Long noncoding RNA TNFRSF10 A-AS1 promotes colorectal cancer through upregulation of HuR
Dan-Dan Wang, Dong-Lei Sun, Shao-Peng Yang, Jia Song, Meng-Yao Wu, Wei-Wei Niu, Mei Song, Xiao-LanZhang
Abstract
Key Words: Colorectal cancer; Long noncoding RNA; TNFRSF10 A-AS1 ; miR-3121 -3 p; HuR
INTRODUCTION
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and the third leading cause of cancer-related deaths around the world[1 ]. Despite marked advances in CRC diagnosis and therapy in recent years, the prognosis remains poor for many patients due to a lack of timely diagnosis and individualized disease management measures. The 5 -year survival rate for CRC patients with distant metastases is approximately 14 %[2 ]. Notably, CRC incidence and mortality rates are rising rapidly in developing countries[3 ]. In addition, epidemiological studies have shown an increasing incidence in adolescents and adults less than 50 years of age[4 ]. Genetic factors and poor lifestyle habits are the main predisposing factors for CRC, but the exact pathogenesis is unknown. Therefore, in-depth studies of the pathogenesis of CRC are of great clinical importance.
Recent evidence from studies of CRC-related pathogenesis has suggested that tumor-specific long noncoding RNAs (lncRNAs) may contribute to early diagnosis and prognostic assessment as well as improve disease treatment outcomes. LncRNAs are long (over 200 nucleotides) RNAs[5 ] that can interact with DNA, RNA and proteins. The function of lncRNAs mainly depends on their subcellular localization. Nuclear lncRNAs tend to bind DNA and proteins to regulate chromatin interactions,transcription and RNA processing, while cytoplasmic lncRNAs modulate mRNA stability or translation to influence cellular signaling cascades and can also act as competing endogenous RNAs (ceRNAs).CeRNAs competitively bind microRNA response elements (MREs) to regulate downstream gene expression[6 ,7 ]. In cells, ceRNAs containing the same MREs can competitively bind to the same microRNA (miRNA) and play posttranscriptional regulatory roles. Recent studies have shown that lncRNA/miRNA/mRNA mechanisms are involved in several aspects of CRC, including tumorigenesis,epithelial-mesenchymal transition (EMT), inflammatory processes and chemo-/radio-resistance[8 ]. For example, lncRNA SNHG6 serves as a ceRNA of miR-26 a/b and miR-214 , thereby regulating their common target, EZH2 , to promote CRC growth and metastasis[9 ]. However, the functions and mechanisms of a vast majority of CRC-associated lncRNAs remain unclear.
LncRNA TNFRSF10 A-AS1 is a novel lncRNA that has been demonstrated to be associated with the mechanism of autophagy in CRC, and Kaplan-Meier survival analysis has indicated that patients with high expression of TNFRSF10 A-AS1 have a better prognosis. However, TNFRSF10 A-AS1 expression in CRC cell lines is relatively high, which is inconsistent with the above findings[10 ]. Another study has identified five lncRNAs, including TNFRSF10 A-AS1 , by the expression of genome-wide lncRNAs in high-throughput RNA sequencing data, which may be useful to predict gastric cancer prognosis[11 ].The current evidence on TNFRSF10 A-AS1 is all based on The Cancer Genome Atlas (TCGA) data analysis, and the role of TNFRSF10 A-AS1 in tumors is inconsistent. Moreover, the exact mechanism of TNFRSF10 A-AS1 is unclear. Therefore, clinical and biological studies are needed to validate these findings. Our work is aimed to study the expression, functional roles and exact mechanisms of TNFRSF10 A-AS1 in CRC. In the present study, our findings suggested that TNFRSF10 A-AS1 was a novel oncogenic lncRNA that promoted CRC progression and might provide new ideas for CRC therapy.
MATERIALS AND METHODS
Database search
The expression of lncRNA TNFRSF10 A-AS1 in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) was analyzed using Gene Expression Profiling Interactive Analysis (GEPIA). GEPIA is an online analysis tool that is available to provide fast and customizable analysis based on TCGA data.We compared the expression of TNFRSF10 A-AS1 between CRC tumor and nontumor tissues by box plots, which used log2(TPM + 1 ) for log-scale (P < 0 .05 compared to control).
Patient tissue samples
We collected 40 pairs of surgically resected CRC tumor tissues and matched adjacent non-tumor tissue samples. These samples were obtained from patients with pathologically diagnosed CRC at the Second Hospital of Hebei Medical University. Patients were required to be free of radiotherapy, chemotherapy and other antitumor treatments prior to surgery, as well as free of any other malignant disease other than CRC. After isolation, some specimens were immediately placed in liquid nitrogen and stored at -80 °C for RNA and protein analysis, and other specimens were fixed in 4 % paraformaldehyde for histological examination. All patients who met the above criteria signed informed consent forms. The study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (2021 -R241 ).
Cell lines
Six human colon cancer cell lines (Caco-2 , DLD1 , HCT116 , HCT15 , HT29 and SW480 ) and one normal colonic epithelial cell line (FHC) were obtained from American Type Culture Collection (ATCC;Manassas, VA, United States). All cell lines were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, NY, United States), containing 10 % fetal bovine serum(FBS; Gibco BRL).
We conducted cell transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States),including small interfering RNA (siRNA), miRNA mimics, miRNA inhibitor and negative control oligonucleotides (Shanghai GenePharma Co., Ltd., Shanghai, China). The stable TNFRSF10 A-AS1 overexpression lentiviral vector, TNFRSF10 A-AS1 knockdown vector and empty vector were designed and synthesized by GenePharma. And finally, cell lines were screened according to the resistance carried by the lentivirus.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated from CRC tissue and cell samples, and then reverse transcribed into complementary DNA (cDNA), and finally quantitative real-time polymerase chain reaction (qRT-PCR) was conducted with cDNA and specific primers. Gene expression was calculated using GAPDH or U6 as internal reference genes. Table 1 lists all the qRT-PCR primer sequences involved in the study.
Cell counting kit-8 proliferation assay
Seeded colon cancer cells into 96 -well cell culture plates at 1000 cells/100 μL per well according to the experimental grouping. At 24 , 48 , 72 , 96 and 120 h, 10 μL of Cell Counting Kit-8 (CCK-8 ) reagent(Shanghai Share-bio Biotechnology Co., Shanghai, China) was added directly to each well. After incubation for 2 h at 37 °C, the optical density (OD) value (450 nm) was estimated.
Colony formation assay
Colon cancer cells were plated in 6 -well cell culture plates at 1000 cells per well according to the experimental grouping and routinely cultured in a cell incubator. Cell culture was terminated once colonies were visible. Cells were then washed, fixed, stained, photographed and counted.
Cell cycle and apoptosis assays
Cell cycle analysis was performed using a Cell Cycle and Apoptosis Analysis Kit (Beyotime Biotechnology Co., Shanghai, China) following the manufacturer’s protocol. Cells were fixed with ethanol,washed and stained with PI staining solution. Cell apoptosis was assayed using an Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology Co.), in which early and late apoptotic/dead cells were labeled using Annexin-V and PI, respectively.
Transwell assay
The upper chamber of Transwells (Corning, Kennebunk, ME, United States) was seeded with cells in 200 μL of culture medium without serum. The bottom chamber of Transwells was filled with medium supplemented with 10 % FBS. The invasion and migration characteristics assay were distinguished by the presence or absence of Matrigel in the upper chamber. Cells in the bottom chambers of Transwells were fixed, stained and photographed following twenty-four hours of routine culture.
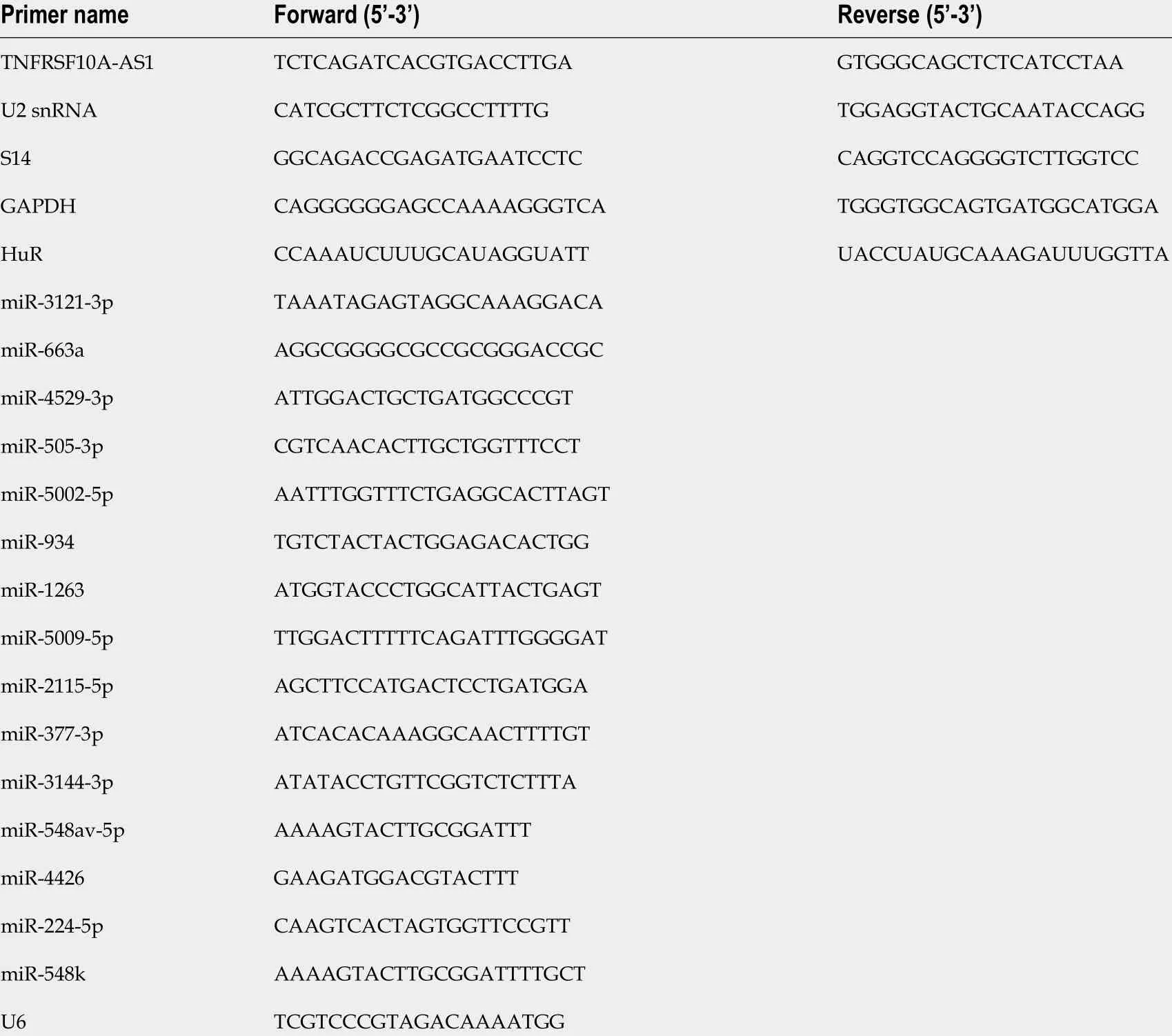
Table 1 Primers for real-time polymerase chain reaction
Western blot assay
Total protein from tissue and cell samples was extracted and the protein concentrations were tested.Different concentrations of polyacrylamide gels were prepared according to the molecular weight of the protein to be detected. Proteins were then electrophoresed and transferred to polyvinylidene fluoride(PVDF) membranes. Finally, the membranes were first incubated with primary antibodies against cyclin D1 , proliferating cell nuclear antigen (PCNA), cleaved caspase-3 , cleaved PARP, HuR as well as GAPDH (Cell Signaling Technology, Danvers, MA, United States) at 4 °C overnight and then the proteins were incubated with secondary antibodies (Cell Signaling Technology) for 1 h.
Immunohistochemistry
Tissue samples were fixed in paraformaldehyde, dehydrated in alcohol, embedded in paraffin as well as sectioned. In addition, immunohistochemical staining of tissue sections was carried out according to the experimental steps. Finally, staining of the sections was observed under a microscope.
Luciferase reporter assay
TNFRSF10 A-AS1 containing wild-type (WT) or mutant (MUT) miR-3121 -3 p-binding sequences was designed, synthesized and sub-cloned into the pmiRGLO vector (Shanghai GenePharma Co., Ltd.).DLD1 cells were cotransfected with reporter plasmid, miR-3121 -3 p mimics or mimics NC. After cotransfection under routine culture conditions for 48 h, the Dual-Luciferase Reporter Assay System (Promega,Madison, WI, United States) was used to evaluate the luciferase activity of all the above reporter vectors.
Subcutaneous xenograft models
We performed animal experiments forin vivovalidation. BALB/c nude mice at four- to six-week-old were subcutaneously injected with DLD1 cells stably transfected with sh-TNFRSF10 A-AS1 (sh-lnc) or sh-negative control vector (sh-NC) (1 × 107 , 200 μL) and HCT116 cells stably overexpressing TNFRSF10 A-AS1 (oe-lnc) or empty vector (control) (1 × 107 , 200 μL) into the right and left dorsal flanks of the nude mice, respectively. After implantation, the tumor volume was observed and measured every 2 d. The tumor volume was calculated using the measured length and width of the tumor according tothe formula: Tumor volume = (length × width2)/2 . At the end of the animal study, subcutaneous tumors were removed and used to assess tumor volume and weight as well as to perform qRT-PCR, Western blot and histological analyses.
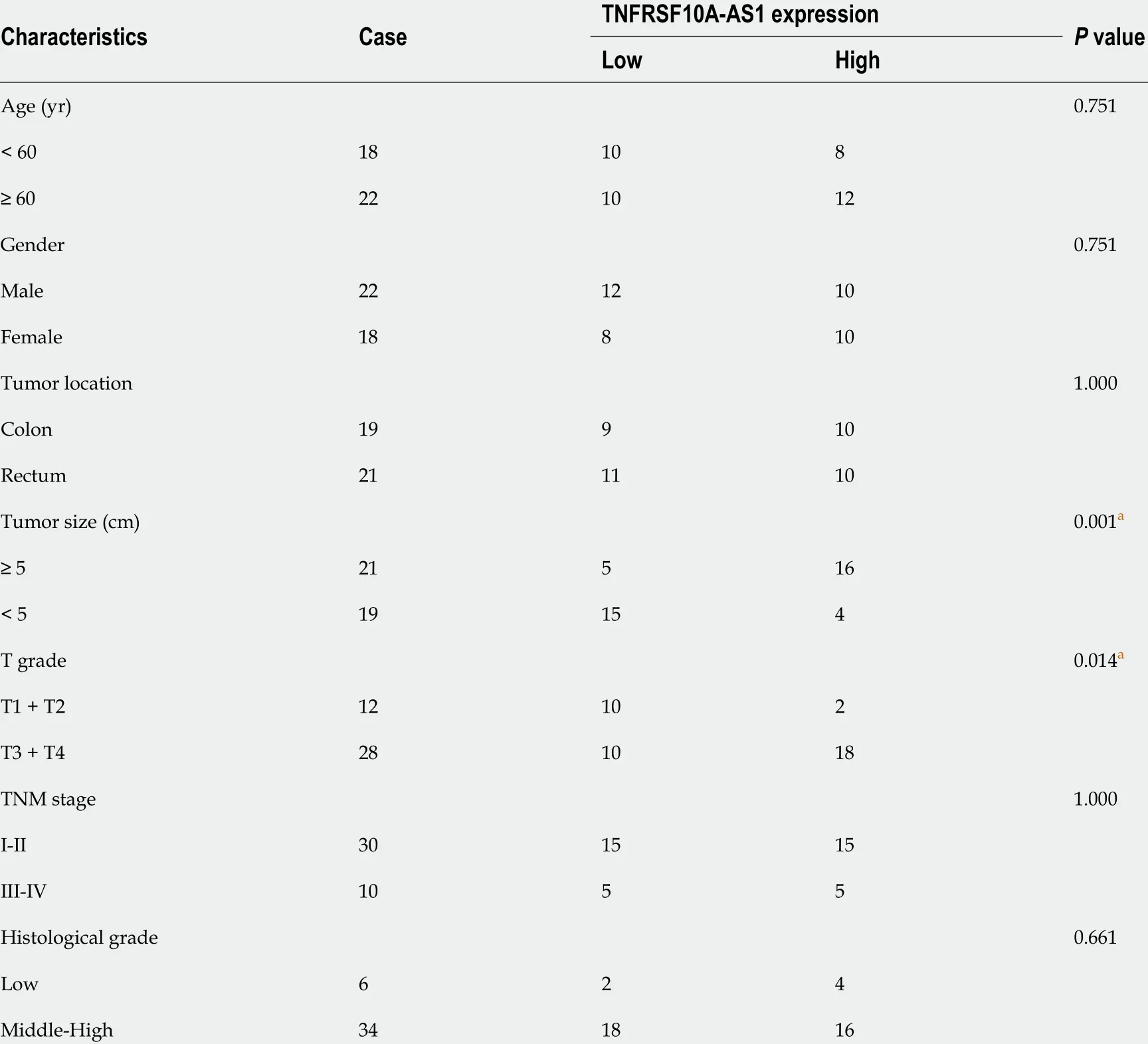
Table 2 Associations between TNFRSF10 A-AS1 levels and clinicopathological features in colorectal cancer patients
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 software or SPSS 22 .0 software. Data were presented as the mean ± SD. TNFRSF10 A-AS1 and miR-3121 -3 p expression levels in CRC patients were compared using the paired-samplettest. Statistical significance between groups was analyzed using theχ2test, Fisher’s exact probability, Student’s t test or one-way Analysis of Variance (ANOVA). P < 0 .05 was considered statistically significant.
RESULTS
TNFRSF1 0 A-AS1 is upregulated in CRC
To identify whether lncRNA TNFRSF10 A-AS1 is differentially expressed in CRC, we first analyzed TNFRSF10 A-AS1 expression in TCGA database. We used the GEPIA online database to analyze TNFRSF10 A-AS1 expression of CRC tissues, and results showed that TNFRSF10 A-AS1 was expressed higher in both colon and rectal carcinoma tissues than that of corresponding adjacent normal tissues(Figure 1 A). We then examined TNFRSF10 A-AS1 expression in 40 pairs of CRC tissue samples.TNFRSF10 A-AS1 expression was significantly increased in 34 of 40 (75 %) tumor samples compared to the adjacent normal mucosa tissues (Figure 1 B). Next, TNFRSF10 A-AS1 expression was verified in multiple colon cancer cell lines, namely, DLD1 , HCT15 , HT29 , HCT116 , SW480 and Caco-2 , and human colon mucosal epithelial FHC cells. TNFRSF10 A-AS1 was significantly upregulated in the colon cancer cell lines compared to FHC cells (Figure 1 D). RT-PCR also confirmed that TNFRSF10 A-AS1 was upregulated in CRC tissues and cells (Figure 1 C). We further explored the cellular localization of TNFRSF10 A-AS1 in DLD1 and HCT116 cells. qRT-PCR results showed that TNFRSF10 A-AS1 was predominantly found in the cytoplasm (Figure 1 E), which indicated that TNFRSF10 A-AS1 might function in the cytoplasm. In addition, we analyzed associations between TNFRSF10 A-AS1 and clinicopathological features of CRC patients and found that the upregulation of TNFRSF10 A-AS1 was associated with advanced T grade and tumor size (Table 2 ). Collectively, the above findings indicated that TNFRSF10 A-AS1 was increased in CRC and might be involved in colorectal carcinogenesis as an oncogene.
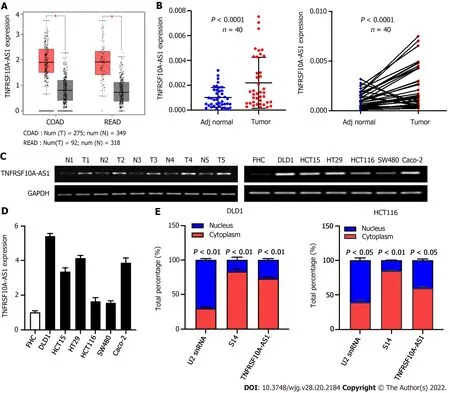
Figure 1 TNFRSF10 A-AS1 is upregulated in colorectal cancer. A: TNFRSF10 A-AS1 was upregulated in tumor tissues compared to normal tissues in TCGA colorectal cancer (CRC) database; B: TNFRSF10 A-AS1 was overexpressed in CRC tumor tissues compared to matched adjacent nontumor tissues (n = 40 );C: Levels of TNFRSF10 A-AS1 in CRC were measured by reverse transcription-polymerase chain reaction (RT-PCR); D: TNFRSF10 A-AS1 was higher in colon cancer cell lines than in colon mucosal epithelial FHC cells; E: The expression levels of TNFRSF10 A-AS1 in the cytoplasm and nucleus of DLD1 and HCT116 cells were detected by quantitative real-time polymerase chain reaction (qRT-PCR). U2 snRNA was used as the internal reference for genes expressed in the nucleus, and S14 was used as the internal reference for genes expressed in the cytoplasm. COAD: Colon adenocarcinoma; READ: Rectum adenocarcinoma.
TNFRSF1 0 A-AS1 promotes colon cancer cell proliferation, migration and invasion as well as inhibits cell apoptosis
To explore the biological function of TNFRSF10 A-AS1 in colon cancer cells, we first designed a siRNA against TNFRSF10 A-AS1 , a short hairpin RNA (shRNA) against TNFRSF10 A-AS1 and a lentiviral overexpression vector for TNFRSF10 A-AS1 . Silencing TNFRSF10 A-AS1 in DLD1 and HT29 cells significantly inhibited cell viability and clonogenicity, whereas ectopic expression of TNFRSF10 A-AS1 in HCT116 and SW480 cells significantly promoted cell viability and clonogenicity (Figure 2 A and B).We also conducted flow cytometry to clarify the role of TNFRSF10 A-AS1 in cell cycle distribution.Knockdown of TNFRSF10 A-AS1 led to a significant increase in G1 phase cells and a decrease in S-phase cells in both DLD1 and HT29 cells. In contrast, the opposite results were exhibited in both HCT116 and SW480 cells with overexpression of TNFRSF10 A-AS1 (Figure 2 C). Moreover, Western blot analysis revealed that downregulation of TNFRSF10 A-AS1 decreased cyclin D1 and PCNA expression, while the upregulation of TNFRSF10 A-AS1 increased cyclin D1 and PCNA expression (Figure 2 E). In general,genes that promote proliferation can suppress cell apoptosis. Therefore, we conducted Annexin VAPC/PI staining and Western blot analysis to test the impact of TNFRSF10 A-AS1 on the apoptosis of colon cancer cells. The results showed that downregulation of TNFRSF10 A-AS1 increased the apoptosis rate and the expression of the cleaved forms of caspase-3 and PARP, which are markers of cell apoptosis, whereas upregulation of TNFRSF10 A-AS1 led to the opposite effects (Figure 2 D and E).
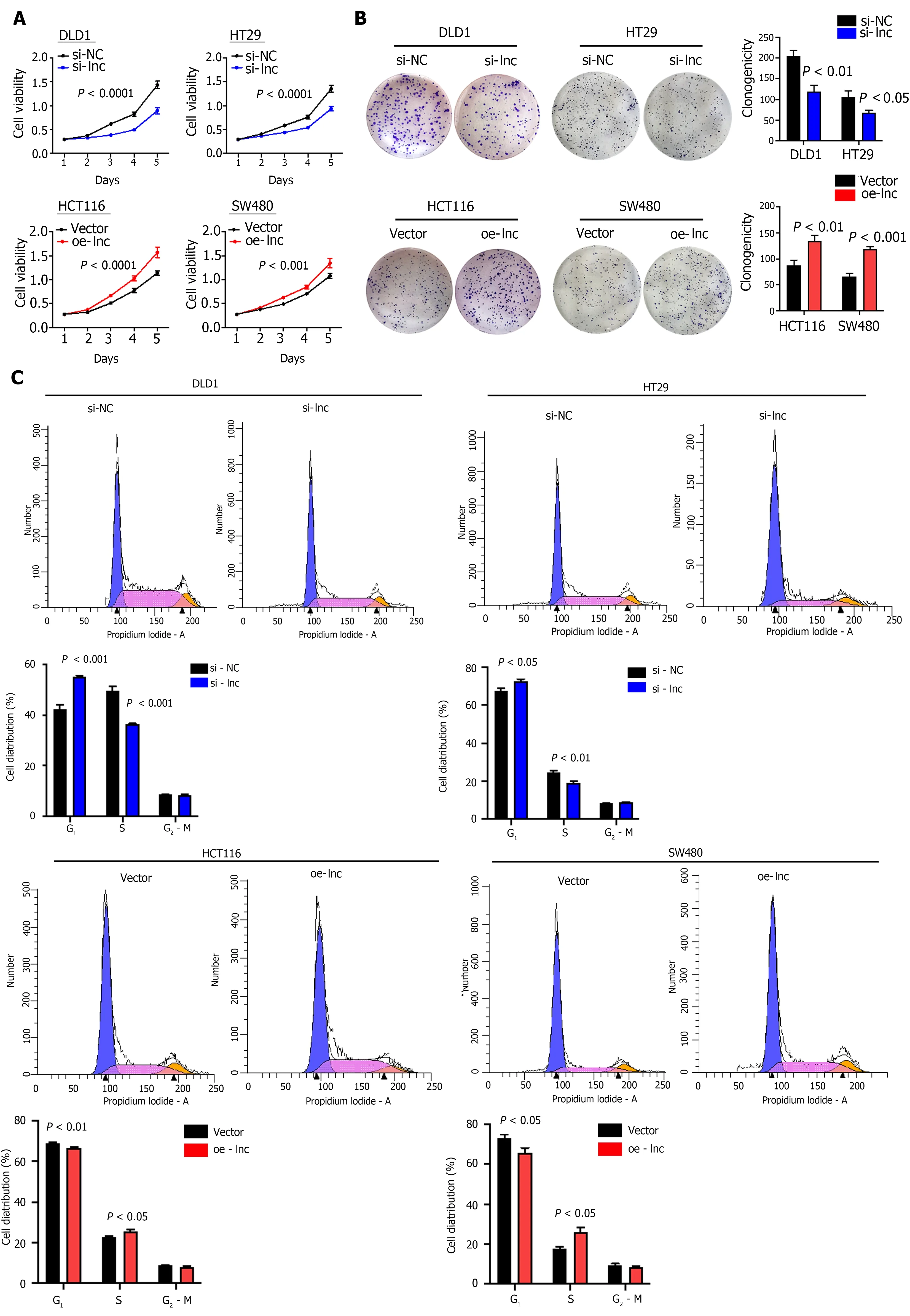
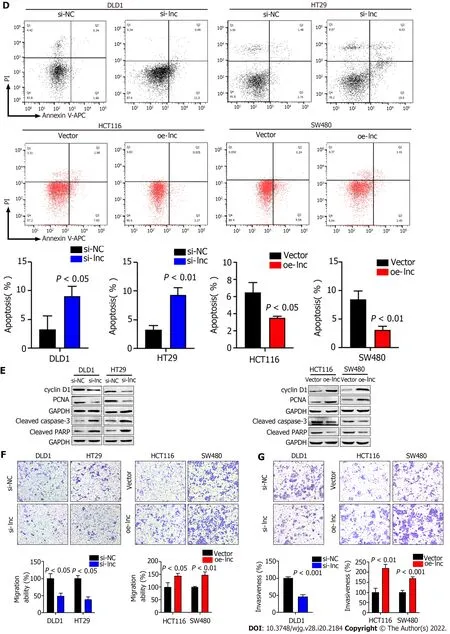
Figure 2 TNFRSF10 A-AS1 promotes colon cancer cell proliferation, migration and invasion as well as inhibits cell apoptosis in vitro. A and B: Downregulation of TNFRSF10 A-AS1 decreased cell viability and clonogenicity in DLD1 and HT29 cells, whereas upregulation of TNFRSF10 A-AS1 greatly increased cell viability and clonogenicity in HCT116 and SW480 cells; C: Silencing TNFRSF10 A-AS1 arrested cells at the G1 /S transition, whereas overexpressing TNFRSF10 A-AS1 facilitated this transition; D: Downregulation of TNFRSF10 A-AS1 enhanced cell apoptosis, whereas upregulation of TNFRSF10 A-AS1 inhibited cell apoptosis; E: Western blot analysis showed that TNFRSF10 A-AS1 knockdown decreased the expression of cyclin D1 and PCNA but increased the activation of caspase-3 and PARP, whereas TNFRSF10 A-AS1 overexpression had the opposite effect; F and G: Silencing TNFRSF10 A-AS1 suppressed DLD1 and HT29 cell migration and invasion abilities, whereas these abilities were significantly enhanced by overexpressing TNFRSF10 A-AS1 in HCT116 and SW480 cells.
In addition to malignant proliferation, tumor cells also exhibit migration and invasion abilities.Therefore, we performed Transwell assays with or without Matrigel to explore the impact of TNFRSF10 A-AS1 on the migration and invasion properties of colon cancer cells. Downregulation of TNFRSF10 A-AS1 markedly suppressed the migration and invasion abilities of DLD1 and HT29 cells,whereas overexpressing TNFRSF10 A-AS1 significantly enhanced the abilities of HCT116 and SW480 cells (Figure 2 F and G). Therefore, these findings demonstrated that TNFRSF10 A-AS1 exerted a cancerpromoting role in CRC, prompting further investigations of its potential regulatory mechanisms.
miR-3 121 -3 p is downregulated in CRC
The above findings suggested that TNFRSF10 A-AS1 might play a tumor-promoting role in the cytoplasm. Thus, we explored the potential molecular mechanism of TNFRSF10 A-AS1 based on the ceRNA mechanism. We first predicted potential target miRNAs of TNFRSF10 A-AS1 by using miRNA target prediction tools, including miRDB, DIANA and LncRNAMAP. The prediction results of the three databases yielded 15 candidate miRNAs (Figure 3 A). Among them, miR-3121 -3 p expression was upregulated in TNFRSF10 A-AS1 -deficient cells and downregulated in TNFRSF10 A-AS1 -overexpressing cells, consistent with the ceRNA mechanism theory (Figure 3 B). Subsequently, we performed qRT-PCR to evaluate miR-3121 -3 p expression in CRC tissues, which indicated significant downregulation of miR-3121 -3 p in colorectal tumor tissues compared to matched nontumor tissues (Figure 3 C). We further confirmed the interaction between TNFRSF10 A-AS1 and miR-3121 -3 p using a dual-luciferase reporter assay (Figure 3 E). Luciferase activity was reduced in cells cotransfected with TNFRSF10 A-AS1 -WT and the miR-3121 -3 p mimics, whereas there was no difference between cells cotransfected with TNFRSF10 AAS1 -MUT and the miR-3121 -3 p mimics and cells cotransfected with TNFRSF10 A-AS1 -MUT and mimics NC (Figure 3 F). Finally, we analyzed the correlation between miR-3121 -3 p levels and the clinicopathological features of CRC patients and discovered that downregulation of miR-3121 -3 p was associated with large tumor size (Table 3 ). These results suggested that miR-3121 -3 p might be a tumor suppressor and exert an inhibitory effect on CRC.
miR-3 121 -3 p inhibits the proliferation, migration and invasion of colon cancer cells by silencing HuR
Because there are few studies related to miR-3121 -3 p and no relevant studies in CRC, we set out to investigate the function and mechanism of miR-3121 -3 p in CRC. We first predicted the downstream proteins of miR-3121 -3 p using four online databases, i.e., miRDB, TargetScan, miRTarBase and RNA22 ,and then obtained 3 candidate genes (ELAVL1 , PLEKHA6 and ATL3 ) after taking the intersection(Figure 4 A). Of the three candidate genes, ELAVL1 (HuR) has been demonstrated to be involved in CRC progression and may act as an oncogene in CRC. qRT-PCR showed that the expression of miR-3121 -3 p was relatively lower in DLD1 cells and higher in HCT116 cells (Figure 3 D). Thus, we selected DLD1 cells and HCT116 cells for transfection with miR-3121 -3 p mimics and inhibitor, respectively, to further identify the effects of miR-3121 -3 p on the malignant phenotype of CRC. Experimental results at both the mRNA and protein levels indicated that upregulation of miR-3121 -3 p decreased HuR expression,whereas downregulation of miR-3121 -3 p increased HuR expression (Figure 4 B), suggesting that HuR might be a downstream target protein of miR-3121 -3 p.
We next studied the impacts of miR-3121 -3 p on the biological function of colon cancer cells. Growth curves based on the CCK-8 assay displayed that upregulation of miR-3121 -3 p suppressed the proliferation and viability of DLD1 cells, whereas downregulation of miR-3121 -3 p enhanced the proliferation and viability of HCT116 cells (Figure 4 C). The colony number of DLD1 cells transfected with the miR-3121 -3 p mimics was significantly lower than that of the control group, whereas the colony number of HCT116 cells transfected with the miR-3121 -3 p inhibitor was significantly higher than the control group(Figure 4 D). In addition, Transwell assays indicated that upregulation of miR-3121 -3 p in DLD1 cells significantly impaired the migration and invasion abilities, whereas downregulation of miR-3121 -3 p in HCT116 cells significantly enhanced these abilities (Figure 4 E). In summary, the above results confirmedthat miR-3121 -3 p played a suppressive effect in the malignant phenotype of CRC, including aberrant proliferation and metastasis.
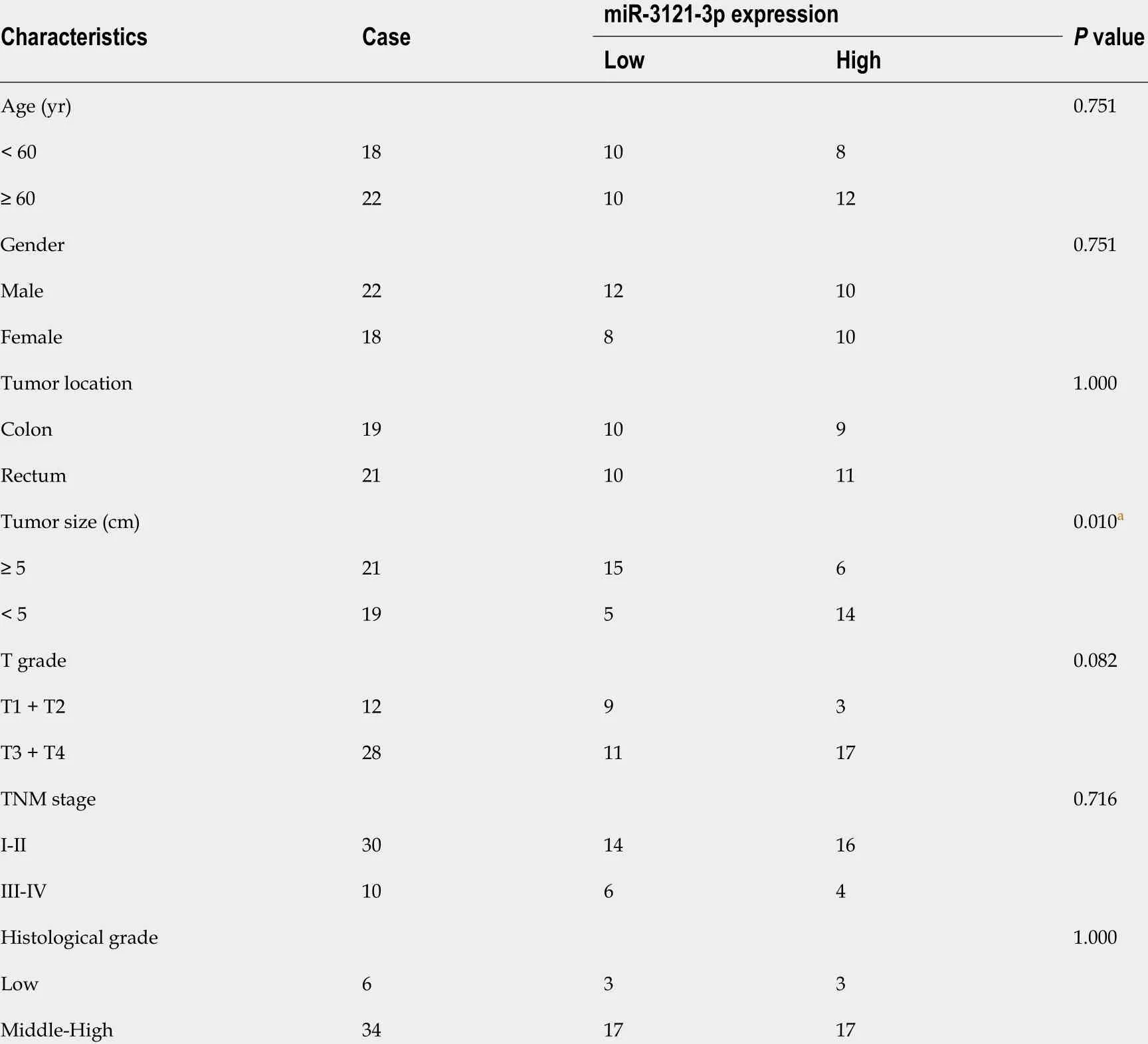
Table 3 Associations between miR-3121 -3 p levels and clinicopathological features in colorectal cancer patients
TNFRSF1 0 A-AS1 accelerates the proliferation, migration and invasion of colon cancer cells via the miR-3 121 -3 p/HuR axis
We performed rescue experiments using miR-3121 -3 p mimics and inhibitor with the aim of verifying whether TNFRSF10 A-AS1 exerts a tumor-promoting effect through the miR-3121 -3 p/HuR axis. We first examined HuR expression at the mRNA and protein levels. Silencing TNFRSF10 A-AS1 decreased the mRNA and protein levels of HuR in DLD1 cells, whereas overexpressing TNFRSF10 A-AS1 in HCT116 cells enhanced HuR expression. In addition, the miR-3121 -3 p inhibitor and mimics reversed the regulatory effects on HuR expression caused by TNFRSF10 A-AS1 silencing and overexpression,respectively (Figure 5 A). Furthermore, we sought to clarify whether the miR-3121 -3 p mimics and inhibitor reverse the impacts of TNFRSF10 A-AS1 on CRC biological functions. All the experiments results, including CCK-8 , colony formation as well as Transwell assays, indicated that the suppressive impacts on DLD1 cells caused by TNFRSF10 A-AS1 knockdown could be balanced by miR-3121 -3 p inhibitor, while miR-3121 -3 p mimics could counteract the promoting effect of TNFRSF10 A-AS1 overexpression on HCT116 cells (Figure 5 B-E). The above results indicated that the miR-3121 -3 p/HuR regulatory axis might be one of the mechanisms by which TNFRSF10 A-AS1 promoted CRC proliferation, migration and invasion.
TNFRSF1 0 A-AS1 promotes CRC growth in vivo
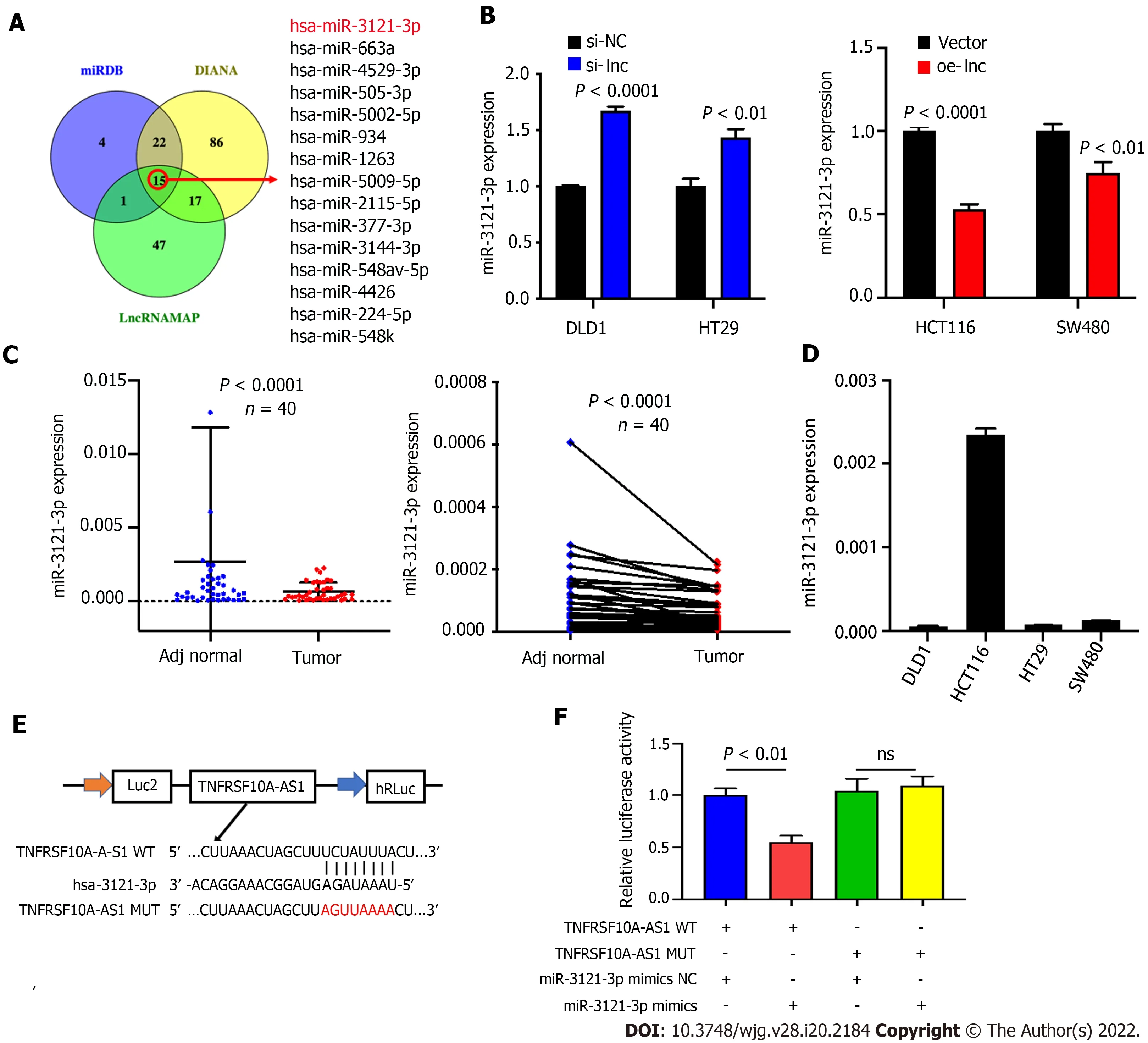
Figure 3 miR-3121 -3 p is downregulated in colorectal cancer. A: We used three websites (miRDB, DIANA and LncRNAMAP) to predict 15 microRNAs with binding sites for TNFRSF10 A-AS1 ; B: miR-3121 -3 p expression was increased in cells transfected with si-lnc and decreased in cells transfected with oe-lnc; C: miR-3121 -3 p was downregulated in colorectal cancer tumor tissues compared to matched nontumor tissues (n = 40 ); D: Levels of miR-3121 -3 p in colon cancer cell lines were detected by quantitative real-time polymerase chain reaction; E: Schematic illustration of the predicted binding sites of miR-3121 -3 p in the TNFRSF10 A-AS1 sequence; F: Relative luciferase activities after cotransfection with TNFRSF10 A-AS1 -WT, TNFRSF10 A-AS1 -MUT, miR-3121 -3 p mimics or NC in DLD1 cells. WT:Wild-type; MUT: Mutant.
Given that the abovein vitroexperimental results suggested that TNFRSF10 A-AS1 had a tumorpromoting effect on colon cancer cells, we selected TNFRSF10 A-AS1 -deficient DLD1 cells and TNFRSF10 A-AS1 -overexpressing HCT116 cells for tumor xenograft experiments to verify the effect of TNFRSF10 A-AS1in vivo. Consistent with the results ofin vitroexperiments, we found that the downregulation of TNFRSF10 A-AS1 significantly reduced the tumor size and weight of DLD1 xenografts(Figure 6 A), whereas upregulation of TNFRSF10 A-AS1 markedly promoted HCT116 xenograft growth(Figure 6 B). Moreover, IHC assays demonstrated that downregulation of TNFRSF10 A-AS1 reduced the expression of the Ki-67 proliferation marker in xenograft tumor tissues, whereas upregulation of TNFRSF10 A-AS1 produced the opposite effect (Figure 6 C). qRT-PCR, Western blot and IHC analyses all revealed that HuR expression was downregulated in TNFRSF10 A-AS1 -deficient xenograft tumor tissues but upregulated in TNFRSF10 A-AS1 -overexpressing xenograft tumor tissues (Figure 6 C-E). Western blot analysis of tissue samples from CRC patients also showed that HuR was increased in tumor tissues compared to paired nontumor tissues (Figure 6 F). Therefore, thesein vivoresults further supported the tumor-promoting role of TNFRSF10 A-AS1 in CRC, which functioned at least in part as a molecular sponge of miR-3121 -3 p to upregulate HuR expression.
DISCUSSION
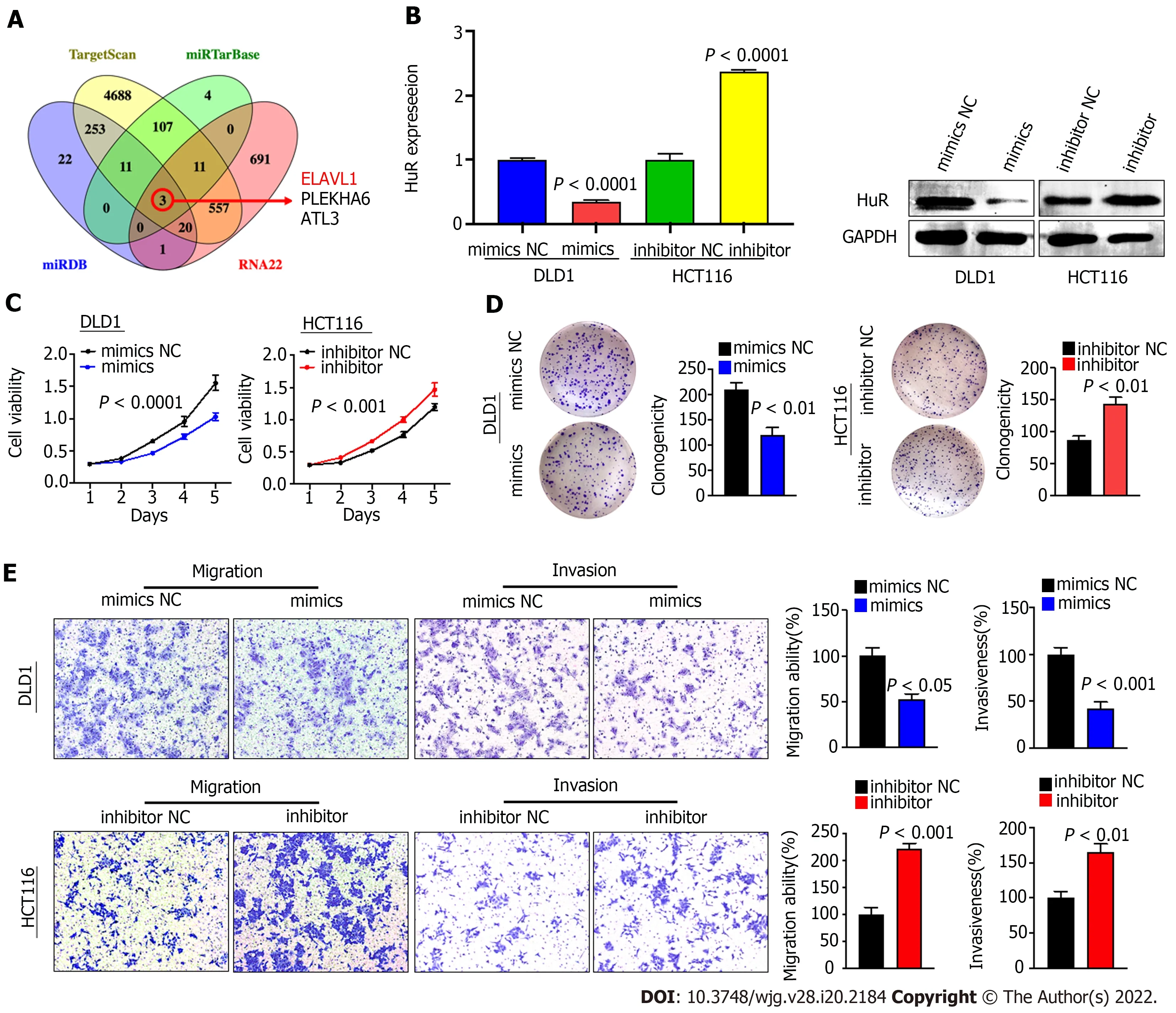
Figure 4 miR-3121 -3 p suppresses colon cancer cell proliferation, migration and invasion in vitro. A: Four websites (miRDB, TargetScan,miRTarBase and RNA22 ) were used to predict 3 downstream target mRNAs with binding sites for miR-3121 -3 p; B: HuR expression was assessed in DLD1 and HCT116 cells transfected with miR-3121 -3 p mimics, miR-3121 -3 p inhibitor or NC using quantitative real-time polymerase chain reaction and Western blot assays; C and D: miR-3121 -3 p mimics inhibited cell viability and clonogenicity, but miR-3121 -3 p inhibitor enhanced cell viability and clonogenicity; E: Upregulation of miR-3121 -3 p inhibited cell migration and invasion capabilities, whereas downregulation of miR-3121 -3 p enhanced these capabilities.
In this study, we first uncovered TNFRSF10 A-AS1 as an oncogene that participated in colorectal carcinogenesis and progression. Although the available findings have demonstrated that TNFRSF10 AAS1 is involved in the autophagic mechanism of CRC as well as gastric cancer prognosis, the current evidence is all based on the analysis of TCGA data, and its functional roles and detailed mechanisms in CRC cannot be determined. Our study showed that TNFRSF10 A-AS1 was upregulated in CRC and the upregulation of TNFRSF10 A-AS1 was associated with advanced tumor size and T grade. In functional experiments bothin vitroandin vivo, we found that TNFRSF10 A-AS1 enhanced the malignant phenotype of colon cancer cells. Regarding the molecular mechanism of TNFRSF10 A-AS1 , we identified miR-3121 -3 p by bioinformatics analyses and verified the direct interaction between TNFRSF10 A-AS1 and miR-3121 -3 p by dual-luciferase reporter assays. In contrast to TNFRSF10 A-AS1 , miR-3121 -3 p was downregulated in CRC, negatively associated with the tumor size as well as exerted inhibitory effects on the malignant phenotype of colon cancer cells, which is consistent with ceRNA mechanism theory.Mechanistically, one of the ways in which TNFRSF10 A-AS1 exerted its pro-cancer effects was through sponging miR-3121 -3 p. By binding to miR-3121 -3 p, TNFRSF10 A-AS1 attenuated the silencing effect of miR-3121 -3 p on its downstream target gene, HuR. Taken together, our findings provide evidence for an important role of TNFRSF10 A-AS1 and the TNFRSF10 A-AS1 /miR-3121 -3 p/HuR axis in CRC progression, thereby providing a theoretical basis for the diagnosis and treatment of CRC.
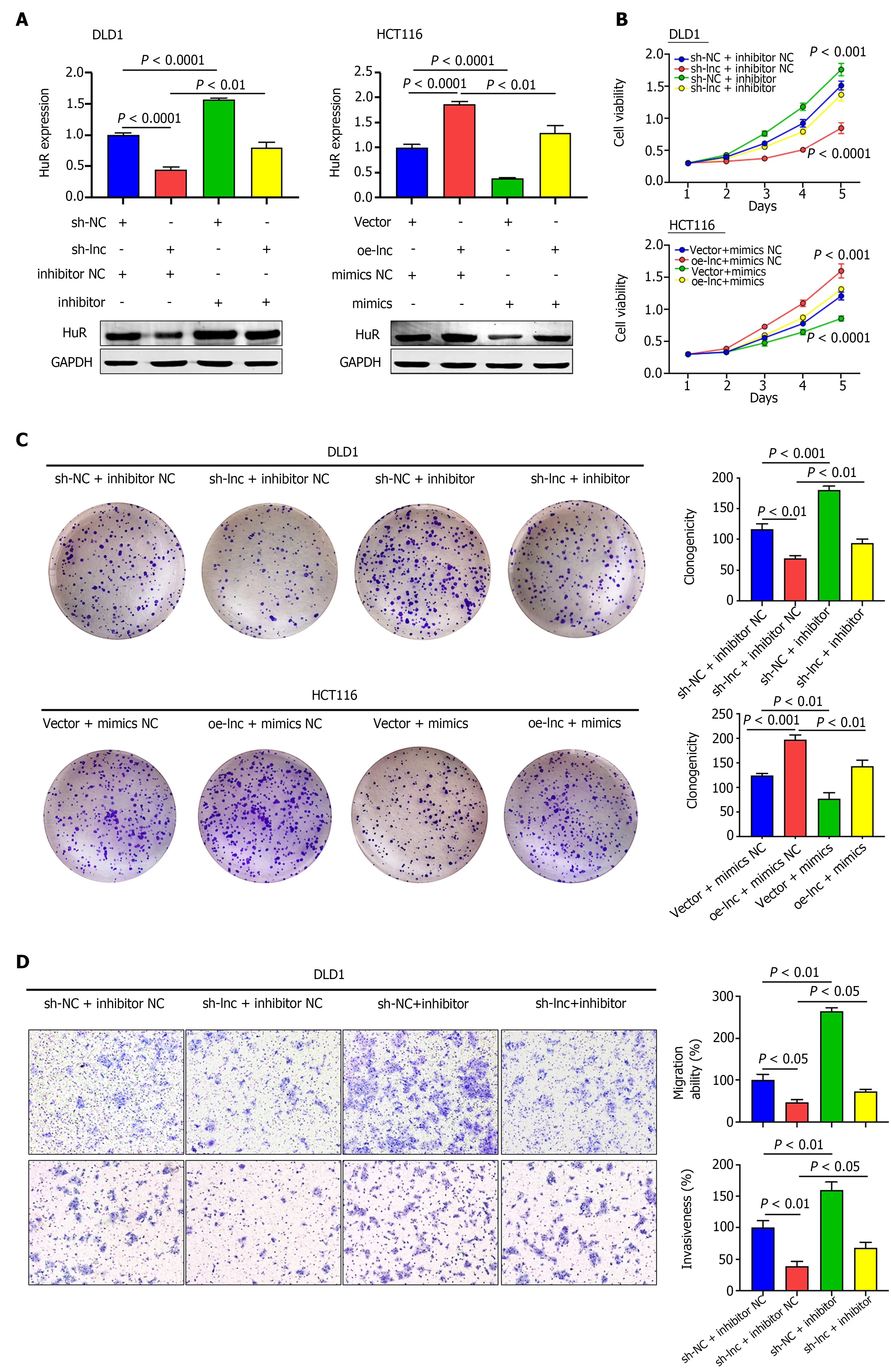
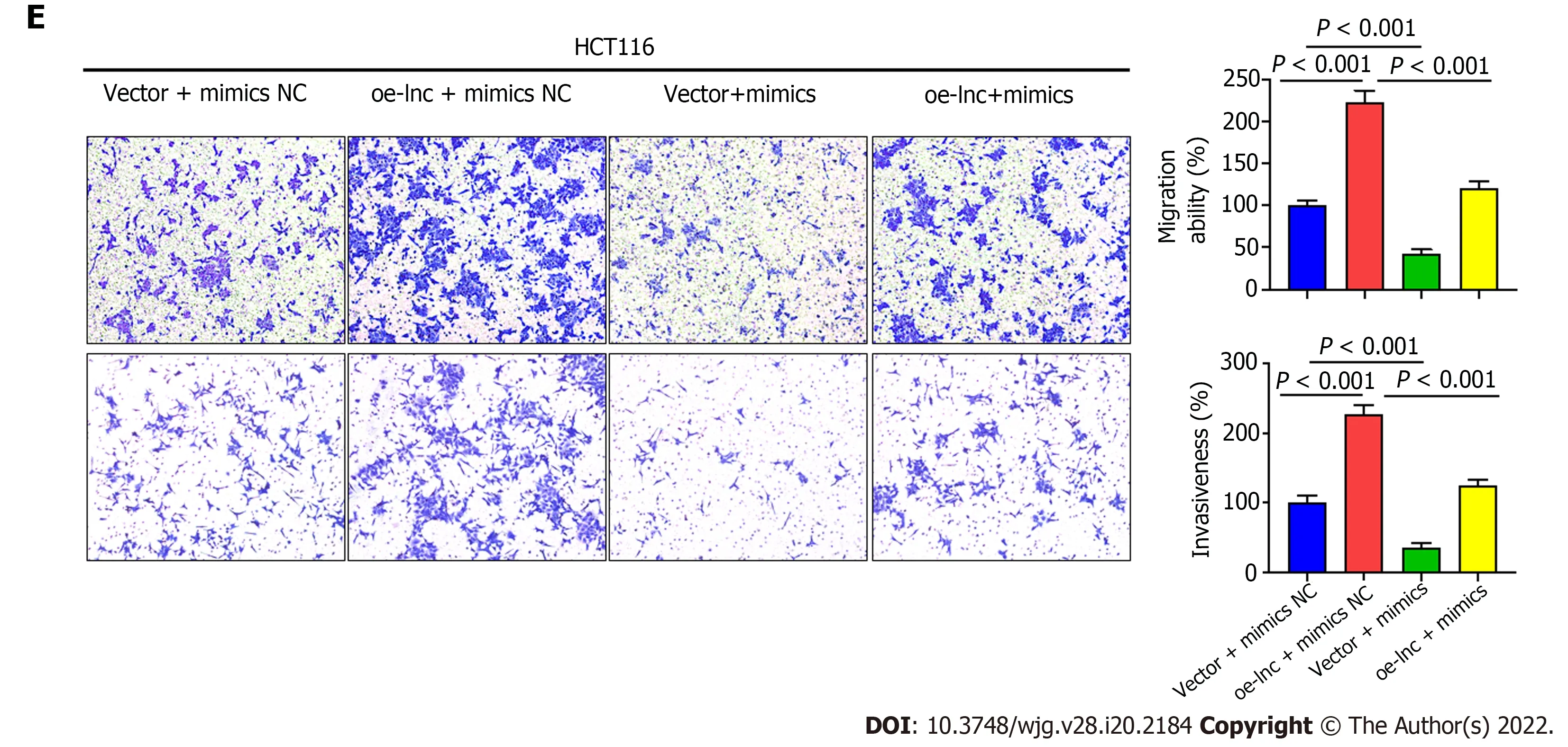
Figure 5 TNFRSF10 A-AS1 promotes colon cancer cell proliferation, migration and invasion via the miR-3121 -3 p/HuR axis. A: HuR was evaluated in DLD1 and HCT116 cells transfected with miR-3121 -3 p mimics, miR-3121 -3 p inhibitor, NC, sh-lnc or oe-lnc; B and C: Cell proliferation abilities were estimated using Counting Kit-8 and colony formation assays in DLD1 and HCT116 cells transfected with miR-3121 -3 p mimics, miR-3121 -3 p inhibitor, NC, sh-lnc or oe-lnc; D and E: Transwell assays were performed to analyze cell migration and invasion abilities in DLD1 and HCT116 cells transfected with miR-3121 -3 p mimics,miR-3121 -3 p inhibitor, NC, sh-lnc or oe-lnc.
ceRNA is an important regulatory mode of cytoplasmic lncRNA. Here, after confirming that TNFRSF10 A-AS1 was more abundant in the cytoplasm of CRC cell lines, we used online tools to predict potential target miRNAs of TNFRSF10 A-AS1 . We further validated the expression relationship between TNFRSF10 A-AS1 and candidate miRNAs, and we discovered that miR-3121 -3 p might be a downstream target miRNA of TNFRSF10 A-AS1 . And the dual-luciferase reporter assay further confirmed that TNFRSF10 A-AS1 might function as the molecular sponge of miR-3121 -3 p. In the lncRNA-miRNAmRNA network, miRNAs, as intermediate mediators, exert biological effects by modulating downstream target proteins. After experimental evidence established that TNFRSF10 A-AS1 could sponge miR-3121 -3 p, we explored its effector genes. Among candidate target genes predicted by target prediction software, only HuR has been shown previously to regulate CRC progression, and our results also verified that miR-3121 -3 p exerted a negative regulatory effect on HuR expression. Rescue experiments also confirmed that miR-3121 -3 p mimics and inhibitor counteracted the regulatory effects of TNFRSF10 A-AS1 overexpression and knockdown, respectively, on HuR expression, cell proliferation,migration ability and invasion ability. Thus, one of the mechanisms by which TNFRSF10 A-AS1 acts may be as a molecular sponge for miR-3121 -3 p, which upregulates HuR expression and ultimately promotes colorectal carcinogenesis and progression.
HuR, a cancer-associated RNA-binding protein (RBP), is a member of the embryonic lethal abnormal vision (ELAV) family, and it increases mRNA stability by binding to conserved AU-rich elements(AREs) within 3 ’ untranslated regions (UTRs), thereby preventing gene degradation[12 ]. HuR expression is upregulated in different tumors, including breast cancer, CRC, gastric cancer and prostate cancer, and it is correlated with advanced clinicopathological parameters, expression of tumorassociated proteins, a low survival rate and poor prognosis[13 -16 ]. CRC-related studies show that HuR is upregulated and promotes colon cancer growth by targeting RNA in the cytoplasm[17 ]. In addition,increased HuR protein in the cytoplasm is correlated with T stage, and importantly, HuR overexpression increases the growth of colon cancer cells in nude mouse models[18 ]. A recent study has revealed that miR-22 , as a tumor suppressive miRNA, directly binds to HuR and downregulates its expression to inhibit proliferation and migration abilities of CRC, as well as xenogeneic tumor growth[19 ]. Through our experimental research, we found that the upstream lncRNA TNFRSF10 A-AS1 sponged an important tumor suppressive miRNA in CRC, miR-3121 -3 p, which negatively regulated HuR expression. Consistent with previous studies, HuR was highly expressed in CRC. Our study results further suggested that the expression of HuR might be positively regulated by the upstream molecular TNFRSF10 A-AS1 , which might be one of the mechanisms by which TNFRSF10 A-AS1 exerted a pro-cancer effect in CRC.
Based on the results of comprehensive analyses, including bioinformatics analyses, cell functional experiments and clinical research tools, we have sufficient evidence that TNFRSF10 A-AS1 is a novel oncogene that promotes tumor malignancy in CRC. Mechanistically, TNFRSF10 A-AS1 regulates the expression of the downstream target, HuR, by sponging miR-3121 -3 p, which further promotes CRC progression. Targeting the TNFRSF10 A-AS1 /miR-3121 -3 p/HuR axis may have potential therapeutic implications for CRC.
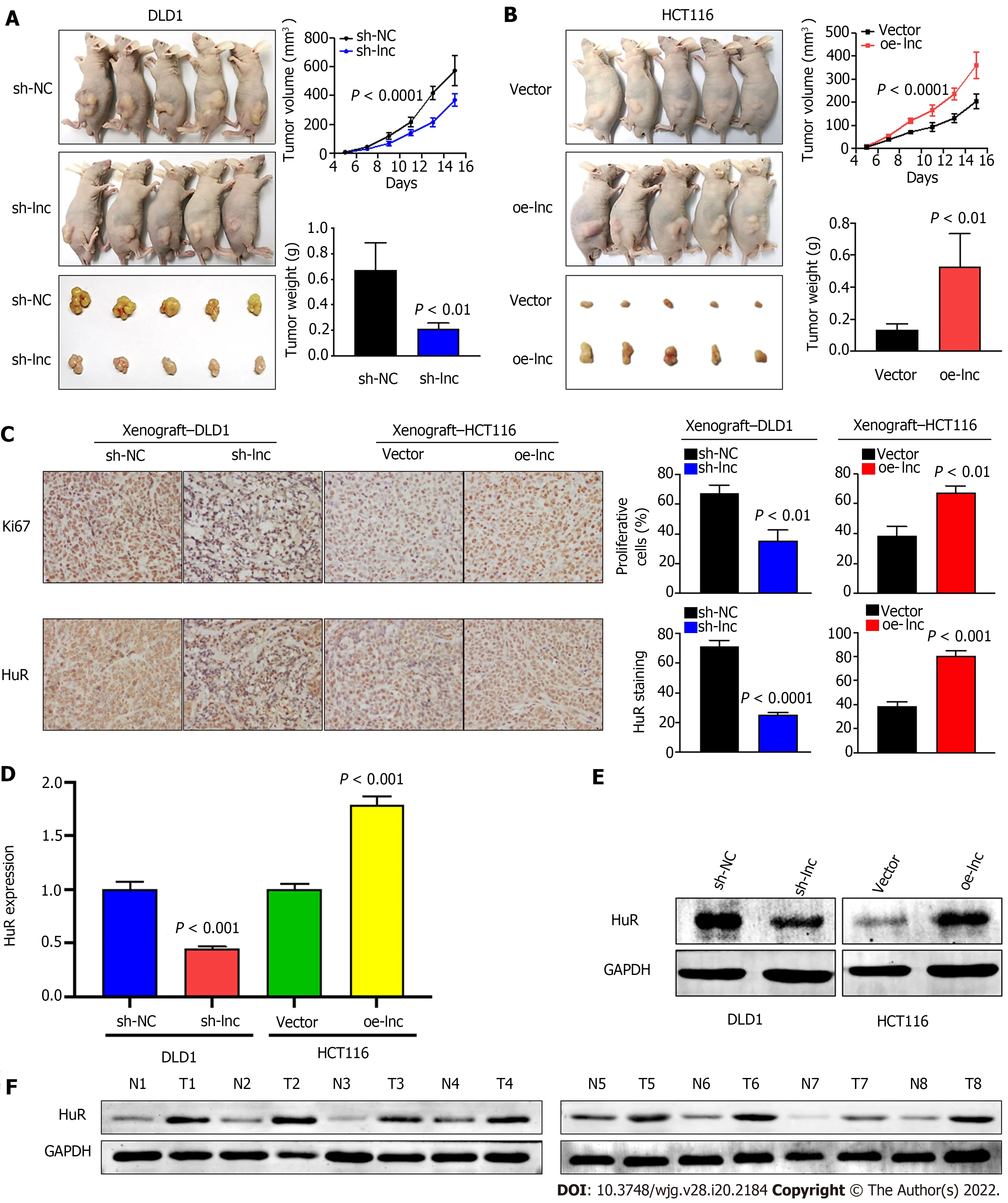
Figure 6 TNFRSF10 A-AS1 promotes the growth of colorectal cancer in vivo. A and B: Tumor volume and weight were significantly decreased in the TNFRSF10 A-AS1 knockdown group but markedly increased in the TNFRSF10 A-AS1 overexpression group; C-E: TNFRSF10 A-AS1 enhanced cell proliferation (as assessed by Ki67 staining) and increased HuR expression (as assessed by HuR staining, quantitative real-time polymerase chain reaction and Western blot) in xenograft tumor tissues; F: HuR was upregulated in colorectal cancer patient tumor tissues compared to matched nontumor tissues by Western blot analysis.
CONCLUSION
TNFRSF10 A-AS1 as a ceRNA promotes tumor malignancy of CRC through the miR-3121 -3 p/HuR axis.TNFRSF10 A-AS1 /miR-3121 -3 p/HuR axis may provide more effective targets for CRC therapy.
ARTICLE HIGHLIGHTS

ACKNOWLEDGEMENTS
The authors acknowledge the support of Dr. Jun Yu (The Chinese University of Hong Kong) for supervising this study and revising the manuscript.
FOOTNOTES
Author contributions:Wang DD, Sun DL and Zhang XL conceived, designed the study; Wang DD, Sun DL, Yang SP and Song J performed most experiments, analyzed the data, wrote the manuscript and edited the paper; Sun DL and Zhang XL helped to supervise the study; Wu MY, Niu WW and Song M helped to perform the experiments and analyzed the data; Sun DL and Zhang XL helped to edit the paper.
Institutional review board statement:The study was reviewed and approved by the Ethics Committee of the Second Hospital of Hebei Medical University (No. 2021 -R241 ).
Institutional animal care and use committee statement:All procedures involving animals were reviewed and approved by the Ethics Committee of the Second Hospital of Hebei Medical University (No. 2021 -AE011 ).
Conflict-of-interest statement:There are no conflicts of interest to report.
Data sharing statement:No additional data are available.
ARRIVE guidelines statement:The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4 .0 ) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4 .0 /
Country/Territory of origin:China
ORCID number:Dan-Dan Wang 0000 -0001 -7809 -7429 ; Dong-Lei Sun 0000 -0002 -7930 -7544 ; Shao-Peng Yang 0000 -0002 -1322 -1609 ; Jia Song 0000 -0003 -4291 -6837 ; Meng-Yao Wu 0000 -0003 -0510 -8654 ; Wei-Wei Niu 0000 -0002 -9243 -3625 ; Mei Song 0000 -0002 -2504 -2480 ; Xiao-Lan Zhang 0000 -0003 -4951 -0952 .
S-Editor:Chen YL
L-Editor:A
P-Editor:Cai YX
 World Journal of Gastroenterology2022年20期
World Journal of Gastroenterology2022年20期
- World Journal of Gastroenterology的其它文章
- Future prospect of “Gut microbiome composition can predict the response to nivolumab in advanced hepatocellular carcinoma patients”
- First prospective European study for the feasibility and safety of magnetically controlled capsule endoscopy in gastric mucosal abnormalities
- Digital single-operator video cholangioscopy improves endoscopic management in patients with primary sclerosing cholangitis-a retrospective observational study
- Radiomics for the detection of microvascular invasion in hepatocellular carcinoma
- Endoscopic management of intramural spontaneous duodenal hematoma: A case report
- Gadolinium-ethoxybenzyl-diethylenetriamine penta-acetic acid-enhanced magnetic resonance imaging for evaluating fibrosis regression in chronic hepatitis C patients after direct-acting antiviral
