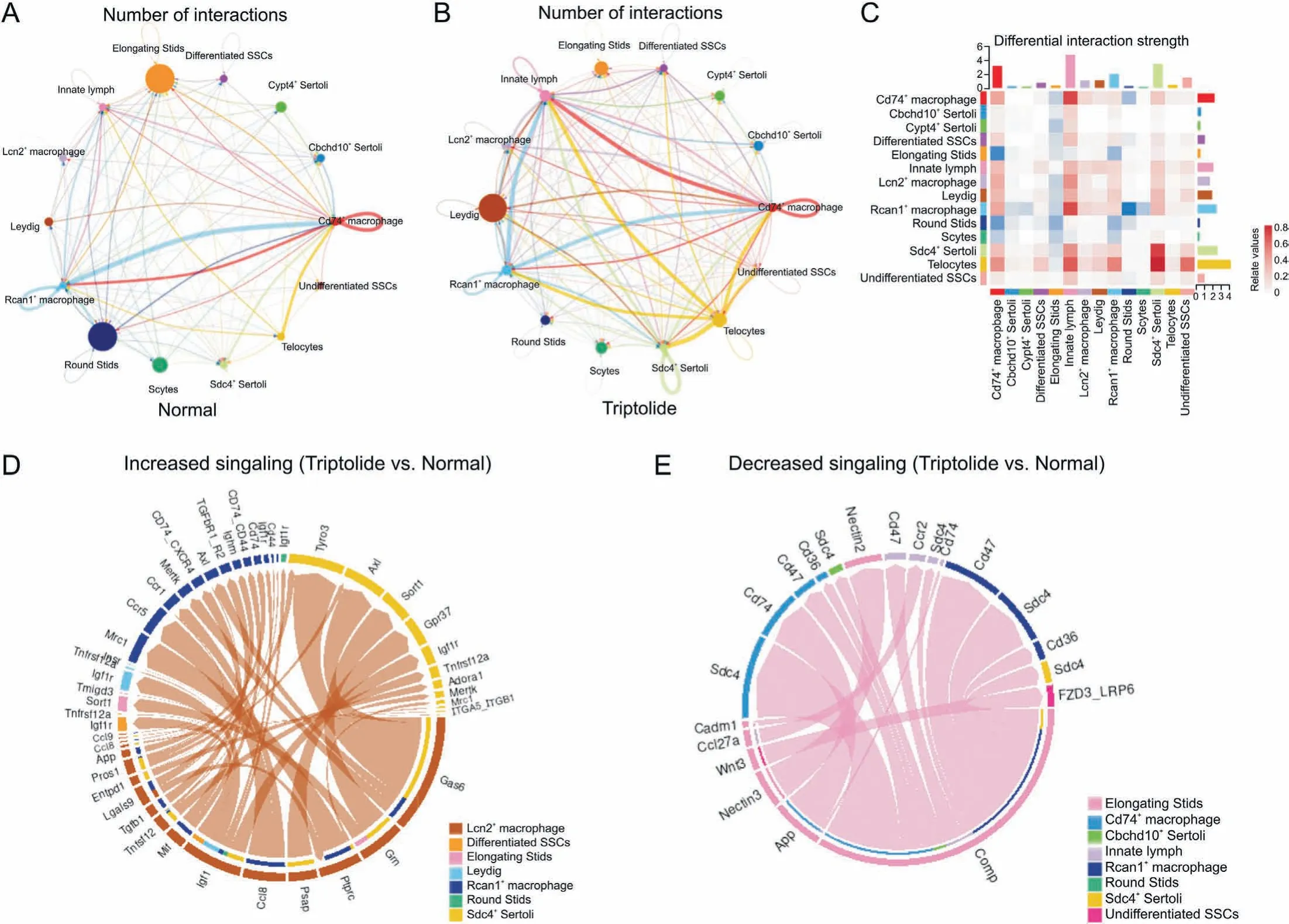
Fig. 8. Triptolide-associated changes in cell-cell interactions. (A, B) Network plots indicating interaction numbers (A) and strengths (B) in testes of normal and triptolide-treated mice. (C) CellChat analysis revealing the differential interaction strength of each cell type after triptolide treatment. (D) Chordal graph showing increased ligand-receptor pair in Lcn2+ macrophage upon triptolide treatment. The edge width represents the communication probability. (E) Chordal graph showing the decreased ligand-receptor pair in elongating STids upon triptolide treatment. The edge width represents the communication probability. SPG: spermatogonia; SSC: spermatogonia stem cells; Scytes: spermatocytes;round Stids: round spermatids; elongating Stids: elongating spermatids.
To further characterize the intercellular communication, we identified increased ligand-receptor pairs of Lcn2+macrophages with other cells (Fig. 8D). For instance, we showed that the interaction of Gas6 in Lcn2+macrophages with TAM receptors Tyro3 and Axl in differentiated SSCs were increased(Fig.8D).TAM receptors are family members of receptor tyrosine kinases that are crucial for inflammation response [39], the increased interaction of Gas6 with TAM receptors suggests that triptolide treatment may promote inflammatory response in testis. We also identified decreased signalings in elongating STids (Fig. 8E), and these changes may also play a role for triptolide-induced testicular injury.
4. Discussion
To our knowledge, we comprehensively reveal the first gene expression profiles in different cell types of mouse testes in triptolide-induced testicular injury using advanced ScRNA-seq technology. Our results showed that in addition to loss of germ cells, triptolide leads to increased proinflammatory macrophages.The dysregulation of several critical signaling pathways, such as ROS, the inflammatory response, and spermatid differentiation in both somatic cell and germ cells, and their interaction may be key events in triptolide-induced male infertility.These high-resolution cellular atlase not only offer novel information to help understand the underlying mechanisms of triptolide-associated male infertility but also provide valuable information for discoveries of novel agents that mitigate triptolide-associated herbal medicine-induced male infertility.
Somatic and germ cell interactions within testes are crucial for spermatogenesis. Although several mechanisms, such as oxidative stress [12] and perturbation of gut microbiome [16], have been proposed to be involved in triptolide-induced testicular toxicology and male infertility, the underlying mechanisms are still not fully understood. Previous studies investigated triptolide-induced toxicity mainly using bulk sequencing technology or focusing on certain cell types in in vitro models. Here, our ScRNA-seq results uncovered the changed genes and pathways associated with triptolide-induced testicular injury in multiple different cell types of testes. As a complex organ with high cellular heterogeneity, we showed that the responses of different cell types to triptolide were distinct.In germ cells,our ScRNA-seq results showed that triptolide increased pathways associated with inflammation and apoptosis,and reduced pathways enriched in germ cell development, and sperm motility. These results indicate that triptolide not only induces germ cells apoptosis but also inhibits the formation of germ cells.Apart from germ cells,we also showed that triptolide induces ROS signaling and reduces genes set-associated with spermatid development in somatic cells. As somatic cells are critical for spermatogenesis, the dysregulated pathways in Leydig cells and Sertoli cells suggest triptolide may also inhibit spermatogenesis via disturbing somatic cell functions of the testes.
Although several pathways were reported for triptolide-induced testicular injury, shared and cell type-specific changes among different cell types of testes after triptolide-treatment have not been identified before. For instance, increased oxidative stress in Sertoli cell lines was reported after triptolide treatment[12].Here,we showed that apart from Sertoli cells,Leydig cells and germ cells also showed elevated ROS signaling, suggesting that elevated ROS signaling in multiple cell types within testes may be a common mechanism for triptolide-induced testicular injury, and targeting ROS may be a potential strategy for alleviating triptolide-induced testicular injury. Furthermore, autophagy has been implicated in multiple diseases, including male infertility [40-42], and autophagy plays a critical role in regulating ROS. Future studies to dissect the roles of autophagy in triptolide-induced male infertility is important. Interestingly, many Chinese medicines, including their active components, have been reported to reduce triptolideinduced toxicity [43], and further dissection of their roles and underlying mechanisms in reducing triptolide-associated testicular injury is urgently needed.
Another important finding is that triptolide-treated mice showed increased macrophages, and exhibited multiple elevated inflammatory response associated genes,suggesting that increased inflammation may provide a hostile microenvironment for triptolide-induced testicular injury.Although triptolide shows antiinflammatory and immunosuppressive activity in autoimmune disorder models such as rheumatoid arthritis [44], it promotes inflammatory response in the testes. These results suggest that inhibition of inflammatory response may alleviate triptolide-induced testicular injury, and to test this possibility is warranted.
5. Conclusions
Collectively,our results provide a large amount of information on triptolide-associated changes in gene expression, pathways, and ligand-receptor pairs for the major cell types of mouse testes. We expect that our dataset will not only help to shed light on how triptolide affects spermatogenesis but may also serve as a resource for better understanding and future investigation of the underlying molecular mechanisms,as well as for developing therapeutic agents that are associated with triptolide associated side effects in testes.Overall,our results contribute to a variety of efforts towarding understanding triptolide-induced male infertility, and may provide potential therapeutic targets for alleviating triptolide-associated testicular toxicity.
CRediT author statement
Wei Zhang:Conceptualization, Data curation, Investigation,Methodology, Visualization, Writing - Original draft preparation;Siyu Xia:Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing - Original draft preparation;Jinhuna Ou:Data curation, Formal analysis, Investigation, Methodology, Writing - Original draft preparation;Min Cao:Data curation, Investigation, Methodology;Guangqing Cheng:Data curation, Formal analysis, Investigation, Methodology, Software,Visualization, Writing - Original draft preparation;Zhijie Li:Conceptualization, Methodology, Project administration, Supervision,Resources,Writing-Original draft preparation;Jigang Wang:Conceptualization, Supervision, Resources, Writing - Reviewing and Editing, Funding acquisition;Chuanbin Yang:Conceptualization, Supervision, Resources, Writing - Reviewing and Editing,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The work was supported by grants from the National Key Research and Development Program of China (Grant Nos.:2020YFA0908000 and 2022YFC2303600),the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (Grant No.: ZYYCXTD-C-202002), the National Natural Science Foundation of China(Grant Nos.:82201786,82141001,82274182,82074098 and 82173914),the CACMS Innovation Fund (Grant Nos.: CI2021A05101 and CI2021A05104), the Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences(Grant No.:CI2021B014),the Science and TechnologyFoundationofShenzhen(GrantNos.:JCYJ20220818102613029,JCYJ20210324114014039,and JCYJ20210324115800001), Guangdong Basic and Applied Basic ResearchFoundation(GrantNos.:2020A1515110549,2021A1515110646), the Science and Technology Foundation of Shenzhen(Shenzhen Clinical Medical Research Center for Geriatric Diseases),the National Key R&D Program of China Key projects for international cooperation on science, technology and innovation(Grant No.: 2020YFE0205100), and the Fundamental Research Funds for the Central Public Welfare Research Institutes(Grant Nos.:ZZ14-YQ-050, ZZ14-YQ-051, ZZ14-YQ-052, ZZ14-FL-002, ZZ14-ND-010 and ZZ15-ND-10).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.04.006.
 Journal of Pharmaceutical Analysis2023年8期
Journal of Pharmaceutical Analysis2023年8期
- Journal of Pharmaceutical Analysis的其它文章
- Single-cell and spatially resolved omics: Advances and limitations
- Preface for Special Issue: Single-Cell and Spatially Resolved Omics
- Single-cell transcriptome analysis uncovers underlying mechanisms of acute liver injury induced by tripterygium glycosides tablet in mice
- Single-cell and spatial heterogeneity landscapes of mature epicardial cells
- Temporal dynamics of microglia-astrocyte interaction in neuroprotective glial scar formation after intracerebral hemorrhage
- Promise of spatially resolved omics for tumor research
