Phylogenetic relationships of the zokor genus Eospalax(Mammalia, Rodentia, Spalacidae) inferred from wholegenome analyses, with description of a new species endemic to Hengduan Mountains
Tao Zhang, Meng-Long Lei, Hao Zhou, Zhong-Zheng Chen,4,*, Peng Shi,5,6,*
1 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming,Yunnan 650204, China
2 Jiangsu Key Laboratory of Neuropsychiatric Diseases and College of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu 215123, China
3 Joint Laboratory of Animal Models for Human Diseases and Drug Development, Soochow University and Kunming Institute of Zoology,Chinese Academy of Sciences, Kunming, Yunnan 650223, China
4 Collaborative Innovation Center of Recovery and Reconstruction of Degraded Ecosystem in Wanjiang Basin Co-founded by Anhui Province and Ministry of Education, School of Ecology and Environment, Anhui Normal University, Wuhu, Anhui 241000, China
5 School of Future Technology, University of Chinese Academy of Sciences, Beijing 100049, China
6 Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
ABSTRACT Zokors in the genus Eospalax, which are endemic to northern and western China, are subterranean rodents that inhabit various niches, including grasslands, high-altitude meadows, forests, and farmlands.Six species in Eospalax were described a century ago but their taxonomy and phylogeny remain controversial.In this study, we performed high-depth whole-genome sequencing of 47 zokor samples, comprising all six previously described species.Genomic analyses revealed a reliable and robust phylogeny of Eospalax and supported the validity of the six named species.According to the inferred phylogenetic relationships, Eospalax first divergent into two clades in the early Pliocene (ca.4.68 million years ago (Ma)), one inhabiting the highaltitude Qinghai-Xizang (Tibet) Plateau (QTP) and adjacent regions, and the another inhabiting the lowaltitude Loess Plateau and Qinling-Daba Mountains.The most recent divergences occurred between E.baileyi and E.smithii and between E.rufescens and E.rothschildi in the late Pliocene (ca.2.09 and 2.19 Ma, respectively).We also collected specimens of zokors in the southern Hengduan Mountains (Muli County, Sichuan Province), far from the known distributions of all other zokors.Morphological andmolecular analyses strongly suggested that the specimens represent a new species, formally described here as Eospalax muliensis sp.nov.The new species belongs to the high-altitude clade and diverged from closely related species (ca.4.22 Ma)shortly after the first divergence in Eospalax.Interestingly, Eospalax muliensis sp.nov.possesses more supposedly plesiomorphic characters, suggesting a possible origin of the genus in the Hengduan Mountains.
Keywords: Zokor; Eospalax; Phylogenomic analyses; New species; Hengduan Mountains
INTRODUCTION
Zokors (subfamily Myospalacinae) are a group of strictly subterranean rodents endemic to East Asia, which belong to the family Spalacidae and are closely related to the Rhizomyinae and Spalacinae subfamilies (Bouckaert et al.,2019; Guo et al., 2021; Lin et al., 2014; Norris et al., 2004).Zokors are cylindrical in shape, with stocky and strong forelimbs and hardened nose pads to excavate their complex burrow systems.Based on morphological and molecular evidence, extant zokors are classified into two genera: i.e.,Myospalax, which includes species with flat occiputs, andEospalax, which includes species with convex occiputs (Su et al., 2014; Zheng, 1994; Zhou & Zhou, 2008).Myospalaxspecies are mainly distributed in the grasslands of northeast China, Mongolia, and Russia, whileEospalaxspecies inhabit various niches in northern and western China (Figure 1A) (Fan& Shi, 1982), including the high-altitude Qinghai-Xizang (Tibet)Plateau (QTP).The phenotypic and genetic adaptations of the plateau zokor (E.baileyi) to the hypoxic conditions on the QTP have been well researched (Wei et al., 2006; Xu et al., 2021;Zhang et al., 2021).
However, despite studies on the physiology, ecology, and genetics of certain species, the taxonomy and phylogeny ofEospalaxremain controversial.Six species ofEospalaxwere described in the late 19thand early 20thcentury, but the taxonomic status ofEospalaxspecies has changed with time,especially forE.baileyi,E.cansus, andE.rufescens(Table 1).Eospalax fontanierii,E.rothschildi, andE.smithiiwere recognized as distinct species by Allen (1940), with this classification adopted by many monographs until recently(Smith & Xie, 2008; Wilson & Reeder, 2005).Based on comprehensive morphological comparisons, Fan & Shi (1982)found thatE.cansusis stably smaller thanE.fontanieriiin body size and skull andE.baileyiis distinct fromE.fontanieriiin nose pad shape and presence of a postero-external lobe on M3.Thus, they elevatedE.baileyiandE.cansusto species level.Subsequent studies using mitochondrial DNA markers have generally validated the status of all sixEospalaxspecies(He et al., 2012; Su et al., 2014; Zhou & Zhou, 2008).However, the classification ofE.baileyihas remained somewhat controversial due to insufficient molecular markers and incomplete sampling (Jiang et al., 2017; Wei et al., 2021).In addition, the potential existence of new undescribed species within theEospalaxgenus may complicate established taxonomy.Here, based on a literature review of species distribution, we found reports of zokors in the southern Hengduan Mountains (Muli County, SichuanProvince) (Liu et al., 2007), far from the known distribution of all other zokors (Figure 1A).Thus, in the current study, we collected 14Eospalaxspecimens from Muli County between October 2020 and October 2021.Externally, these zokors differed considerably from all other known species due to their small size and long tail (Figure 1B), indicating that they may represent a distinct taxon, which we name herein asEospalax muliensissp.nov.
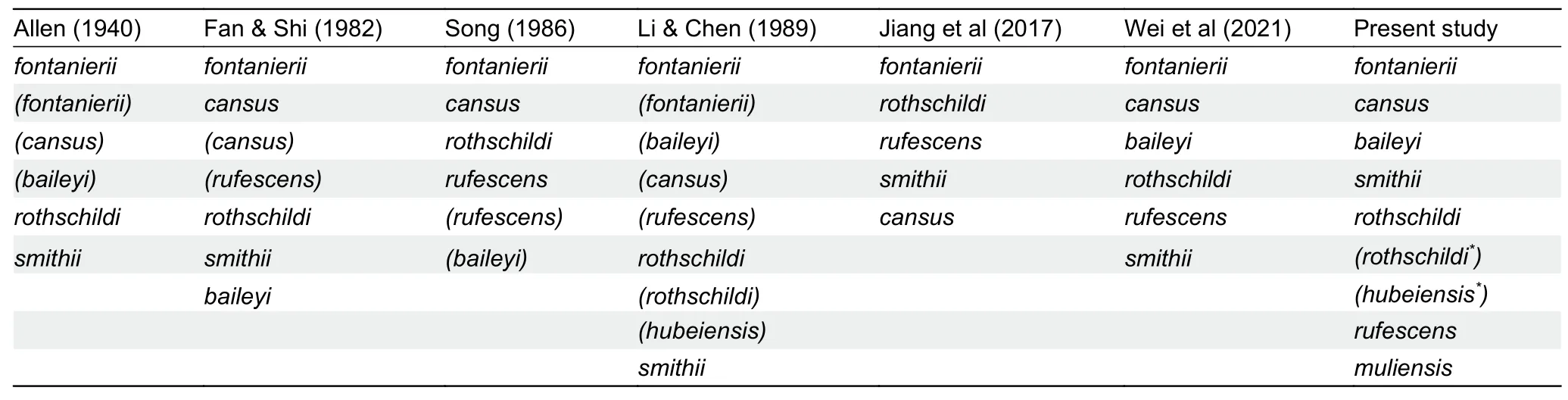
Table 1 Previous and proposed classifications of Eospalax genus
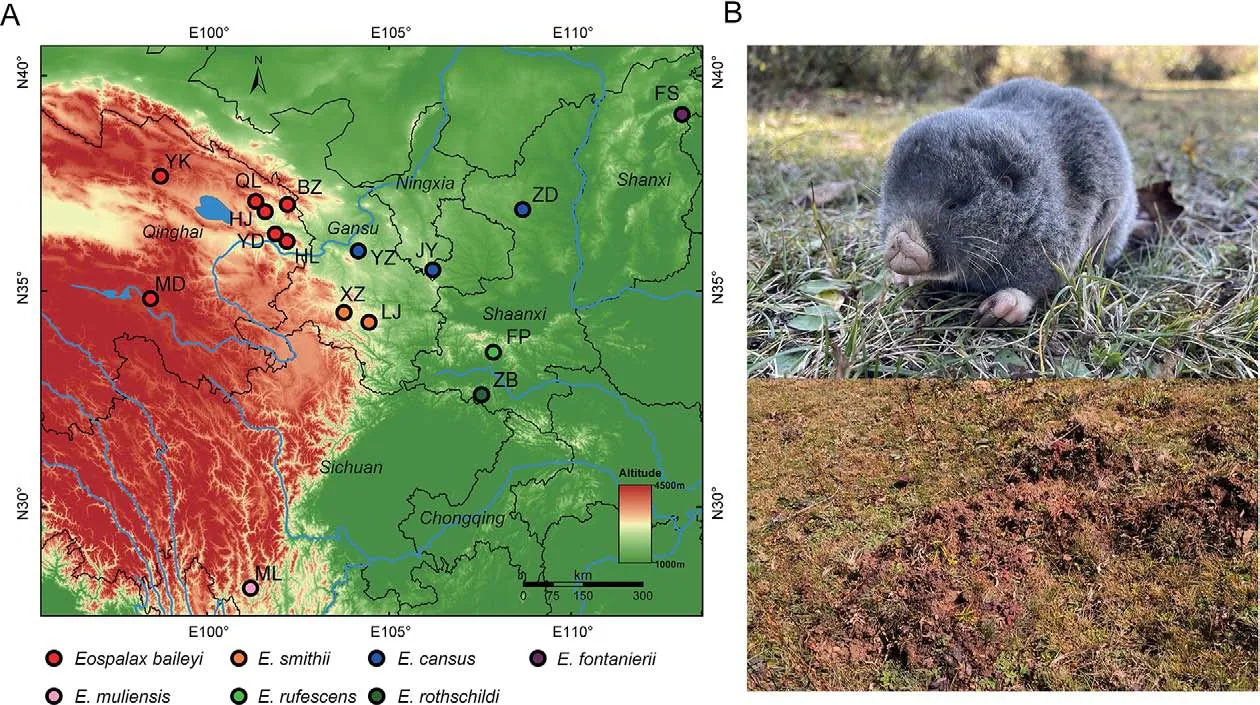
Figure 1 Sampling of species in Eospalax
In addition to taxonomic controversies, the phylogeny ofEospalaxremains incomplete and inconsistent.Conflicting phylogenies amongEospalaxspecies have been reported in previous studies based on both mitochondrial DNA markers(Cai et al., 2020; Su et al., 2014; Zhou & Zhou, 2008; Zou et al., 2020) and morphological traits (Li & Chen, 1986; Li &Wang, 1996).Notably, phylogenetic relationships inferred from mitochondrial markers are unstable when different markers or tree-construction methods are applied (Su et al., 2014; Zhou &Zhou, 2008).Therefore, the ambiguous and conflicting phylogenetic relationships reported inEospalaxhamper studies of this remarkable lineage and underscore the need for a reliable and robust phylogeny.
In the current study, we aimed to clarify the taxonomy and phylogeny ofEospalax.All six known species ofEospalaxwere sampled and whole genomes were sequenced to high depth.Whole-genome analysis revealed reliable and robust phylogenetic relationships and validated the status of all six previously described species.Furthermore, by integrating morphological and genomic evidence, the zokor specimens collected from Muli County were found to represent a new species.Thus, the phylogeny and diversification of the new species were also studied.
MATERIALS AND METHODS
Sample collection
Fieldwork was carried out from April 2017 to October 2021.Skeletal muscle was collected for DNA extraction.All collections followed the animal use protocols approved by the Animal Care and Ethics Committee of the Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS)(Approval No.SMKX-SQ-2021-067).
Sampling sites of each species were located in their generally acknowledged distribution areas (Figure 1A;Supplementary Table S1).The seven sampling sites ofE.baileyiwere reported in our previous work (Zhang et al.,2021).Because the distribution ofE.cansusis wide and adjacent to several other species inEospalax, three sampling sites ofE.cansuswere chosen across Shaanxi, Ningxia, and Gansu provinces to ensure representativeness.For other species, one or two sampling sites were selected.Two zokors collected near Hulun Lake (Inner Mongolia Autonomous Region, China) were recognized asMyospalax psilurusaccording to their skull and hairless tails.Thus, these two zokors were used as an outgroup.
Whole-genome sequencing and de novo assembly-based variant calling
In this study, 47 zokor samples were collected and whole genomes were sequenced to a high depth, with an average of 137.2 Gb of data for each sample (Figure 1A; Supplementary Table S1).Combined with previously reported whole-genome sequences of 38E.baileyisamples (Zhang et al., 2021), our dataset covered all six previously describedEospalaxspecies,as well asEospalax muliensissp.nov.and two samples of closely relatedM.psilurus.
DNA extraction, resequencing library construction, and quality control of raw sequencing reads were performed following Zhang et al (2021).Briefly, the resequencing library was constructed following standard protocols and sequenced using Illumina HiSeq or BGISEQ paired-end mode.Raw sequencing reads were trimmed using Trimmomatic v0.36(Bolger et al., 2014) to remove low-quality reads.Although aE.baileyigenome assembly was recently published (Zhang et al., 2021), all other zokor genomes are currently unavailable.Thus, we sequenced all samples to high depth and appliedde novoassembly-based variant calling following the FermiKit pipeline (Li, 2015b).For comparability, variant calling of allE.baileyisamples was also applied using this pipeline.First,clean reads of each sample were error corrected using BFC v.r181 (Li, 2015a).Second, Fermi2 v.r178 (Li, 2012) was used tode novoassemble the genome of each sample.Third, the assembly of each sample was mapped to the plateau zokor genome using minimap2 v.2.17 (Li, 2018) with parameters “-ax asm5/asm10/asm20 -k 23 -w 11 -f 1 000 -n 2 -s 100 --endbonus 5”.According to the genetic distance between samples and plateau zokors, different parameters of -ax were applied to balance the sensitivity and specificity of mapping.Specifically, asm5, asm10, and asm20 were applied forsamples ofE.baileyi,E.smithii, and all other species,respectively.After mapping, all sample variants were called jointly using the “pileup” functions in HTSbox v.r310 with parameters “-d -V 0.15 -S 300 -q 20 -Q 3 -s 5”.Raw genotypes were first filtered according to allele balance.For homozygous genotypes, at least 90% of reads should support these genotypes.For heterozygous genotypes, the two types of alleles should have 25%-75% of read-support simultaneously.If the above criteria were not satisfied, the genotypes were set as missing.After allelic balance filtering,we only retained those biallelic single nucleotide variants(SNVs) with a missing rate of no more than 20% using VCFtools v.0.1.17 (Danecek et al., 2011).
Phylogenomic analyses
Phylogenetic relationships among all zokor samples were inferred using whole-genome SNVs and maximum-likelihood(ML) algorithms.Considering the computational resources required, we pruned high-quality SNVs by randomly choosing one SNV in each 100 bp window across the genome.ML analyses were conducted in RAxML v.8.2.12 (Stamatakis,2014) with the nucleotide substitution model of GTRGAMMA.To test the reliability of inferred phylogenetic relationships, 100 bootstraps were applied.
In addition to the construction of phylogenetic trees based on whole-genome pruned SNVs, we also applied a summary of sliding-window ML trees.First, we split the whole genomes into over 50 000 non-overlapping windows (each 50 kb long).For each window, ML trees were constructed using RAxML based on all high-quality SNVs.Weighting and summary of all sliding-window ML trees across whole genomes were explored using topology weighting by iterative sampling of subtrees (Twisst) (Martin & van Belleghem, 2017).
Divergence time among all species in the genusEospalaxwas determined using SNAPP v2.6.2 (Bryant et al., 2012) in BEAST2 (Bouckaert et al., 2019).Secondary calibration was applied following He et al (2020), which estimated that the divergence betweenMyospalaxandEospalaxoccurred 8.8 million years ago (Ma), with a 95% highest posterior density(HPD) interval from 7.43 to 10.23 Ma.The estimate from He et al (2020) is congruent with fossil records of the genusMyotalpavus, which occurred in the Late Miocene and may be the ancestor of bothEospalaxandMyospalax(Zheng, 1994).Thus, we constrained the divergence time betweenMyospalaxandEospalaxwith a lognormal distribution centered at 8.8 Ma and a standard deviation of 0.085 in SNAPP analysis.For each species, the individual with the lowest proportion of missing data was used to represent that species.The SNVs were first pruned as described previously, with those identified as monomorphic or containing missing data then filtered.Finally, 100 000 randomly selected SNVs were used, and 10 million Markov Chain Monte Carlo (MCMC) iterations were set for SNAPP analysis.Convergence was assessed using Tracer v1.6 (Rambaut et al., 2018) with 10% burn-in.DensiTree v.2.6.2 (https://www.cs.auckland.ac.nz/~remco/DensiTree/)was used to visualize all posterior trees sampled by SNAPP.TreeAnnotator v2.6.2 (https://beast.community/treeannotator)was used to quantify the posterior probabilities of clades as node support in a maximum clade credibility tree with 10%burn-in.Divergence time and phylogenetic relationships were visualized in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).SNAPP analysis was implemented three times independently and obtained nearly identical results.
Absolute genetic distance (DXY) among species was estimated in 50 kb windows with a 50 kb step size along the genome using popgenWindows.py in genomics_general(https://github.com/simonhmartin/genomics_general).We excluded windows located in scaffolds <1 Mb due to possible poor assembly of short scaffolds.
Mitochondrial genome (mitogenome) analysis
For all whole-genome sequenced samples, we assembled their mitogenomes.We randomly selected 5 Gb of resequencing reads for each sample.Assembly was performed using MitoZ v.2.3 (Meng et al., 2019) or GetOrganelle v.1.7.2b (Jin et al., 2020) with default parameters.The mitogenome of a publishedM.psilurussample (JX014234) (Li et al., 2016) served as a reference in GetOrganelle analysis.Annotation of the assembled mitogenomes was performed with MitoZ.The quality of each mitogenome was carefully checked based on length (>16 kb),gene count (37 genes), and circularity.
In addition to the mitogenomes assembled in this study, we downloaded all other mitogenomes inEospalaxandMyospalaxreported in previous studies.Thirteen proteincoding genes and two ribosomal RNA (rRNA) genes were included in the following analyses.Gene alignments were performed by CLUSTAL v.2.1 (Larkin et al., 2007) and then concatenated.The best partition schemes for the dataset were identified using PartitionFinder v.2.1.1 (Lanfear et al., 2017)with recommended corrected Akaike Information Criterion(AICc).This analysis partitioned 15 mitochondrial genes into eight subsets.Mitochondrial phylogenomic analyses were inferred using RAxML with 1 000 bootstrap replicates.
Morphological data and analyses
A total of 62 specimens ofEospalaxwere collected.All specimens and tissue samples were deposited in KIZ, CAS.Specimens were identified following Fan & Shi (1982) and Luo et al (2000).Forty-eight specimens were assigned to the six recognized species, includingE.baileyi(n=34),E.cansus(n=5),E.fontanierii(n=1),E.rothschildi(n=2),E.rufescens(n=2), andE.smithii(n=4).Fourteen specimens collected from Muli County, Sichuan Province, were assigned to the putative new species (Eospalaxmuliensissp.nov.).
The body weight (BW) of each specimen was weighed up to 0.01 g using an electronic scale.External measurements,including head and body length (HB), tail length (TL), and hindfoot length (HF), of some individuals were taken in the field using a ruler to the nearest 0.1 mm.Twenty-one cranial and dental measurements were taken using a digital caliper graduated to 0.01 mm following Pan et al (2007).These measurements included greatest length of skull (GLS),condylobasal length (CBL), basal length (BL), palatal length(PL), zygomatic width (ZMW), interorbital breadth (IOB),foramen infraorbital breadth (FIB), rostrum width (RSW),mastoid width (MTW), maximum width across upper second molars (M2-M2), length of upper tooth row (LUTR), length of upper molars (LUM), nasal length (NSL), braincase height(BCH), greatest breadth of foramen magnum (GBFM), length of auditory bulla (LAB), distance between auditory bulla(DAB), mandibular length (MDL), length of below toothrow(LBTR), length of lower molars (LLM), and length of lower incisor (LLI).We performed principal component analysis(PCA) to analyze morphometric variation of these specimens in SPSS v19.0 (SPSS Inc., USA).PCA was only based on the 21 craniodental measurements of adult specimens.All craniodental variables were log10-transformed before PCA.We considered any component with an eigenvalue exceeding 1.0 to be meaningfully interpretable.We further examined specimen morphology as per Fan & Shi (1982), Song (1986),and Luo et al (2000), and followed their terminologies for morphological descriptions.
RESULTS
Phylogenetic relationship and molecular dating
A total of 85 whole-genome sequenced samples were included in this study (Figure 1A; Supplementary Table S1),covering all six previously describedEospalaxspecies as well asEospalax muliensissp.nov.and two samples of closely relatedMyospalax psilurus.
After stringent quality control of genome alignment and variant calling, ~227 million high-quality SNVs were identified.Based on the genome-wide SNVs, ML analysis yielded a highly resolved phylogeny.All nodes that separated species within the genusEospalaxexhibited 100% bootstrap support(Figure 2A).Accordingly, species inEospalaxcould be divided into two major clades (i.e., high- and low-altitude clades).The high-altitude clade, comprised ofE.baileyi,E.smithii, andEospalax muliensissp.nov., generally inhabited the highaltitude QTP and adjacent regions (2 700-4 300 m a.s.l.),while the low-altitude clade, comprised of the other fourEospalaxspecies, generally inhabited the relatively lowaltitude Loess Plateau and Qinling-Daba Mountains.Within the high-altitude clade,Eospalax muliensissp.nov.was found in a basal position and inhabited extremely high altitudes (3 700 m a.s.l.).Within the second clade,E.fontanieriisplit first from the other species.Notably, threeE.cansuspopulations located remotely from each other formed a tight monophyletic cluster.
The genetic distances between theM.psilurusoutgroup and species inEospalaxranged from 3.5% to 3.7%(Figure 2B).Within the genusEospalax, the genetic distances among species ranged from 1.0% to 2.2% (Figure 2B).For each species pair separated by the same internal node, their genetic distances were comparable (Figure 2B),demonstrating the reliability of the inferred phylogenetic relationships and that the applied methods enabled phylogenomic analysis.
Phylogenetic relationships inferred from the mitogenomes were also constructed (Supplementary Figure S1).Consistent with nuclear genomic analyses,Eospalax muliensissp.nov.was also identified as a distinct genetic lineage based on mitogenomic analyses (Supplementary Figure S1).However,significant incongruences among phylogenies inferred from nuclear genomes and mitogenomes were found, especially in the positions ofEospalax muliensissp.nov.andE.fontanierii(Figure 2A; Supplementary Figure S1) (see Discussion).
To confirm the inferred phylogenetic relationships and investigate the alternative topology, we split the genomes into>50 000 windows (50 kb in size) and constructed window trees based on the corresponding SNVs in each window.Most window trees (75.8%) were consistent with the whole-genome-based phylogenetic tree (Figure 3A, B).In addition,four alternative topologies were supported by at least 1% of windows (Figure 3A).The top two alternative topologies differed from the majority tree in the position ofEospalax muliensissp.nov.(Figure 3C,D).The other two lesssupported topologies revealed an uncertain relative position ofE.fontanieriiandE.cansuswithin the second clade(Supplementary Figure S2).
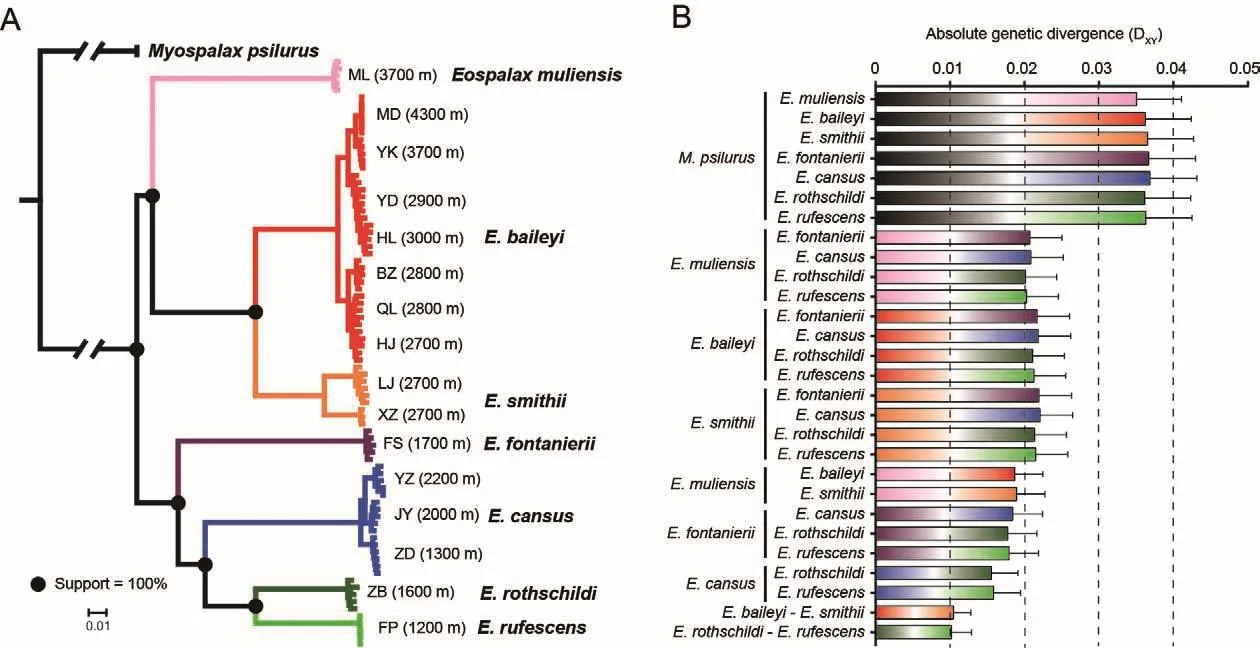
Figure 2 Phylogenetic relationships among species of Eospalax

Figure 3 Alternative topologies and divergence times of genus Eospalax
In divergence time analysis, all posterior trees sampled by SNAPP were identical to the whole-genome-based phylogenetic tree (Supplementary Figure S3).The two major high- and low-altitude clades diverged from each other ~4.68 Ma (95% CI=3.94-5.48 Ma) (Figure 3E).Shortly after,Eospalax muliensissp.nov.diverged fromE.baileyiandE.smithii(~4.22 Ma; 95% CI=3.57-4.97 Ma).Within the lowaltitude clade,E.fontanieriifirst diverged from other species~3.87 Ma (95% CI=3.26-4.54 Ma).Thus, the speciation events within the genusEospalaxgenerally occurred in the Pliocene (Figure 3E).
Morphometric comparison and morphological diagnosis
The external and craniodental measurements of each species are given in Table 2.A total of 57 intact skulls of adult specimens were used for PCA.The first three principal components (PC1-3) had eigenvalues greater than 1.0, and together accounted for >79.78% of total variance(Supplementary Table S2).PC1 accounted for 44.08% of the variation (eigenvalue=9.26) and was strongly loaded with most variables, except IOB, GBFM, DAB, and M2-M2, indicating it may be a marker of size.PC2 accounted for 21.57% of the variation (eigenvalue=4.53) and was strongly dominated by RSW, IOB, FIB, and LLI (loading>0.62), while PC3 accounted for 14.13% of the variation (eigenvalue=2.97) and was dominated by GBFM, DAB, and M2-M2.
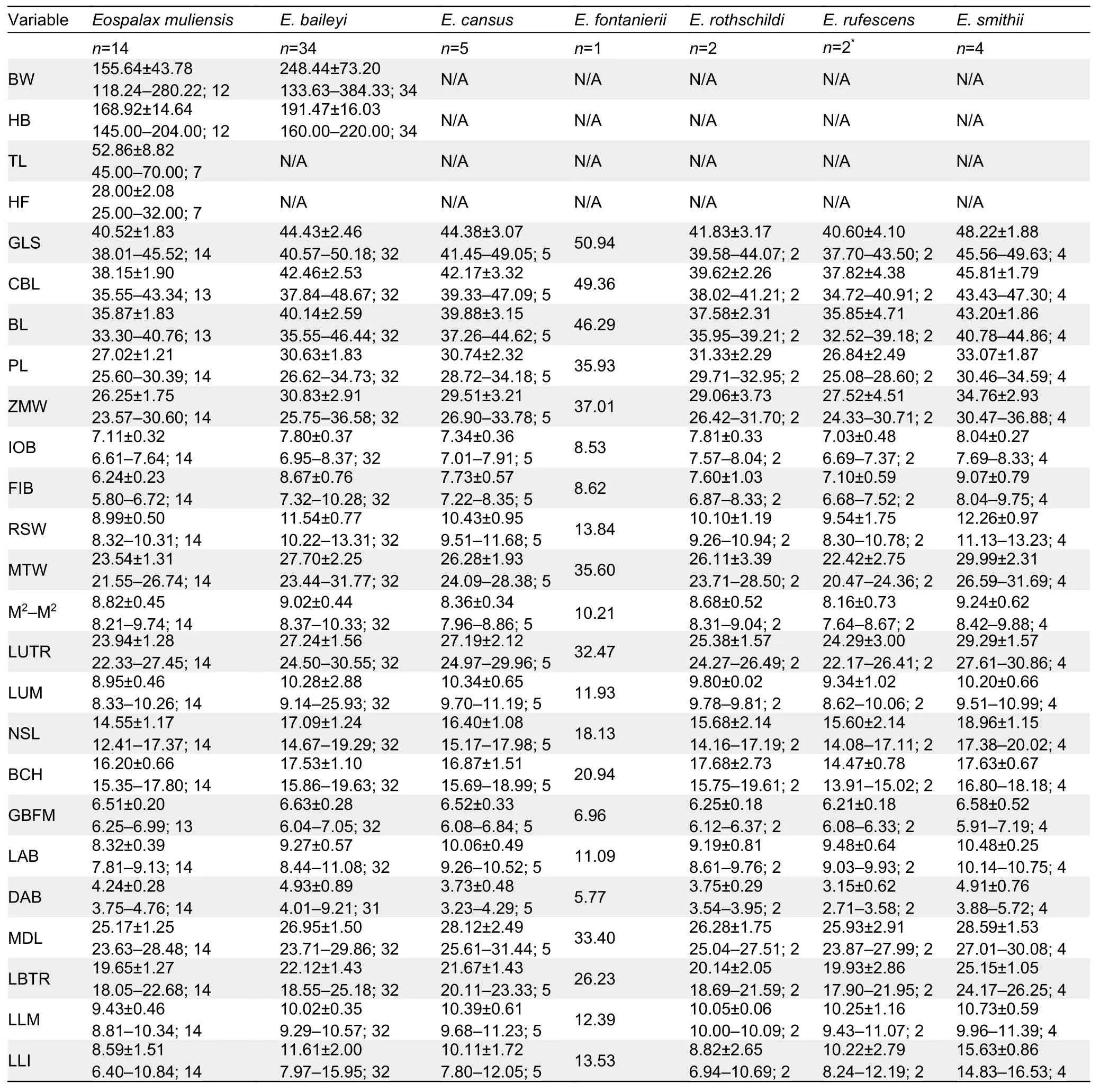
Table 2 Body weight and external and cranial measurements (mm) (mean±SD and ranges) of Eospalax specimens (n) examined in this study
The PC1-2 plot (Figure 4) showed thatEospalax muliensissp.nov.occupied the lower-left corner and was well separated from the other species.Specimens ofEospalax muliensissp.nov.were plotted on the negative field of both PC1 and PC2, reflecting its generally smaller size, narrower RSW, IOB, and FIB, and shorter LLI.Specimens ofE.baileyioccupied a large area, from -1.34 to 1.45 in PC1 and -0.49 to 1.70 in PC2, indicating great variation in size, with a relatively wide RSW, IOB, and FIB, and long LLI.Specimens ofE.cansus,E.fontanierii,E.rufescens, andE.smithiiwere mostly plotted on the positive region of PC1, indicating relatively large skulls.
Both molecular and morphological evidence supportedEospalax muliensissp.nov.as a new species, which is formally described below.
Taxonomic account
Eospalax muliensis Zhang, Chen & Shi, sp.nov.
Suggested common name: Muli zokor; 木里鼢鼠(Muli Fenshu)
Holotype:KIZ 040324 (field No.: SC 2110385).Adult female collected by Tao Zhang, Meng-Long Lei, Zhong-Zheng Chen,and Hao Zhou on 21 April 2021.Dried skin, cleaned skull, and alcohol-preserved carcass are deposited in KIZ.Measurements (mm): BW=133.00 g, HB=172.0, TL=56.5,HF=28.0, GLS=39.41, CB=37.46, BL=35.40, PL=27.27,ZMW=25.58, IOB=7.27, FIB=6.12, RSW=8.84, MTW=22.95,M2-M2=9.30, LUTR=23.93, LUM=9.11, NSL=14.65,BCH=15.84, GBFM=6.25, LAB=8.33, DAB=4.12, MDL=24.75,LBTR=20.36, LLM=9.63, LLI=10.51.
Type locality:Kangwu Ranch (N28.135°, E101.196°), Muli County, Sichuan Province, China, adjacent to Shangri-La Lake at an altitude of ~3 700 m a.s.l.
Paratypes:Seven specimens were collected from the type locality.Four specimens (KIZ 040325-040328) were deposited in KIZ, CAS; three specimens (SC 2110384,2110386, and 2110451) were deposited in the Biological Museum of Anhui Normal University.All specimens were prepared as dried skins with cleaned skulls and alcoholpreserved carcasses.
Etymology:The species namemuliensisis derived from MuliCounty, the type locality of the new species in Sichuan Province, China, and the Latin adjectival suffix -ensismeans“belonging to”.
Diagnosis:Eospalax muliensissp.nov.can be distinguished from other described species ofEospalaxby a combination of the following characters: size small (mean BW=155.64 g;mean GLS=40.52 mm); tail relatively long, densely hairy;nose-pad trifoliate; rostrum weak, nearly rectangular, nasals small, posterior border of nasals transverse, anterior halves of premaxilla nearly aligned with nasals; braincase well-domed,temporal ridges not conspicuous, parallel in front, lambdoid crests only present on side of skull; occipital shield with developed occipital ridges, extending well posteriorly, almost forming plane with occipital condyle; about 1/3 of incisive foramina included within maxillae, remaining 2/3 of incisive foramina included within premaxillae; palate and pterygopalatine fossa large, M2-M2almost equal to LUM; M3with two reentrant angles on outer side.
Description:Small-sized zokor (Table 2), with head and body length <175 mm (range: 145.0-175.0 mm) and skull length<42.20 mm (range: 38.01-42.17 mm).Only one elderly male(KIZ 0403327) significantly larger, with head and body length of 204 mm and skull length of 45.52 mm.Dorsal pelage dark grayish brown, with slightly cinnamon-colored tips, ventral pelage slightly paler; most individuals without white blaze on forehead, small when present.Lips and muzzle white,surrounded by short white hairs.Nose-pad trifoliate.Eyes very small and external ears absent (Figure 1B).Tail relatively long, densely hairy, proximal 2/3 grayish-brown, and distal 1/3 white.Forepaws strong and powerful; third claw longest and stoutest; second and fourth claws almost equal in length,about 2/3 of third claw; fifth claw stout, about half length of fourth finger; thumb very small.Dorsal surfaces of hands and feet covered with short white hair.
Skull relatively weaker than other zokors (Figure 5).Rostrum relatively weak, thin, and nearly rectangular; nasals relatively small and trapezoidal, extending far beyond incisors,posterior border of nasals transverse.Premaxillaundeveloped, anterior half not expanded, nearly aligned with nasals.Zygomatic arches slightly expanded and thin;zygomatic plates narrow and simple.Interorbital region slightly constricted, almost same width as rostrum.Braincase welldomed, rounded ventrally.Temporal ridges not conspicuous(but much more developed in old male KIZ 0403327), parallel in front.Lambdoid crests sharp, but only on side of skull;occipital shield and occipital ridges developed, extending well posteriorly, almost forming plane with occipital condyle; small medial occipital ridge present between occipital ridges.Incisive foramina small, about 2/3 included within premaxilla,posterior 1/3 within maxillaries.Palate and pterygopalatine fossa broad, relatively wider than otherEospalaxspecies.Body of mandible curved.Coronoid process weak, pointed,and curved to posterior.Condyloid process stout and squared.Angular process small, curved upward almost joining with ramus.
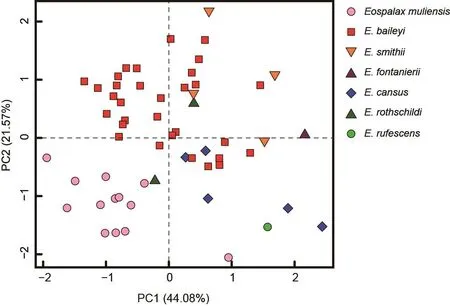
Figure 4 Plot of principal components 1 and 2 from analysis of 21 craniodental measurements of Eospalax genus
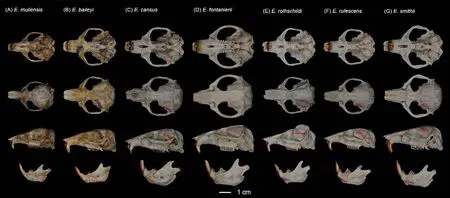
Figure 5 Dorsal, ventral, and lateral views of skull and mandible of Eospalax muliensis sp.nov.(KIZ 040324, holotype) (A), E.baileyi (DF 004) (B), E.cansus (LM 001) (C), E.fontanierii (FS 001) (D), E.rothschildi (ZB 001) (E), E.rufescens (FP 001) (F), and E.smithii (XZ 001) (G)
Teeth relatively light.Incisor teeth stout with tips curving slightly backward, outer side orange.Upper molar tooth rows slightly divergent to posterior; M1with two reentrant angles on outer and inner side; M2and M3with two reentrant angles on outer side, and one inside, no postero-external lobe on M3;lower incisor short, slightly curved, outer side orange; M1with two reentrant angles on outer side and three on inner side; M2and M3with two reentrant angles on outer and inner sides.Comparison: According to the measurements,Eospalax muliensissp.nov.is similar in size toE.rothschildi, but much smaller than other species in all dimensions (Table 2;Supplementary Table S3).The new species has some features unique in the genus.The skull ofEospalax muliensissp.nov.is obviously weaker than that of the otherEospalaxspecies, especially in the rostrum region (Figure 5).Therefore,the rostrum-related measurements (i.e., RSW, FIB, and NSL)ofEospalax muliensissp.nov.are the smallest amongEospalaxspecies (Table 2).The nasals of the new species are very short and narrow, the smallest in the genus.The premaxilla ofEospalax muliensissp.nov.is depressed and very thin, with the anterior half parts nearly aligned with the nasals, in contrast with other species, in which the premaxillae are well-developed and broader than the nasals.The palate and pterygopalatine fossa are relatively wider inEospalax muliensissp.nov than in other species, thus M2-M2is almost equal to LUM in the new species but is much smaller than LUM in other species.The braincase ofEospalax muliensissp.nov.are well-domed and temporal ridges being ambiguous, characteristics only seen in some individuals ofE.cansusand different from the flattened braincase with developed temporal ridges seen in other species.
Eospalax muliensissp.nov.can be distinguished from small-sizedE.rothschildiby its strong and powerful forepaws(vs.slender); longer tail TL=45-70 mm (vs.short tail TL=23-38 mm, Supplementary Table S3); and proximal 2/3 of tail grayish-brown and distal 1/3 white (vs.gray above white below).Eospalax muliensissp.nov.can be distinguished fromE.fontanieriiandE.cansusbased on trifoliate nose-pad and densely hairy tail (vs.oval nose-pad and nearly naked tail).Eospalax muliensissp.nov.can be clearly distinguished fromE.baileyiandE.smithiibased on incisive foramina about 1/3 expanding premaxillary boundaries (vs.incisive foramina wholly contained in premaxillary).Eospalax muliensissp.nov.can be distinguished fromE.baileyiby the postero-external lobe on M3; the parallel temporal ridges ofE.muliensissp.nov.is distinct from the temporal ridges meeting in the median line ofE.smithii.The zygomatic arches ofE.rufescensare much stouter and flare wider thanEospalax muliensissp.nov.
Distribution and habitat:Eospalax muliensissp.nov.is currently known only from the Kangwu Ranch, Muli County,Sichuan Province, China (elevational range ~3 700 m a.s.l.).The habitat is alpine meadow, surrounded by shrubs(Supplementary Figure S4).This species may be adapted to high-elevation habitats, and thus may occur in other alpine meadows in the southern Hengduan Mountains.
DISCUSSION
Taxonomic implications
Both morphological and molecular analyses strongly suggested that the zokors inhabiting Muli County represent a new species (Eospalax muliensissp.nov.) ofEospalax.The new species can be diagnosed from all others withinEospalaxby the combination of small size, long tail, weak rostrum and nasal, and large palate and pterygopalatine fossa.Genomic analyses also revealed thatEospalax muliensissp.nov.diverged from other recognized zokor species ~4.22 Ma, with high genetic distances in nuclear and mitochondrial genomes.
Our results also strongly supported the status of all six previously describedEospalaxspecies.The most recent divergences inEospalaxoccurred betweenE.baileyiandE.smithii, and betweenE.rufescensandE.rothschildi.Evidence revealed substantial differences among the two pairs of sister species and supported their species status.First, based on whole-genome analysis, the two species pairs diverged 2.09 Ma and 2.19 Ma (Figure 3E), with relatively high genetic distances of 1.05% and 1.02% (Figure 2B), respectively.Genetic distances of the mitochondrial cytbgene between sister species of these pairs reached 11.9% and 12.5%,respectively (Supplementary Table S4) (Bradley & Baker,2001).Second, the morphological characteristics that distinguish these sister species have been long recognized.For example,E.smithiican be distinguished fromE.baileyibased on temporal ridges meeting at the median line(Figure 5) (Fan & Shi, 1982); andE.rothschildican be distinguished fromE.rufescensbased on temporal ridges conspicuous and raised (Figure 5) (Fan & Shi, 1982).Third,the distribution of habitats differs among these sister species.Eospalax baileyiis dominant on the QTP (altitudinal range of 2 700-4 300 m a.s.l.), whileE.smithiiinhabits a small area around the Min Mountains (eastern edge of QTP) at an altitude of 2 700 m a.s.l.and possibly around the Liupanshan Mountains (within the Loess Plateau) at much lower altitudes(Fan & Shi, 1982).In the second sister species pair,E.rufescensoccurs in the Qinling Mountains, whileE.rothschildioccurs in the Daba Mountains and probably the Min Mountains.Although the forest habitats of these two species are similar, their local climate and flora differ, and they are likely geographically isolated by the Hanjiang River.In summary, genomic, morphological, and biogeographicalevidence well supports the delimitation of these two pairs of recently diverged sister species.Furthermore, the status of other species withinEospalaxthat were divergent longer before should also be retained.
Phylogenetic implications
Utilizing whole-genome sequencing data, our study revealed reliable and robust phylogenetic relationships among species in the genusEospalax.Before discussing the implications of the whole-genome-based phylogenetic relationships, we first explored the significant incongruences among phylogenies inferred from nuclear genomes, mitogenomes, and previous studies.Regarding phylogenies inferred from mitogenomes and mitochondrial genes, our results and previous studies show general similarity in the close relationship betweenE.baileyiandE.smithiiand the basal position ofE.fontanierii(Supplementary Figure S1) (Cai et al., 2020; He et al., 2012,2020; Su et al., 2014).However, extensive inconsistencies appear in mitochondrial analyses, which may be due to the limited informative sites of mitochondrial genes and complex speciation and post-speciation processes in the genusEospalax.For the latter, some clues may be present in our mitogenomic analysis that integrates the mitogenomes in our and previously published data.Our mitogenomic analyses revealed a robust phylogeny but embedded oneE.cansus(KC514112) individual tightly withinE.baileyi(Supplementary Figure S1).This individual was reportedly sampled from Lintan County, Gansu Province (around Min Mountains) (Su et al.,2013), which contains the type localities ofE.cansus,E.rothschildi, andE.smithii.Furthermore,E.baileyihabitats are also adjacent to this region.Besides the possible misidentification of the specimen or habitat expansion ofE.baileyiin Lintan, another plausible explanation may be postspeciation gene flow betweenE.cansusandE.baileyiaround the Min Mountains.If this hypothesis is true, the evolutionary history of zokors around the Min Mountains may be more complex, which requires further research.
Several incongruences in the phylogenetic topologies inferred from nuclear and mitochondrial genomes were also found.Notably, nuclear genomic analysis placedE.fontanieriiwithin the low-altitude clade (Figure 2A), whereas mitogenomic analysis and most previous studies placedE.fontanieriiin a basal position, whileEospalax muliensissp.nov.were put aside (Supplementary Figure S1).In addition,nuclear genomic analysis placedEospalax muliensissp.nov.within the high-altitude clade (Figure 2A), while mitogenomic analysis placed it in the basal position of the genus(Supplementary Figure S1).Nuclear and mitochondrial discordance appears to be prevalent in phylogenetic studies and the possible roles of rapid speciation and incomplete lineage sorting have been discussed extensively (He et al.,2010; Mckay & Zink, 2010).Rapid speciation and incomplete lineage sorting may have contributed to the inconsistent relationships found amongE.cansus,E.rufescens, andE.rothschildi, given the rapid cladogenesis of these three species (Figures 2A, 3E).Other factors may also have contributed to the inconsistent positions ofE.fontanieriiandEospalax muliensissp.nov.For example, the clear pattern of mitochondrial phylogenomic analyses is the relevance between geographical distributions and phylogenetic positions of the species inEospalax.Eospalax fontanieriiandEospalax muliensissp.nov.are distributed in the remote northeast and southwest regions (Figure 1A), and their phylogenetic positions are basal.Furthermore, other species are geographically and phylogenetically adjacent.The possible explanation for this pattern may be ancient cross-species hybridization between geographically adjacent species or some other geographically relevant factors (Toews &Brelsford, 2012).However, these hypotheses lack solid evidence and require further comprehensive investigations.Regardless of the actual source of incongruence, the phylogeny inferred from nuclear genomes was more robust and thus serves as our current ‘‘best” estimate (Figure 2A).
From the inferred nuclear whole-genome-based phylogeny,Eospalaxwas first divided into high- and low-altitude clades.Because uplift of the QTP may predate the separation of these two clades (Spicer et al., 2021) and no other species in the subfamily Myospalacinae or family Spalacidae inhabit the QTP, adaptation to high altitude may have occurred in the common ancestor ofEospalax muliensissp.nov.,E.baileyi,andE.smithii.Nowadays,E.smithiiinhabits areas at relatively low altitudes, implying possible re-adaptation to low elevations.Notably, given the possible existence of other unrecognized species in the Hengduan Mountains, adaptation to high altitude could have occurred in the common ancestor of allEospalaxspecies.Within the low-altitude clade,successful habitation of forest niches appeared inE.rufescensandE.rothschildi.Because these two species are closely related and separated more recently (Figures 2A, 3E),forest adaptation likely occurred in their common ancestor.Thus, although the dispersal capacity of zokors is extremely limited (Wei et al., 1997), species inEospalaxsuccessfully and rapidly adapted to diverse ecological niches in the Pliocene, and further investigations on their adaptive mechanisms are warranted.
Implications of systematics and macroevolution from Muli zokors
The discovery ofEospalax muliensissp.nov.in the southern Hengduan Mountains is unexpected and of great interest because the habitats ofEospalax muliensissp.nov.are southernmost of extant zokors and it is the second extant zokor species that inhabit extremely high altitudes (~3 700 m a.s.l.).The Hengduan Mountains are considered a global biodiversity hot spot (Myers et al., 2000).The extremely diverse ecological habitats in three dimensions of Hengduan Mountains (Sky islands) protected species from climate fluctuations and are known inhabited by some relict species(Chen et al., 2017; Chen et al., 2021; He & Jiang, 2014).The discovery ofEospalax muliensissp.nov.in such a biodiverse area suggests the possible existence of other unknown zokor species in the Hengduan Mountains.On the other hand, the discovery could also provide clues for deciphering the origin and evolutionary history of the genusEospalax.
Based on the paleontological rules and evolutionary trajectories of zokor fossils, Li & Chen (1986) proposed nine plesiomorphic characters of ancientEospalaxspecies.Strikingly, of the nine characters, six are found inEospalaxmuliensissp.nov., in contrast to the five or fewer in other species inEospalax.For example, body size trajectory in zokor fossils is from small- to large-sized (Zheng, 1994),whereasEospalax muliensissp.nov.is among the smaller ones inEospalax.Eospalax muliensissp.nov.also possesses other plesiomorphic characters such as: long tail;weak rostrum and nasal; slightly expanded and thin zygomatic arch; ambiguous temporal ridge; and separated temporal ridge.Absent ofEospalax muliensissp.nov., previous studies proposed the Loess Plateau or Qinling-Daba Mountains as the possible origin center ofEospalax(Li & Chen, 1986; Li &Wang, 1996).Considering the plesiomorphic characters and relatively basal phylogenetic position ofEospalax muliensissp.nov., it is possible thatEospalaxmay have originated from the Hengduan Mountains.This hypothesis seems to contradict fossil evidence, as the earliest knownEospalaxfossils are recorded from the Nihewan Basin, Hebei Province (Zheng,1994) and are dated to ~2 Ma (MN18) (Li et al., 1984).However, the estimated diversification ofEospalaxspecies occurred ~4.68 Ma, and thus the contradiction may be caused by an incompleteEospalaxfossil record, especially in the Hengduan Mountains.Therefore, the origin and evolutionary history ofEospalaxremain obscure and require further interdisciplinary evidence.
SCIENTIFIC FIELD SURVEY PERMISSION INFORMATION
All collections followed the animal use protocols approved by the Animal Care and Ethics Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences (Approval No.SMKX-SQ-2021-067).Permission for field sampling in each sampling sites was granted by the local Forestry Bureau.
DATA AVAILABILITY
All sequences reported in this study were deposited in the Genome Sequence Archive database (http://gsa.big.ac.cn/)under Accession ID (CRA005933) and in Science Data Bank(https://www.scidb.cn/) under DOI: 10.11922/sciencedb.o00023.00002.Mitogenomes were deposited in GenBank under Accession Nos.OM329951-OM330035.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
P.S.conceived and supervised the study.T.Z., M.L.L., H.Z.,and Z.Z.C.collected samples.Z.Z.C.and T.Z.performed morphometric comparison and morphological diagnosis.T.Z.analyzed whole-genome resequencing data.T.Z., Z.Z.C., and P.S.wrote the paper.All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We are grateful to Bao-Guo Li (Northwest University) for sharing sample sites and helpful discussion.We are grateful to Jia-Tang Li (Chengdu Institute of Biology, Chinese Academy of Sciences), Zheng-Jun He (Animal Husbandry Bureau of Tibetan Autonomous County of Muli, Sichuan Province), and Shi-Yan Ding (Forestry Bureau of Zhenba County, Shaanxi Province) for help in field sampling.
- Zoological Research的其它文章
- Adult hippocampal neurogenesis and its impairment in Alzheimer’s disease
- Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities
- Combinational benefit of antihistamines and remdesivir for reducing SARS-CoV-2 replication and alleviating inflammation-induced lung injury in mice
- Characterization of two novel knock-in mouse models of syndromic retinal ciliopathy carrying hypomorphic Sdccag8 mutations
- Scan of the endogenous retrovirus sequences across the swine genome and survey of their copy number variation and sequence diversity among various Chinese and Western pig breeds
- Novel astrovirus and paramyxovirus in Mongolian gerbils (Meriones unguiculatus) from China

