Cassava MeRS40 is required for the regulation of plant salt tolerance
MA Xiao-wen, MA Qiu-xiang, MA Mu-qing, CHEN Yan-hang, , GU Jin-bao, , LI Yang, , HU Qing, LUO Qing-wen, WEN Ming-fu, ZHANG Peng, LI Cong, #, WANG Zhen-yu, #
1 Institute of Nanfan & Seed Industry, Guangdong Academy of Sciences, Guangzhou 510316, P.R.China
2 National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 200032, P.R.China
3 Zhanjiang Research Center, Institute of Nanfan & Seed Industry, Guangdong Academy of Sciences, Zhanjiang 524300, P.R.China
Abstract Soil salinity affects the expression of serine/arginine-rich (SR) genes and isoforms by alternative splicing, which in turn regulates the adaptation of plants to stress.We previously identified the cassava spliceosomal component 35 like (SCL) and SR subfamilies, belonging to the SR protein family, which are extensively involved in responses to abiotic stresses.However, the post-transcriptional regulatory mechanism of cassava arginine/serine-rich (RS) subfamily in response to salt stress remains to be explored.In the current study, we identified 37 genes of the RS subfamily from 11 plant species and systematically investigated the transcript levels of the RS40 and RS31 genes under diverse abiotic stress conditions.Subsequently, an analysis of the conserved protein domains revealed that plant RS subfamily genes were likely to preserve their conserved molecular functions and played critical functional roles in responses to abiotic stresses.Importantly, we found that overexpression of MeRS40 in Arabidopsis enhanced salt tolerance by maintaining reactive oxygen species homeostasis and up-regulating the salt-responsive genes.However, overexpression of MeRS40 gene in cassava reduced salt tolerance due to the depression of its endogenous gene expression by negative autoregulation of its own pre-mRNA.Moreover, the MeRS40 protein interacted with MeU1-70Ks (MeU1-70Ka and MeU1-70Kb) in vivo and in vitro, respectively.Therefore, our findings highlight the critical role of cassava SR proteins in responses to salt stress in plants.
Keywords: cassava, alternative splicing, serine/arginine-rich proteins, salt stress
1.Introduction
Alternative splicing(AS)is determined by spliceosome, which is constituted of five small nuclear ribonucleoproteins (snRNPs, U1, U2, U4/U6, and U5), and hundreds of non-snRNPs, including the serine/arginine-rich proteins of nuclear phosphoproteins (SR proteins), SR-related proteins, and the heterogeneous nuclear ribonucleoproteins (hnRNPs) (Matera and Wang 2014; Fica and Nagai 2017).A series of recent studies have illuminated the mechanisms of spliceosome formation and splicing in remarkable detail (Yanet al.2017; Kastneret al.2019; Plaschkaet al.2019).Among these spliceosomal proteins, SR proteins are the highly conserved family of RNA-binding proteins involved in the regulation of AS (Moranteet al.2010).SR proteins influence the splice site selection in a phosphorylationdependent manner and facilitate the AS process, in which they seem to be regulated in a tissue-specific, developmentally-regulated, and stress-responsive pattern (Duque 2011).The typical features of SR proteins are the presence of one or two N-terminal RNA recognition motifs (RRMs) and a C-terminal arginine/serine-rich (RS) domain (Graveley 2000; Bourgeoiset al.2004).The RRM domain mediates the binding of SR proteins to the pre-mRNA by recognizing short sequences of pre-mRNA, including exonic splicing enhancers (ESE) and silencers (ESS) (Graveleyet al.1999).The RS domain promotes the recruitment of core splicing components to nearby splice sites by participating in the protein–protein and protein–RNA interactions (Graveley 2000).Six subfamilies (SR, SC, RSZ, SCL, RS2Z and RS) have been identified in plant SR proteins, and three of the subfamilies (SCL, RS2Z and RS) are plant-specific while the other three subfamilies are the orthologs of human ASF/SF2, SC35, and 9G8(Bartaet al.2010).
Several studies have revealed that SR proteins participate in regulating the AS of plant responses to environmental stress (Palusaet al.2007; Tanabeet al.2007; Duque 2011; Yoshimuraet al.2011; Chenet al.2013; Mortonet al.2019; Liet al.2021; Kumaret al.2022).AnArabidopsisSR-like protein, AtSR45a, interacting with other splicing factors, is involved in the regulation of high-light stress-responsive alternative splicing, and serves as a novel player in signaling pathwaysviaabscisic acid (ABA) in the adaptation to environmental sugar levels (Carvalhoet al.2010; Yoshimuraet al.2011).A recent study showed that AtSR45a is required for the AS and mRNA maturation of several salt-tolerance genes, and it is involved in post-transcriptional regulation of salinity tolerance inArabidopsis thaliana(Liet al.2021).The expression ofAtSR34bis induced by cadmium (Cd) and asr34bmutant is moderately sensitive to Cd (Zhanget al.2014).AtSR45negatively regulates glucose (Glc) signaling during early seedling development by down-regulating both Glc-specific ABA accumulation and ABA biosynthesis and signaling gene expression (Carvalhoet al.2010).The transcript level ofBrSR45ais enhanced by drought stress, andBrSR45aregulates the drought stress responseviathe AS of target genes in a concentrationdependent manner inBrassicarapa(Muthusamyet al.2020).Overexpression ofOsSCL30diminishes the resistance to low temperature, drought and salt stresses in transgenic rice, and results in greater accumulation of reactive oxygen species (Zhanget al.2022).
In addition, alternative isoforms of SR proteins imposed by environmental stress resulted in changing the AS of target genes, leading to an increase in the complexity of the transcriptome and proteome in plant stress adaptation (Duque 2011).For AtRSZ33, an RSZ subfamily protein, autoregulated AS events resulted in an extremely truncated protein (Kalynaet al.2006).SC35 and SC35-like (SCL) proteins preferentially bind to a specific RNA sequence containing the AGAAGA motif, and the loss of these proteins results in pleiotropic changes in plant morphology and development (Yanet al.2017).Therefore, the functional characterization of plant SR proteins would provide insights into the molecular mechanisms of post-transcriptional regulation in plants.
Cassava (ManihotesculentaCrantz) is widely cultivated in the tropical and sub-tropical areas.It serves as an important staple crop for over 750 million people (Janssonet al.2009; Bredesonet al.2016).Cassava has been recognized as a drought and low-soil fertility resistant crop, however, the molecular mechanisms of its adaption to environmental stress require further exploration (Anet al.2012).Cassava is frequently challenged with a variety of environmental stresses during its growth and development, which ultimately leads to decreases in yield and productivity.Therefore, a comprehensive understanding of the plant stress response signaling and the molecular mechanism would be beneficial for determining the plant resistance to stresses (Zhu 2016).Our previous study revealed that environmental stresses affect the expression of SR genes and isoforms by alternative splicing, which in turn regulates the adaptation of plants to stress (Guet al.2020).We further characterized the roles of SCL, RSZ and SR family members, which have identical roles in the responses to drought and salt stress (Huet al.2021; Wenget al.2021; Chenet al.2022).Overexpression ofMeSR34inArabidopsisenhances salt tolerance by maintaining reactive oxygen species (ROS) homeostasis (Guet al.2020).Moreover,MeSCL30andMeRSZ21bcan enhance plant drought tolerance by maintaining ROS homeostasis and the ABA-dependent signaling pathway (Wenget al.2021, Chenet al.2022).It is well known that the RS subfamily is widespread throughout the animal and plant kingdoms (Bartaet al.2010).However, there is less information on the cassava RS subfamily genes, and their involvement in post-transcriptional regulation mechanisms under salt stress conditions remains unclear.In this study, we first performed a phylogenetic comparison of the RS subfamily genes in plants, and then one cassava RS subfamily gene,MeRS40, was comprehensively analyzed in response to salt stress.Moreover, the overexpression ofMeRS40inArabidopsisincreased salt tolerance by maintaining ROS homeostasis and increasing the expression of salt stress-responsive genes.Unexpectedly, the overexpression ofMeRS40in cassava reduced salt tolerance due to the depression of the endogenous gene expression ofMeRS40, which was caused by the negative autoregulation of its own pre-mRNA.Interestingly, the MeRS40 protein interacted with MeU1-70Ks, suggesting that the MeRS40 protein might be involved in the connection of spliceosome assembly between 5′ and 3′ splice sites in cassava.Consequently, our research lays the foundation for an indepth understanding of the role of RS subfamily genes in the positive regulation of salt stress tolerance in plants.
2.Materials and methods
2.1.Plant materials and growth conditions
Two cassava cultivars, South China 8 (SC8) and TMS60444, were cultured on cassava basic medium (CBM) (Murashige and Skoog (MS) salts, 2% sucrose, 2 mmol L–1CuSO4, 0.3% Gelrite, pH 5.8) in a phyto-chamber under long-day (LD) conditions (16 h light/8 h dark).The average temperature was at (27±1)°C and the relative humidity was 70%.Seedlings ofA.thalianaon MS medium agar plates (1/2 MS salts, 1% sucrose, 1.2% agar, pH 5.7) were routinely grown at (23±1)°C under LD conditions.Soilgrown plants were grown at (23±1)°C under LD conditions.Tobacco cultivar,Nicotianabenthamiana, was also grown at (23±1)°C under LD conditions.One-month-old seedlings were used for injection.
2.2.Sequence identification of plant RS subfamily genes
FourArabidopsisprotein sequences (AT3G61860, AT2G46610, AT5G52040, and AT4G25500) were downloaded from the TAIR databases (https://www.arabidopsis.org).These four protein sequences were used as queries to carry out BLAST searches with an e-value of <1e–10against all of the available plant protein sequences from the PHYTOZOME 12.1.6 database (https://phytozome.jgi.doe.gov/pz/portal.html).From the BLAST search results, 37 putative RS subfamily protein sequences from 11 plant species were selected for analysis (Appendix A).
2.3.Evolutionary and muti-sequence alignment analysis
The protein sequences obtained were used to construct a neighbor-joining (NJ) phylogenetic tree using Clustal X 2.0 and MEGA 7.0 Software (Thompsonet al.1997; Kumaret al.2016).The RS subfamily proteins were used to perform multiple sequence alignments using DNAMAN 8.0 Software (Lynnon Biosoft, USA).
2.4.Protein structure and subcellular localization
Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) was used to predict the protein domain (Letunicet al.2021), and the LOCALIZER Program (http://localizer.csiro.au/) was used to predict the subcellular localization (Sperschneideret al.2017).
2.5.Expression analysis
Expression data for the plant RS subfamily genes in different stress responses and different tissues were downloaded from the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov).All published RNA-seq datasets were available in the GEO data repository (https://www.ncbi.nlm.nih.gov/guide/genes-expression/) and corresponded to a transcriptional analysis of diverse plants subjected to different abiotic stresses.The expression profiles of different tissues were downloaded from the Bart Lab Cassava Atlas database (http://shiny.danforthcenter.org/cassava/), TAIR database, SoyBase database (https://www.soybase.org/soyseq/), and RGAP (Rice Genome Annotation Project) database (http://rice.plantbiology.msu.edu/).Heat maps of fragments per kilobase per million (FPKM) values in theRS40/31genes were generated by GraphPad Prism Software.The FPKM values of the control were set to 1.
2.6.Plasmid construction and generation of transgenic plants
The coding DNA sequence (CDS) ofMeRS40(1 119 bp) was amplified from the cDNA of young leaves of cassava using MeRS40-attB-F and MeRS40-attB-R primers.The CDS obtained was then cloned into the pDONR207 vector by the BP recombination reaction (Thermo Fisher Scientific, Waltham, USA).The resulting entry clone (pDONR207-MeRS40) was introduced into the Gateway destination vectors pGWB505, pGWB514, and pMDC43 by LR recombination (Magnaniet al.2006).The CDS ofMeRS31(738 bp) was amplified using the MeRS31-attB-F and MeRS31-attB-R primers and inserted into the pDONR207 vector by the BP recombination reaction (Thermo Fisher Scientific, Waltham, USA).pDONR207-MeRS31was then introduced into the Gateway destination vector pGWB505 (Magnaniet al.2006).The pGWB514-MeRS40(P35S:MeRS40-3HA) construct was introduced intoAgrobacteriumtumefaciensstrain GV3103viachemical transformation and then transformed into Col-0 throughAgrobacterium-mediated plant transformation (Clough and Bent 1998).Two single-copyT3transgenic lines were confirmed by quantitative real-time PCR (qRT-PCR) and tested for salt tolerance.The pMDC43-MeRS40(P35S:GFPMeRS40) construct was introduced intoA.tumefaciensstrain LBA4404viachemical transformation and then transformed into TMS60444 throughAgrobacteriummediated cassava transformation (Bullet al.2009), and two transgenic lines were confirmed by qRT-PCR and tested for salt tolerance.
2.7.Sample collection, RNA extraction and qRT-PCR
The root, stem, and leaf tissues of three-week-old cultured cassava plants were harvested for gene expression analysis ofMeRS40andMeRS31.To analyze the gene expression ofMeRS40in response to salt in the TMS60444 and transgenic cassava plants, three-weekold cultured cassava plants grown on CBM media were subjected to 0.3 mol L–1NaCl for indicated time intervals, and the leaves were harvested from the plants for subsequent analysis.In addition, two-week-old seedlings of Col-0 and transgenicArabidopsisplants grown on 1/2 MS solid media were treated with either water (Control) or 0.15 mol L–1NaCl for 1 h prior to the gene expression analysis ofMeRS40and the salt-responsive genes.Total RNA was extracted using the FastPure Plant Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Nanjing, China).For qRT-PCR, first-strand cDNAs were synthesized from DNase-treated total RNA (2.5 μg, reaction total volume 20 μL) using the EasyScipt?RT/RI enzyme (TransGen Biotech Co., Ltd., Beijing, China).qRT-PCR was performed using a Roche LightCycle?96 System and the TransStart Tip Green qPCR SuperMix (TransGen Biotech Co., Ltd.).TheUbiquitingene was used as an internal control, and for normalization, each qRT-PCR was repeated independently at least three times.The primer sequences are given in Appendix B.
2.8.Subcellular localization of MeRS40 and MeRS31 and bimolecular fluorescence complementation (BiFC) assays
The pGWB505-MeRS31(P35S:MeRS40-GFP) and pGWB505-MeRS40(P35S:MeRS40-GFP) fusion protein vectors were individually transformed intoA.tumefaciensstrain GV3101.Subcellular localization images were captured under a Zeiss LSM780 confocal laser scanning microscope and processed using ZEN 2009 Light Edition Software.To detect the interactions between MeRS40 and the MeU1-70Ks (MeU1-70Ka and MeU1-70Kb)in vivo, the CDS ofMeRS40was ligated into pEarlygate201YN.TwoU1-70K(MeU1-70KaandMeU1-70Kb) coding sequences were inserted separately into pEarlygate202YC.The constructs were individually transformed intoA.tumefaciensstrain GV3101 and grown at 30°C under dark conditions.Then the cells were harvested by centrifugation, re-suspended and incubated in induction medium (0.01 mol L–1MES, pH 5.6, 0.01 mol L–1MgCl2and 150 μmol L–1acetosyringone) for 2 h.The bacteria were then re-suspended in the induction medium to a final concentration of OD600=0.5 before inoculation.The suspensions were infiltrated into young but fully expandedN.benthamianaleaves using a needleless syringe (Chenet al.2018).After 48 h of incubation at 23°C, the YFP fluorescent signals were detected under a confocal microscope.
2.9.Yeast two-hybrid assays
To detect the interactions between MeRS40 and the two MeU1-70Ksinvitro, the CDS ofMeRS40was ligated into the pGBKT7 vector.TwoU1-70K(MeU1-70KaandMeU1-70Kb) coding sequences were cloned separately into the pGADT7 vector.Yeast transformation was performed according to the manufacturer’s instructions (Clontech, CA, USA).These constructs were cotransformed into yeast stain Y2H gold and then plated on –tryptophan/–leucine (–L–W, for transformation control) and –leucine/–tryptophan/–histidine–adenine (–L–W–H–Ade, for selection) media.The pGADT7 empty vector was used as a negative control.
2.10.Physiological assays
For the growth and development assay, Col-0 and two transgenic lines ofMeRS40were grown on MS medium under LD conditions at 23°C.Seedlings were photographed and fresh weights were measured at the indicated times.
For the seed germination assay, seeds of Col-0 and two transgenic lines ofMeRS40were sown horizontally on a half-strength MS medium with or without NaCl.In each experiment, at least 81 seeds per genotype were stratified at 4°C for 2 days, and radicle emergence was used as an indication of seed germination.
For assessing salt resistance in soil, Col-0 and two transgenic lines ofMeRS40were grown in soil under LD conditions for two weeks.Subsequently, the soil was irrigated with 0.3 mol L–1NaCl every four days, and the plants were allowed to grow for an additional 1 to 3 weeks before they were photographed, and the survival rate was determined.
The hydrogen peroxide (H2O2) content was determined by the potassium iodide method (Thordal-Christensenet al.1997).For the detection of H2O2staining,Arabidopsisleaves were stained with 3,3′-diaminobenzidine (DAB) for 24 h, then decolorized with ethanol (Guet al.2020).Peroxidase (POD) activity was determined by the guaiacol method (Guet al.2020).Ascorbate peroxidase (APX) activity was determined by monitoring the decrease in ascorbate concentration within 180 s at 290 nm (Guet al.2020).Catalase (CAT) activity was determined by the oxidation reaction of ascorbic acid (Guet al.2020).Proline content was determined based on the method of Bates (Guet al.2020).The K+and Na+contents were determined by atomic absorption spectrometry as described previously (Guet al.2020).
For assessing salt resistance in soil, three-week-old TMS60444 and two cassava transgenic lines ofMeRS40were transplanted into grey plastic pots with bottom holes.An air pump was used to maintain normal ventilation.Soil-grown cassava seedlings were grown in the growth room at 27°C under LD conditions for two weeks.Subsequently, the soil was irrigated with 0.3 mol L–1NaCl every four days, and the plants were allowed to grow for an additional 27 days before they were photographed, and the survival rate was determined.
2.11.RNA immunoprecipitation assays (RlP)
The RIP assay was performed according to Guet al.(2020) with minor modifications.Briefly, three grams of four-week-old seedlings of TMS60444 and the cassava overexpression lines were harvested for crosslinking by 1% formaldehyde, and the reaction was stopped by adding 0.125 mol L–1glycine.The lysed supernatant was sonicated to produce fragments of 300–500 bp and treated with DNase to remove any DNA before immunoprecipitation.RNA-IP was then performed using an anti-GFP antibody (Abcam, Shanghai, China) coupled with Dynabeads protein A/G (Thermo Fisher Scientific, Waltham, USA).After washing four times, the protein on the crosslinked complex was digested with proteinase K, and the RNA was isolated by adding 1 mL of TRIzol RNA extraction reagent (Thermo Fisher Scientific, Waltham, USA) for subsequent analysis.
3.Results
3.1.Phylogenetic analysis of the RS subfamily in cassava
We previously identified 18 SR proteins in cassava, and MeRS40 and MeRS31 belong to the RS subfamily (Guet al.2020).To comprehensively characterize the evolutionary and phylogenetic relationships of RS subfamily members between cassava and other plant species, the phylogenetic tree was constructed with MEGA7.0 using the NJ method.A total of 37 amino acid sequences were obtained from 11 plant species, which were roughly classified into seven monocots and four eudicots, includingArabidopsis, soybean (Glycinemax), cassava, purple false brome (Brachypodiumdistachyon), rice (Oryzasativa), foxtail millet (Setariaitalica), sorghum (Sorghumbicolor), maize (Zeamays), wheat (Triticumaestivum), barley (Hodeumvulgare), and potato (Solanumtuberosum) (Appendix A).Eleven RS proteins were identified in dicots, including six RS proteins in soybean (GmRS31a, GmRS31b, GmRS40a, GmRS40b, GmRS41a, and GmRS41b), two RS proteins in cassava (MeRS40 and MeRS31), and three RS proteins in potato (StRS31, StRS41a, and StRS41b).Meanwhile, 22 RS proteins were recognized in monocots, including three RS proteins in purple false brome (BdRd41, BdRd31a, and BdRd31b), two RS proteins in rice (OsRS41 and OsRS31), two RS proteins in foxtail millet (SiRS31 and SiRS41), three RS proteins in sorghum (SbRS41, SbRS31a, and SbRS31b), four RS proteins in maize (ZmRS41a, ZmRS41b, ZmRS31a, and ZmRS31b), six RS proteins in wheat (TaRS31d, TaRS31b, TaRS31a, TaRS41a, TaRS41b, and TaRS41d), and two RS proteins in barley (HvRS31 and HvRS41).In general, the 37 RS subfamily sequences were clustered into two major clades: RS31/31a and RS40/41 (Fig.1).Notably, among these two major clades, the RS31/31a and RS40/41 proteins were separately classified into different branches which is consistent with the classification of monocot and eudicot plants.These results suggested that the expansion of RS subfamily members was associated with gene duplication events which occurred after the divergence of the monocots and eudicots (Fig.1).The phylogenetic tree analysis also revealed that cassava RS subfamily showed closer relationships to those ofArabidopsisand soybean, but somewhat more distant relationships to rice and maize (Fig.1).
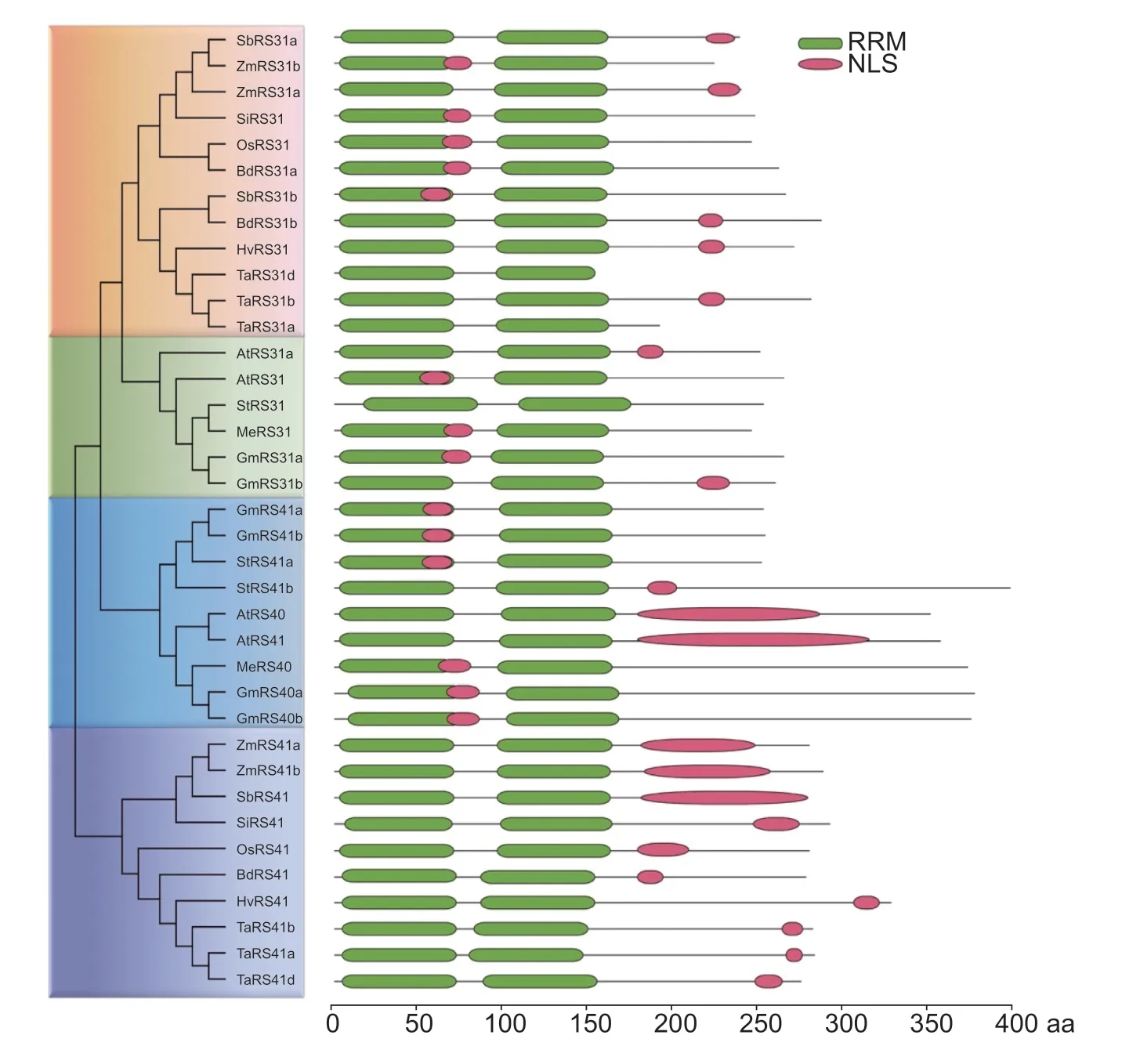
Fig.1 Phylogenetic tree and structure domain analysis of the arginine/serine rich (RS) subfamily proteins from 11 species of plants.The Arabidopsis, soybean, cassava, purple false brome, rice, foxtail millet, sorghum, maize, wheat, barley and potato RS subfamily proteins were used in phylogenetic tree construction.The complete protein sequences were taken from the Phytozome databases (https://jgi.doe.gov/).Protein alignment of the RS subfamily proteins was performed with ClustalX 2.0.The phylogenetic tree was constructed with MEGA5.1 using the neighbor joining (NJ) method (left panel).Protein structure and nuclear localization signal (NLS) predictions were performed by SMART (http://smart.embl-heidelberg.de/) and the LOCALIZER database (http://localizer.csiro.au/) (right panel).The green boxes are the RNA recognition motif (RRM) domains, and red boxes are the NLS.
To further understand the structural features of RS subfamily proteins, we predicted the RS subfamily protein domains using the SMART database.Domain analysis revealed that these RS proteins adopt a similar domain organization with two RRMs in the N-terminal, which bind to the RNA targets, and an RS domain in C-terminal (Fig.1; Appendix C).Furthermore, amino acid alignment results indicated that the RRM domain of RS40/41 showed very strong similarity between monocots and eudicots, suggesting that the RS40/41 proteins were highly conserved during plant evolution (Fig.1).Additionally, the subcellular localization of the RS subfamily proteins was predicted by LOCALIZER Program.One or two putative nuclear localization signals (NLS) were identified in each of the RS subfamily proteins except for TaRS31a, TaRS31d, and StRS31, implying that RS subfamily proteins were mainly localized in the nucleus (Fig.1).Subcellular localization assays further confirmed that the two cassava RS proteins were localized in the nucleus (Appendix C).
3.2.Expression profiles of RS genes in cassava
To further investigate the functional characterization ofMeRS31andMeRS40in cassava, the expression patterns ofMeRS31andMeRS40were first performed by qRT-PCR.The results showed thatMeRS31andMeRS40were expressed at higher levels in leaves, which is consistent with the RNA-seq data.These results demonstrated thatMeRS40andMeRS31exhibited tissue-specific expression patterns in cassava seedlings (Fig.2-A and B).Our previous study showed that the transcript level ofMeRS40was distinctly induced, while the expression ofMeRS31was downregulated in cassava leaves under salt stress conditions, indicating thatMeRS40might have a positive role in the regulation of the salt stress response in cassava (Guet al.2020).We then compared the expression profiles of plantRS40/41genes under various abiotic stresses conditions according to the RNA-seq data from the GEO data repository.This comparison showed that in monocotyledonous plants, the expression ofRS41was not noticeably affected by abiotic stresses, except forOsRS41andBsRS41, whose transcript levels were inhibited by drought or high temperature (Appendices D and E).However, in eudicot plants, the expression ofAtRS40was markedly induced by drought stress while the expression ofGmRS40was highly induced by cold stress, showing thatRS40/41might play different biological roles in monocot and eudicot plants under abiotic stresses (Appendices D and E).As expected, the qRT-PCR results demonstrated that the expression ofMeRS40was reduced at 1 h, and then increased at 6 h under salt stress conditions, suggesting thatMeRS40might participate in the adaptation of cassava to salt stress (Fig.2-C).

Fig.2 Expression analysis of two arginine/serine rich (RS) subfamily genes. A, heat map of tissue-specific expression of the MeRS31 and MeRS40 genes, including friable embryogenic callus (FEC), fibrous root, storage root, stem, leaf, lateral bud, mid vein,organized embryogenic structures (OES), petiole, root apex meristem (RAM), and shoot apex meristem (SAM).Transcript levels of the MeRS31 and MeRS40 genes were obtained from the Bart Lab Cassava Atlas database and were calculated as averages for three species.The expression levels of MeRS31 and MeRS40 genes were arbitrarily defined as those with fragments per kilobase of exon per million fragments mapped (FPKM)>1.B, tissue-specific expression levels of the MeRS31 and MeRS40 genes determined by quantitave real-time PCR (qRT-PCR).C, expression level of MeRS40 in response to salt stress for the indicated times.Three independent biological experiments with 12 individual plants (3-week-old-cultured cassava seedlings) were treated with 0.3 mol L–1 NaCl for the indicated times, and leaves were collected for RNA extraction and qRT-PCR analysis.Data (mean±SD of n=3 biological replicates) are normalized for the transcript levels relative to ubiquitin.
3.3.Overexpression of MeRS40 improved salt tolerance in Arabidopsis
To investigate whether theMeRS40gene has biological functions in response to salt, six single-copy transgenic lines were obtained by overexpressing theMeRS40gene inArabidopsis.The expression ofMeRS40gene in two overexpressing lines (OX-5 and OX-6) was strongly enhanced by more than four-fold as compared with that of Col-0 (Appendix F).To further confirm the role ofMeRS40gene in response to salt, seeds of two overexpressing lines were directly sown in the MS plates with or without 0.125 mol L–1NaCl, and there was no obvious difference in subsequent seed germination between the two overexpressing lines and Col-0 under normal conditions.However, the germination rates of two overexpressing lines were significantly higher than that of Col-0 under salt stress conditions, indicating their potential role in improving plant salt tolerance (Fig.3-A and B).Surprisingly, a post-germination assay showed that the two overexpressing lines displayed root lengths and fresh weights that were around 20% greater than Col-0 under normal conditions, suggesting that theMeRS40gene might have a potential role in the regulation of plant growth (Appendix F).To further elucidate the biological role ofMeRS40gene in response to salt, two-week-old soil-grown plants were treated with 0.3 mol L–1NaCl for three weeks.About 75% of the overexpressing lines survived while only 20% of the Col-0 plants survived under salt stress conditions, indicating that overexpression of theMeRS40gene significantly improved the plant survival ratio under salinity conditions inArabidopsis(Fig.3-C and D).

Fig.3 Phenotype analysis in two MeRS40 overexpression lines of Arabidopsis under salt stress conditions.A, representative images of Col-0 and two MeRS40 overexpression lines grown on Murashige and Skoog (MS) medium with or without 0.125 mol L–1 NaCl.B, germination rates of Col-0 and two MeRS40 overexpression lines under salt stress conditions.The germination percentage was observed over 1 to 7 days of continuous incubation at 23°C in 60% relative humidity with a long-day photoperiod (16 h light/8 h dark) in a growth chamber.The value of each biological replicate was the average calculated over three technical replicates by analyzing more than 100 seeds.Data are shown as mean±SD (n=3).C, representative images of Col-0 plants and MeRS40 overexpressing lines subjected to salt stress treatment.Two-week-old plants were treated with 0.3 mol L–1 NaCl solution, and then grown for an additional 3 weeks.Representative plants were photographed.D, survival ratios (shown as the percentage of survival in normal conditions) of plants in the Col-0 plants and MeRS40 overexpressing lines under salt stress conditions.E, the K+/Na+ ratios of Col-0 plants and MeRS40 overexpressing lines under salt stress conditions.Data are shown as mean±SD (n=3).Asterisks indicate significant differences as: *, P<0.05; **, P<0.01.
Salt stress is widely known to lead to an excessive accumulation of Na+and a deficiency of K+in plants (Zhu 2003).Therefore, maintaining cellular K+/Na+levels in saline environments is essential for plant growth.We found that the K+/Na+ratio was reduced under salt conditions in both Col-0 and the two overexpressing lines, which is consistent with the results of a previous study (Zhu 2003).Notably, the K+/Na+ratios of the two overexpressing lines were significantly higher than that of Col-0 under salt conditions, indicating that the overexpression ofMeRS40could relieve the damage from salinity on plant growth by maintaining K+/Na+homeostasis in plant cells (Fig.3-E).
3.4.Overexpression of MeRS40 in Arabidopsis led to the accumulation less reactive oxygen species under salt conditions
To understand whether the two overexpressing lines maintained better ROS homoeostasis than Col-0 plants, we examined the accumulation of H2O2using DAB staining.Meanwhile, the content of H2O2and activities of ROS detoxification enzymes were then analyzed.There was no significant difference between the overexpressing lines and Col-0 plants in the content of H2O2under normal conditions (Fig.4-A).However, the contents of accumulated H2O2were less in the two overexpressing lines than that in Col-0 under salt conditions (Fig.4-B).Moreover, the activities of CAT, APX and POD were higher in the two overexpressing lines than in Col-0 plants under salt conditions, indicating that the overexpression ofMeRS40could reduce the superoxide free radicals that are triggered by ROSviaa detoxification system which may in turn improve salt tolerance in plants (Fig.4-C).Consistent with previous studies, the proline content was significantly increased in response to salt stress, and the two overexpressing lines accumulated more proline than Col-0 plants under salt treatment, revealing that the overexpression ofMeRS40could improve plant salt tolerance by maintaining osmotic pressure (Fig.4-C).
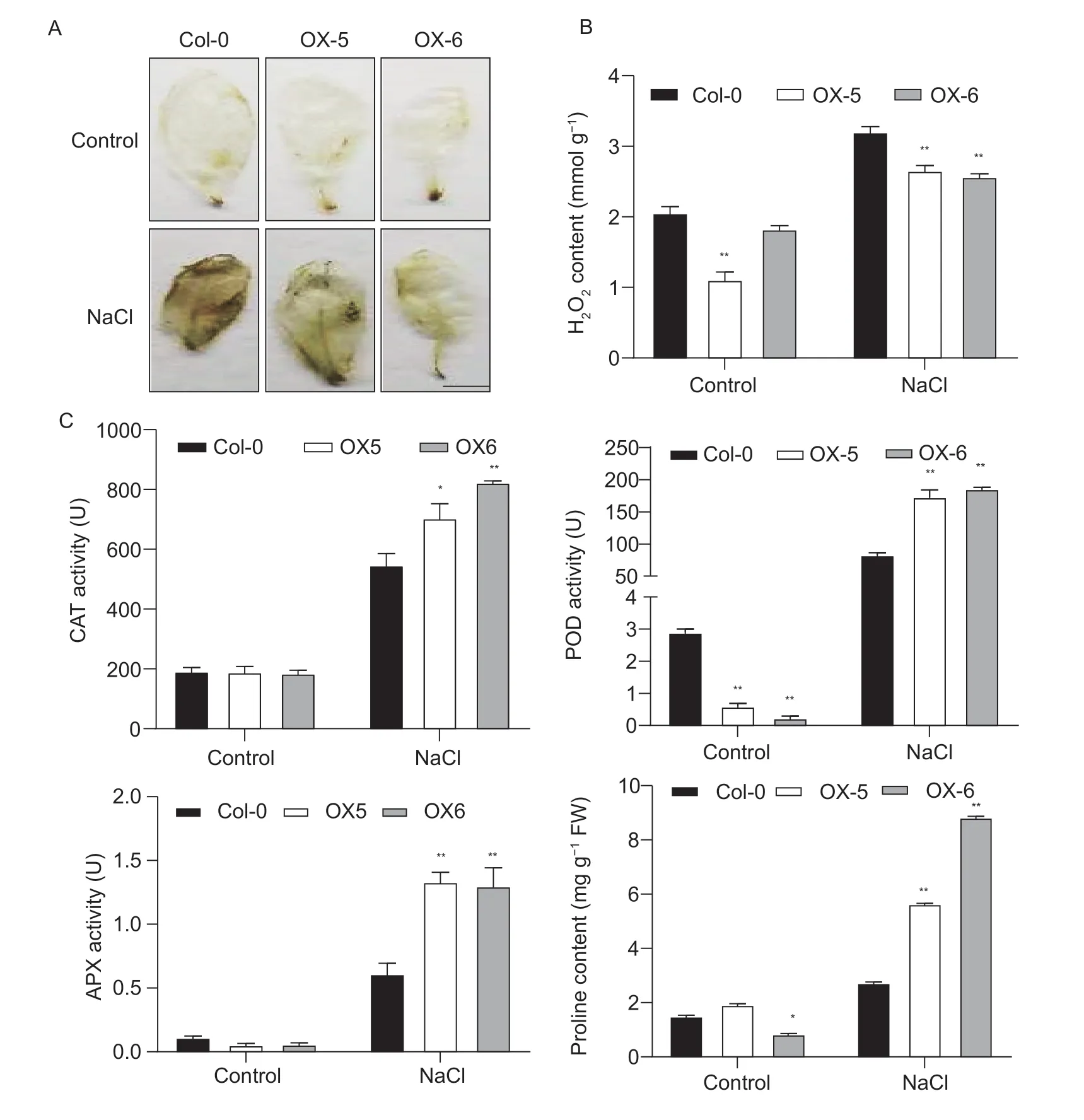
Fig.4 Overexpression of MeRS40 improved plant salt tolerance by maintaining reactive oxygen species (ROS) homeostasis.A, hydrogen peroxide detection in the leaves of Col-0 and MeRS40 overexpressing lines under normal conditions and 0.3 mol L–1 NaCl treatment.Leaves stained with 3,3′-diaminobenzidine (DAB) were used to assess the H2O2 accumulation.Scale bar=1 cm.B, hydrogen peroxide contents of Col-0 and two MeRS40 overexpressing lines under salt stress conditions.C, physiological index values of Col-0 and two MeRS40 overexpressing lines under salt stress conditions.Soil-grown Col-0 and two MeRS40 overexpressing lines treated with 0.3 mol L–1 NaCl for 7 d, and then the activities of catalase (CAT), peroxidase (POD) and ascorbate peroxidase (APX) and the proline content were determined.Error bars indicate ±SD (n=3).Asterisks indicate significant differences as: *, P<0.05; **, P<0.01.
3.5.Overexpression of MeRS40 in Arabidopsis enhanced the expression of salt-related genes
To determine whether improved salt tolerance in the two overexpressing lines might be contributing to the increased expression of salt stress-responsive genes and explore its possible mechanism, the expression levels of four stressresponsive genes were observed by qRT-PCR.Total RNA was extracted from Col-0 and two overexpressing lines treated with 0.15 mol L–1NaCl for 1 h.Under normal conditions, the expression levels ofAtSOS3in the two overexpressing lines were significantly lower than that of Col-0.However, the transcript levels ofAtSOS3were remarkably higher in two overexpressing lines than that of Col-0 under salt stress conditions, suggesting that the twoMeRS40overexpressing lines accumulated higher K+/Na+ratios by regulating the expression of SOS signal pathway genes (Fig.5).To further test whether improved salt tolerance inMeRS40overexpressing lines was achieved by affecting the gene expression of osmotic stress-related, three genes (AtRD29A,AtRD29BandAtRD17) were chosen.Consistent with a previous study, the expression levels ofAtRD29A,AtRD29BandAtRD17were greatly induced by salt, and the transcript levels ofAtRD29A,AtRD29BandAtRD17were higher in two overexpressing lines in comparison with those of Col-0 plants under salt treatment, indicating thatMeRS40might be involved in regulating the response to osmotic stress (Fig.5).
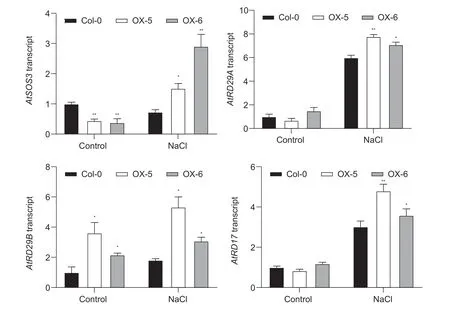
Fig.5 The relative expression levels of salt stress- or osmotic stress-related genes in Col-0 and two overexpressing lines. qRTPCR analysis was performed for four genes in Col-0 and two MeRS40 overexpressing lines for 1 h under salt stress (0.15 mol L–1 NaCl).The expression of AtUBQ served as an internal control.Error bars indicate ±SD (n=3).Asterisks indicate significant differences as: *, P<0.05; **, P<0.01.
3.6.Overexpression of MeRS40 in cassava reduced the salt tolerance by negatively autoregulating its own pre-mRNA
To determine the function ofMeRS40gene in cassava under salt treatment, 20 transgenic lines were obtained by overexpressing theMeRS40gene in cassava, and six single-copy transgenic lines were chosen for further analysis.The expression levels ofMeRS40gene in these lines were greatly enhanced by more than 80-fold as compared with that of TMS60444 plants, and two overexpressing lines (OX-6 and OX-16) reached 400-fold expression ofMeRS40gene compared with the TMS60444 plants (Fig.6-A).TheMeRS40overexpressing lines exhibited similar growth and development when compared with TMS60444 plants under LD conditions (Fig.6-B).Two-week-old soilgrown plants were treated with 0.3 mol L–1NaCl for 27 d.Surprisingly, only 25% of the cassava overexpression lines survived while about 80% of the TMS60444 plants remain healthy under salt stress conditions (Fig.6-B and C).Previous studies revealed that RNA-binding proteins AtGRP7 and AtGRP8 negatively autoregulate their own pre-mRNAs, resulting in the depression of endogenous gene expression (Schoninget al.2007; Schmalet al.2013).Therefore, we tested whether endogenousMeRS40gene expression was reduced in these lines.As expected, the endogenousMeRS40gene expression was remarkably reduced in these overexpression lines (Fig.6-D; Appendices G and H).We then determined whether the MeRS40 protein could negatively autoregulate its own pre-mRNA by thein vivoRIP assay.After IP with GFP antibody, the coimmunoprecipitated RNA pools were reverse transcribed with an adaptor primer and further amplified with specific primers as indicated in Fig.6-E.The results showed that the MeRS40 protein did not bind to the 5′ and 3′ untranslated region (UTR) regions, but it did bind to the exon in the C-terminal region, indicating that the MeRS40 protein could autoregulate its own pre-mRNA at the post-transcriptional level (Fig.6-F).
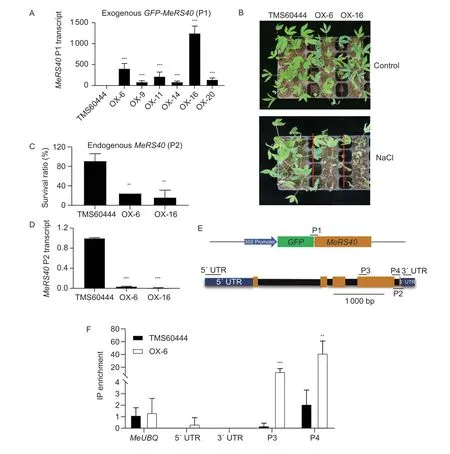
Fig.6 MeRS40 negatively autoregulates its own pre-mRNA. A, transcript levels of exogenous MeRS40 in cassava overexpressing lines.B, representative images of TMS60444 plants and two cassava MeRS40 overexpressing lines subjected to salt stress treatment.Two-week-old plants were treated with 0.3 mol L–1 NaCl solution, and then grown for an additional 27 d.Representative plants were photographed.C, survival ratios (shown as a percentage of survival in normal conditions) of plants in the TMS60444 plants and two cassava MeRS40 overexpressing lines under salt stress conditions.D, transcript levels of endogenous MeRS40 in cassava overexpressing lines.E, the P35S:MeRS40-GFP constructions diagram and gene structure of MeRS40 protein.The yellow boxes are exons, the blue boxes are 5′ and 3′ UTR, and the solid line is the intron.The primer position is shown in above or below the gene structure.F, RNA immunoprecipitation assays (RIP) followed by qRT-PCR to confirm the binding of the MeRS40 protein to its own pre-mRNA in vivo.The TMS60444 plants served as a negative control for the cassava overexpression line 6.Error bars indicate ±SD (n=3).Expression of MeUBQ was served as an internal control in A and D.Asterisks indicate significant differences as: **, P<0.01; ***, P<0.001.
3.7.MeRS40 interacted with MeU1-70K in vitro and in vivo
Several studies have revealed that SR proteins can interact with the U1-70k protein in plants (Graveleyet al.1999; Aliet al.2008; Tanabeet al.2009).We therefore tested whether MeSR40 may interact with MeU1-70kin vitroorin vivo.We first identified two genes homologous toArabidopsisU1-70K, which were referred to asMeU1-70KaandMeU1-70Kb.A yeast two hybrid assay was performed to clarify the physical interaction between MeRS40 and MeU1-70K, and the results revealed that MeRS40 interacted with MeU1-70Ka and MeU1-70Kb (Fig.7-A).To further confirm their interactionsin vivo, the BiFC was performed in tobacco leaves by transientAgrobacteriuminfiltration.The BiFC results showed that MeRS40 interacted with MeU1-70Ka and MeU1-70Kbin vivobut it did not interact with the empty vector (Fig.7-B; Appendix I).These results indicated that the MeRS40 protein might participate in the connection between the 5′ and 3′ splice sites in the spliceosome assembly in cassava.
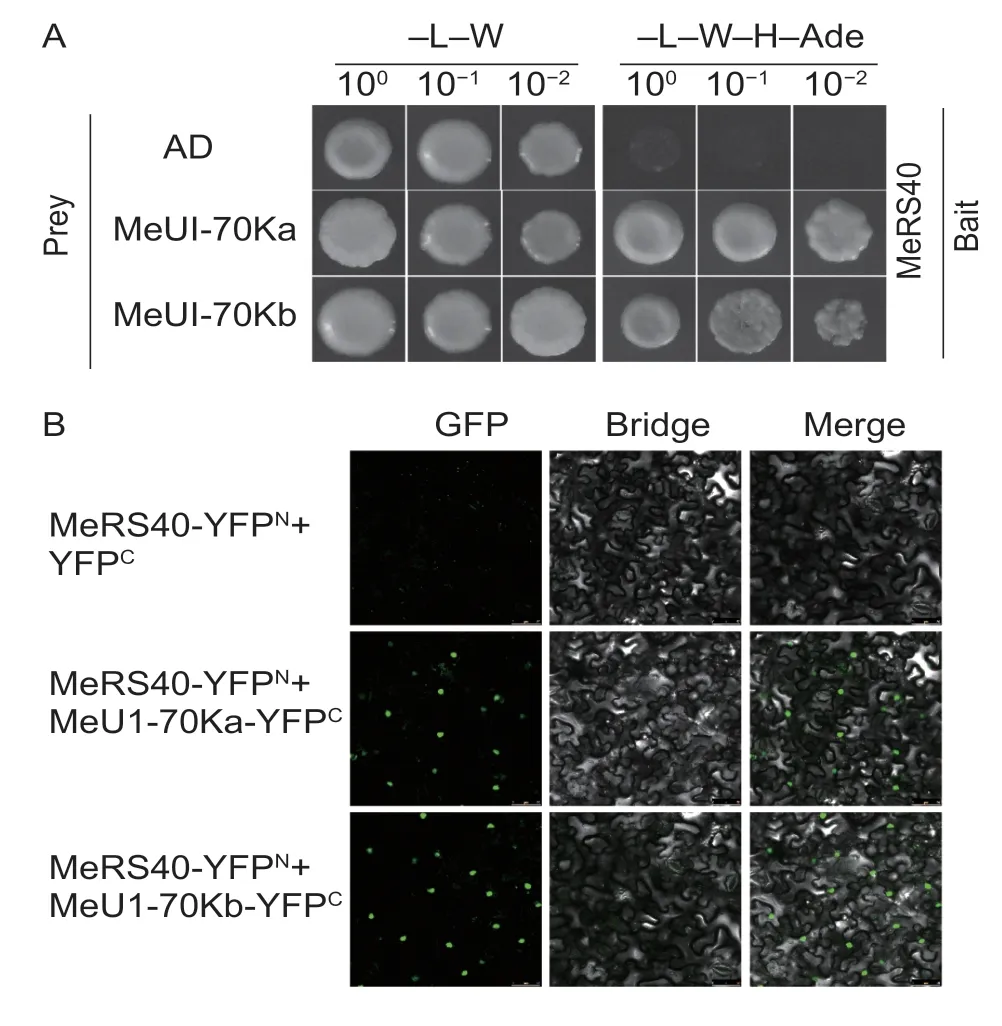
Fig.7 Protein interaction analysis of MeRS40 and MeU1-70Ks.A, identification of the interacting proteins of MeRS40 and MeU1-70Ks based on yeast two-hybrid screening.“Y2H Gold” yeast cells were co-transformed with BD-MeRS40 as bait, and AD-MeU1-70Ka or AD-MeU1-70Kb as prey were diluted to the order of 100, 10–1, and 10–2, and drops were deposited onto SD/–Leu–Trp or SD/–Leu–Trp–His–Ade media.The combination of BD-MeRS40 with empty AD was used to detect self-activation.B, bimolecular fluorescence complementation (BiFC) analyses in tobacco transient assays, where tobacco was co-transformed with YFPN and YFPC.The BiFC signals were observed under a fluorescence microscope.Bars=50 μm.
4.Discussion
Cassava is utilized worldwide for the production of starch, biofuel, and livestock feed.During the growth stage, cassava plants are frequently challenged with various environmental stresses which ultimately lead to a reduction in yield (McCallumet al.2017).Many efforts have been made to improve cassava stress resistance in modern crop breeding.We previously found that SR proteins are extensively involved in the responses to abiotic stresses in cassava.For example, MeSCL30 and MeSCL30A in the SCL subfamily, and the SR subfamily protein MeSR34 are involved in regulating drought or salt stress responses (Guet al.2020; Huet al.2021; Wenget al.2021).Here, we focused on the functional characterization of RS subfamily members in response to abiotic stress in cassava.The RS subfamily is a type of nucleoprotein that was highly conserved during evolution.An extensive analysis revealed that the cassava RS subfamily showed closer relationships with those ofArabidopsisand soybean, and more distant relationships with those of rice and maize.Moreover, overexpression ofMeRS40inArabidopsisenhanced salt tolerance through maintaining reactive oxygen species homeostasis and up-regulating the salt-responsive genes inArabidopsis.However, overexpression of theMeRS40gene in cassava reduced the salt tolerance due to the depression of its endogenous gene expression by the negative autoregulation of its own pre-mRNA.The MeRS40 protein might have a potential function in the spliceosome assembly between 5′ and 3′ splice sites.Therefore, our findings highlight the critical role of cassava SR proteins in responses to salt stress in planta.
AS pathways provide an important mechanism for generating proteome diversity and functional complexity, as well as regulating gene expression by selectively joining different exons and generating different transcripts from a single gene (Duque 2011).AS changes the structures of transcripts and can influence almost all aspects of protein function, such as binding properties, enzymatic activity, intracellular localization, posttranslational modification or protein stability.The SR protein family is an essential component of spliceosomes and has been reported to play important roles in constitutive and alternative splicing of pre-mRNA (Reddy and Shad Ali 2011).Several studies have identified 18, 20, 21, and 18 SR genes inArabidopsis, rice, maize, and sorghum, respectively (Lorkovi? and Barta 2002; Isshikiet al.2006; Rauchet al.2013).We have identified the 37 RS subfamily genes from 11 plant species, and those genes were clustered into two major clades (Fig.1).Furthermore, the expansion of RS subfamily members was associated with gene duplication events which occurred after the divergence of monocots and eudicots.In cassava, two RS subfamily proteins, MeRS40 and MeRS31, were cloned and identified, and they had two conserved functional RRM structural domains in the N-terminal region and a C-terminal RS domain (Fig.1; Appendix C).Further study revealed that these two genes had identical tissue-specific expression patterns, with higher expression levels in leaves (Fig.2-A and B).However, both genes were differentially expressed in response to salt stress (Guet al.2020), indicating that these two proteins could have distinct functions in the adaptation to environmental stress.
Furthermore, our previous study revealed that salt stress strongly alters the AS and produces different splicing variants ofMeRS40in cassava (Guet al.2020).These results indicated thatMeRS40plays a specific role in response to salt stress by affecting the turnover and transportation of specific splice variants or by reducing the stability of the spliceosome complex to accurately recognize splice sites under stress conditions.We found that the expression ofMeRS40was upregulated after a 3 h treatment with 0.3 mol L–1NaCl (Fig.2), suggesting that theMeRS40gene might participate in the regulation of salt stress response.Further study revealed that the overexpression ofMeRS40inArabidopsisimproved basal salt tolerance during seed germination (Fig.3), positively regulated ROS accumulation (Fig.4), up-regulated the transcriptional level of salt-related genes (Fig.5), and ultimately contributed to increasing plant salt tolerance.These results indicated that SR proteins play a pivotal role in regulating the salt stress response.Consistent with this conclusion,ArabidopsisSR45ahas a critical role in regulating plant salt tolerance through post-transcriptional regulation (Albaqamiet al.2019).Furthermore, two spliceosomal proteins, U1-70K and U2AF35b, interact with the SR45 protein and help to accurately recognize the 5′ and 3′ splice sites during the spliceosome assembly process (Golovkin and Reddy 1999), and SR45 negatively regulates glucose and abscisic acid signaling by boosting the proteasomal degradation of SnRK1 (Carvalhoet al.2016).Further study revealed that the SR45 interacting protein SKIP (SNW/Ski-interacting protein) participates in the manipulation of the efficient recognition or cleavage of 5′ and 3′ splice sites, which in turn reduces salt and osmotic stress tolerance inArabidopsis(Albaqamiet al.2019).Ectopic overexpression of the cassava SCL30A and SR34 proteins could improve the salt tolerance in plants (Guet al.2020; Huet al.2021).A more extensive analysis of SR proteins would provide insights on their essential roles in the proper gene expression underlying salt stress in planta.
In contrast to the positive regulation of salt tolerance in ectopic overexpression ofMeRS40inArabidopsis, overexpression of theMeRS40gene in cassava reduced the salt tolerance which might be due to the depression of its endogenous gene expression by the negative autoregulation of its own pre-mRNA (Fig.6).It is wellestablished that the autoregulatory loop is an alternative pathway for precisely regulating several key proteins at the transcript level of specific genes (Johnson and Vilardell 2012).Several RNA binding proteins are involved in the post-transcriptional autoregulation of gene expression which might affect the normal functions of their proteins.For example, AtSRp30 protein autoregulates its expression by controlling the splice-site selection of its own pre-mRNAs (Lopatoet al.1999).The SCL33 protein autoregulates its own pre-mRNA by generating premature termination codon-containing isoforms, which might contribute to controlling the levels of functional SCL33 transcript and protein (Thomaset al.2012).The RNAbinding protein FCA autoregulates its own pre-mRNA by the removal of introns which might limit FCA activity and thus controls flowering time (Quesadaet al.2003).The mammalian spliceosome protein U1A autoregulates its expression by inhibiting the polyadenylation of its own premRNA, and co-transcriptionally suppresses transcriptionterminating premature 3′ end cleavage and polyadenylation (PCPA) from cryptic polyadenylation signals (PASs) in introns, which is termed as telescripting (Kaidaet al.2010; Berget al.2012; Soet al.2019).However, our previous study found that plant U1A did not autoregulate its own pre-mRNA, suggesting that there might be differential functions between the mammalian and plant systems (Guet al.2018).Therefore, the autoregulation of RNA-binding proteins is an important feature in gene regulation that can be shown by systematically analyzing the functional transcripts and proteins in eukaryotes.
As an important component of the splice body, several SR proteins interact with U1 and U2 snRNPs, including U1-70k (Fenget al.2015).Complex interaction networks between SR proteins have been reported (Lopatoet al.2002).RZ-1B and RZ-1C, which contain RNA recognition motifs, have the potential to interact with almost all SR proteins inArabidopsis(Wuet al.2016).Plant SC35/SCL proteins address the potential interaction with a specific subunit of RNA polymerase II, NRPB4 (Yanet al.2017).The presumed mature forms of AtSR45a (the AtSR45a-1a and AtSR45a-2a proteins) not only interact with other SR and SR-like proteins (AtSR45 and AtSCL28) but also with U1-70K and U2AF35b (Tanabeet al.2009).Moreover, AtSR33 and AtSR45 interact individually with AtU1-70K in a yeast two-hybrid assay (Golovkin and Reddy 1999).In this study, we found that MeRS40 interacted with MeU1-70Ka and MeU1-70Kbin vivoandinvitro(Fig.7), respectively, suggesting that MeRS40 and MeRS31 could participate in the formation of the U1 snRNP which is then involved in the splicing of pre-mRNA.However, whether MeRS40 interacts with other SR and SR-like proteins, or the U1 and U2 subunits of the spliceosome remains unknown, so more investigations are needed to elucidate the relationship between the RS subfamily and splicingrelated proteins in cassava.
5.Conclusion
Collectively, 37 RS subfamily genes from 11 plant species were identified, and the transcript levels of RS subfamily genes were systematically investigated under diverse abiotic stresses conditions.Furthermore, we reported a cassava salt stress-related gene,MeRS40, and its overexpression enhanced the tolerance to salt stress inArabidopsisby maintaining ROS homeostasis and upregulating stressrelated genes.However, overexpression ofMeRS40in cassava reduced the salt tolerance, which might be due to negative autoregulation of its own pre-mRNA.Furthermore, the MeRS40 protein interacted with MeU1-70Ks, suggesting that the MeRS40 protein may participate in the spliceosome assembly in cassava, although this requires further investigation.Therefore, we propose an important role ofMeRS40in response to salt stress, which helps us to understand the functions of SR proteins in the development and stress responses in cassava, and SR proteins may play a potential role in increasing the salt tolerance of cassava crops in the future.
Acknowledgements
This work was supported by grants from the Talent Program of Guangdong Academy of Sciences, China (2021GDASYL-20210103038, 2020GDASYL-2020102011, and 2021GDASYL-20210103036), the National Natural Science Foundation of China (32171292 and 32100294), the Guangdong Pearl River Talents Program, China (2021CX02N173), the China Postdoctoral Science Foundation (2020M682629), and the Zhanjiang Plan for Navigation, China (211207157080997).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available at https://doi.org/10.1016/j.jia.2023.04.003
 Journal of Integrative Agriculture2023年5期
Journal of Integrative Agriculture2023年5期
- Journal of Integrative Agriculture的其它文章
- Herbicidal activity and biochemical characteristics of the botanical drupacine against Amaranthus retroflexus L.
- Developing a duplex ARMS-qPCR method to differentiate genotype l and ll African swine fever viruses based on their B646L genes
- The effects of maltodextrin/starch in soy protein isolate–wheat gluten on the thermal stability of high-moisture extrudates
- Elucidation of the structure, antioxidant, and interfacial properties of flaxseed proteins tailored by microwave treatment
- Effects of planting patterns plastic film mulching on soil temperature, moisture, functional bacteria and yield of winter wheat in the Loess Plateau of China
- lnversion tillage with straw incorporation affects the patterns of soil microbial co-occurrence and multi-nutrient cycling in a Hapli-Udic Cambisol
