NOVA1 promotes SMN2 exon 7 splicing by binding the UCAC motif and increases SMN protein expression
Li-Li Du , Jun-Jie Sun , Zhi-Heng Chen , Yi-Xiang Shao , Liu-Cheng Wu
Abstract Spinal muscular atrophy (SMA) is a rare hereditary neuromuscular disease with a high lethality rate in infants.Variants in the homologous genes survival of motor neuron (SMN)1 and SMN2 have been reported to be SMA pathogenic factors.Previous studies showed that a high inclusion rate of SMN2 exon 7 increased SMN expression, which in turn reduced the severity of SMA.The inclusion rate of SMN2 exon 7 was higher in neural tissues than in non-neural tissues.Neuro-oncological ventral antigen (NOVA) is a splicing factor that is specifically and highly expressed in neurons.It plays a key role in nervous system development and in the induction of nervous system diseases.However, it remains unclear whether this splicing factor affects SMA.In this study, we analyzed the inclusion of SMN2 exon 7 in different tissues in a mouse model of SMA (genotype smn-/-SMN22tg/0) and littermate controls (genotype smn+/-SMN22tg/0).We found that inclusion level of SMN2 exon 7 was high in the brain and spinal cord tissue, and that NOVA1 was also highly expressed in nervous system tissues.In addition, SMN2 exon 7 and NOVA1 were expressed synchronously in the central nervous system.We further investigated the effects of NOVA1 on disease and found that the number of neurons in the anterior horn of spinal cord decreased in the mouse model of SMA during postnatal days 1-7, and that NOVA1 expression levels in motor neurons decreased simultaneously as spinal muscular atrophy developed.We also found that in vitro expression of NOVA1 increased the inclusion of SMN2 exon 7 and expression of the SMN2 protein in the U87MG cell line, whereas the opposite was observed when NOVA1 was knocked down.Finally, point mutation and RNA pull-down showed that the UCAC motif in SMN2 exon 7 plays a critical role in NOVA1 binding and promoting the inclusion of exon 7.Moreover, CA was more essential for the inclusion of exon 7 than the order of Y residues in the motif.Collectively, these findings indicate that NOVA1 interacts with the UCAC motif in exon 7 of SMN2, thereby enhancing inclusion of exon 7 in SMN2, which in turn increases expression of the SMN protein.
Key Words: exon 7 inclusion; motor neuron; neuro-oncological ventral antigen 1; SMN2 splicing; spinal cord; spinal muscular atrophy; splicing factors; UCAC motif
Introduction
Spinal muscular atrophy (SMA) is a rare hereditary neuromuscular disease that is primarily characterized by degeneration of alpha motor neurons in the anterior horn of the spinal cord and affects innervation of skeletal muscles, leading to muscle weakness, muscle atrophy, paralysis, or even death (Arnold and Fischbeck, 2018).SMA is caused by deletion of the survival of motor neuron 1 (SMN1) gene on chromosome 5 or by point mutations inSMN1leading to a lack of expression of functional full-length SMN (SMN FL) (Brzustowicz et al., 1990; Lefebvre et al., 1995).SMN is a housekeeping protein that is part of the SMN-Gemin multiprotein complex (Borg et al.,2015).It is involved in assembly of small nuclear ribonucleoprotein and regulation of precursor mRNA (pre-mRNA) splicing, and also participates in a variety of physiological processes, including stress response, axon transport,cytoskeletal dynamics, mitochondrial and bioenergy pathways, and ubiquitin pathways (Chaytow et al., 2018).Thus, SMN is an important molecule that is involved in multiple activities that are essential for human life.
In addition toSMN1, humans have a second, homologous, gene calledSMN2.The key difference between them is that the sixth nucleotide in exon 7 of theSMN1gene is C, whileSMN2contains a T at the same location.Only 10% of the transcripts produced fromSMN2are full-length and generate a stable SMN protein, while the rest are missing exon 7 (SMN?7) and generate a truncated protein that has no biological function and is extremely unstable(Anderton et al., 2013).Hence, variation in the sixth nucleotide is the key factor that influences the inclusion of exon 7 (Lefebvre et al., 1995).Clinical studies have shown that the severity of SMA is negatively correlated with the amount of functional SMN protein, as well asSMN2copy number (Lefebvre et al., 1997; Wirth, 2000).The amount of SMN FL protein generated bySMN2is not sufficient to compensate for defects inSMN1; therefore, promoting theinclusion ofSMN2exon 7 can effectively ameliorate the severity of SMA and improve patient survival (Hua et al., 2011; Wu et al., 2017; Son and Yokota,2018).SMN2transcripts are thus increasingly important for the treatment of SMA.
Alternative splicing is an important mechanism that regulates protein diversity and gene abundance, and is involved in the occurrence and development of many diseases.Two types of splicing factors are involved in the regulation of pre-mRNA splicing.A typical type I serine-rich (SR) protein that promotes pre-mRNA splicing contains an RNA recognition motif and an RS region with repetitive sequences of serine and arginine (Long and Caceres, 2009;?nk?, 2014).Type II splicing factors, such as the heterogeneous nuclear ribonucleoproteins (HNRNPs) proteins, which contain KH domains, inhibit premRNA splicing (Martinez-Contreras et al., 2007).Previous studies have shown that the splicing factor neuro-oncological ventral antigen (NOVA), which is specifically expressed in neurons, promotes or inhibits splicing depending on which site the protein binds to on the pre-mRNA (Ule et al., 2006; Leggere et al., 2016).The SR and HNRNP protein families play important roles in the regulation ofSMN2splicing (Hua et al., 2008; Wee et al., 2014).For example,Hua et al.(2010) showed that blocking HNRNP A1/A2 binding to the intron splicing silencer located inSMN2intron 7 significantly increased the inclusion of SMN2 exon 7 and up-regulated the expression of SMN FL.Moreover, NOVA plays a key role in nervous system development, as well as the development of neurological diseases (Buckanovich et al., 1993).NOVA1 regulates alternative splicing by binding the YCAY motif in the target gene mRNA precursor via its own KH domain (Ule et al., 2003, 2006).Mutation of YCAY to YAAY can eliminate the regulatory effect of NOVA1 on alternative splicing (Ludlow et al.,2018), and the UCAC sequence was found in exon 7 ofSMN2.However, the relationship between the NOVA family and SMA remains unclear.Here, we screened for splicing factors that regulateSMN2exon 7 inclusion in SMA mice,which harbor two copies ofSMN2(Hsieh-Li et al., 2000), and investigated the mechanism by which they promoteSMN2exon 7 inclusion.
Materials and Methods
Animals
SMA mice (genotypesmn-/-SMN22tg/0) (RRID: SCR_005570) and littermate control mice (genotypesmn+/-SMN22tg/0) on postnatal day 1 (P1), P4, and P7 were used in this study.The background strains were FVB inbred mice.The parental mice were donated by the Hua lab at the Institute of Neuroscience of Soochow University and were housed in the specific pathogen-free barrier facility at the Experimental Animal Center of Nantong University, where the animal experiments were also carried out.All mice were permitted free access to water and food, and the light cycle was 12 hours light and 12 hours dark.The use of experimental animals adhered to the 3R principles.The animal experiments were approved by the Experimental Animal Ethics Management Committee of Nantong University (approval No.20181008-088) on October 8,2018.For tissue sample collection, mice were sacrificed by CO2asphyxiation.In brief, after providing a normal supply of oxygen, the concentration of CO2was continuously increased until respiratory and cardiac arrest occurred.The CO2replacement rate was 30-70%, which ensures that the mice lost consciousness before experiencing any pain.After the 35-minute CO2asphyxiation was complete, the spinal cord was immediately removed from the spinal canal with tweezers and snap-frozen in liquid nitrogen for storage at -80°C for subsequent extraction of total RNA and protein.
Plasmid construction
TheSMN2mini-gene was constructed using pCI-SMN1, pCI-SMN2, pEGFPSMN1, and pEGFP-SMN2 according to a previous method described by the Hua lab (Hua et al., 2018).The pCGT7 plasmid was used to overexpressNOVA1.First,NOVA1was amplified using the following primers:hNOVA1Nhe1-forward: 5′-ATG CTA GCA TGA TGG CGG CAG CTC CC-3′,hNOVA1Kpn1-reverse: 5′-ACG GTA CCT CAA CCC ACT TTC TGA GG-3′.The polymerase chain reaction (PCR) mixture included 4 μL 5× Phusin high fidelity buffer, 0.4 μL 10 mM deoxy-ribonucleoside triphosphates, 1.2 μL forward primer, 1.2 μL reverse primer, 1-10 ng cDNA, 1.2 μL dimethyl sulfoxide, 0.3 μL Phusion DNA polymerase (New England Biolabs, NEB, Ipswich, MA, USA), and ddH2O to 20 μL.The reaction procedure was 35 cycles, including 95°C for 15 seconds,60°C for 20 seconds, and 72°C for 30 seconds.The PCR product was purified with a PCR clean-up kit (Axygen, Union, CA, USA, Cat# AP-PCR-250) and then digested withNheI andXbaIand ligated into theXbaI andKpnIsites in pCGT7(Additional Figure 1).The finalNOVA1overexpression plasmid was obtained through transformation, amplification, plasmid extraction, and sequencing to verify the amplification results.
Cell culture and transfection
The human glioma cell line U87MG (Cat# TCHu216) and human embryonic kidney (HEK293T; Cat# GNHu17) cells (purchased from the Chinese Academy of Sciences Cell Bank and donated by the Hua lab at the Institute of Neuroscience in Soochow University) were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Omaha, UT, USA, Cat# SH30243.01) supplemented with 10% (v/v) fetal bovine serum (Lonsera, Ciudad de la Costa, Uruguay,Cat# S711-001S), 100 U/mL penicillin, and 100 μg/mL streptomycin (HyClone,Cat# SV30010), at 37°C in a humidified 5% CO2atmosphere.One microgram of theSMN2mini-gene plasmid and theNOVA1overexpression plasmid or empty vector were transfected into cells that had been seeded into a 6-well plate one day in advance.After 48 hours of transfection, the cells were collected for RNA or protein extraction.For the mutation experiments, 1 μg of a plasmid carrying a mutated version of theSMN2mini-gene was also transfected.A petroleum-based transfection reagent (Sigma, San Jose, CA,USA, Cat# 408727) was used for all transfections.To knock downNOVA1,small interfering RNA (siRNA) sequences were generated using the following primers:NOVA1-sense: 5′-AGA CAA UUG UUC AGU UGC A-3′, antisense: 5′-UGC AAC UGA ACA AUU GUC U-3′.The negative control sequences were:NOVA1 siNC-sense: 5′-UUC UCC GAA CGU GUC ACG UTT-3′, antisense: 5′-ACG UGA CAC GUU CGG AGA ATT-3′.siRNAs were designed using online software (http://biodev.extra.cea.fr/DSIR/DSIR.html), synthesized by Shanghai Genepharma, and transfected using Lipofectamine 2000 (Life Technologies).
Point mutation
The PCR reaction mixture used to construct theSMN2mutation mini-genes was similar to that used to construct theNOVA1 overexpression plasmid.The reaction conditions were 94°C for 10 seconds, 60°C for 20 seconds, and 72°C for 210 seconds, for 30 cycles total.The annealing temperature was adjusted according to the primer Tm value or determined by gradient PCR.The mutation primers are listed in Additional Table 1.Agarose electrophoresis was used to detect the products after PCR amplification (usually 25 μL of the reaction mixture).One microliter ofDpnI was added to the remaining PCR product, which was then allowed to digest in a 37°C water bath for 4 hours.After digestion, 8 μL of the product was transformed, and a single colony was selected for amplification, after which the plasmid was extracted and sequenced to verify whether the mutation was successful.
Reverse transcription-polymerase chain reaction
Total RNA was extracted from spinal cord tissues at P4 or from U87MG and HEK-293T cells using TRIzol reagent according to the manufacturer’s instructions (Vazyme, Nanjing, China; Cat# R401-01).In brief, 1 μg of each RNA sample was used per 20 μL reaction for first-strand cDNA synthesis with oligo (dT)18 and M-MLV reverse transcriptase.The reverse transcriptionpolymerase chain reaction (RT-PCR) reaction mixture used to detect the level ofSMN2exon 7 inclusion was as follows: 12.5 μL 2× Taq master mix (Vazyme,Cat# P112), 1 μL forward primer, 1 μL reverse primer, 1 μL complementary DNA (cDNA), and 9.5 μL ddH2O.The PCR conditions included 28 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 45 seconds, which was used to detect SMN2 exon 7 inclusion level in SMA mice.The primers used were as follows: E6-F: Cy5-5′-ATA ATT CCC CCA CCA CCT CCC-3′, E8-467R: 5′-TTG CCA CAT ACG CCT CAC ATA C-3′.The RT-PCR reaction conditions used to detectSMN2exon 7 inclusion in the cell lines were 26 cycles of 95°C for 15 seconds,60°C for 30 seconds, and 72°C for 25 seconds.The primers used for RT-PCR were as follows:SMNT7F2: 5′-TAC TTA ATA CGA CTC ACT ATA GGC TAG CCT CG-3′, Ex8-29to52-R: Cy5-5′-TCT GAT CGT TTC TTT AGT GGT GTC-3′.
The PCR products were separated on 2% agarose gels, followed by signal quantitation using ImageJ software (National Institutes of Health, Bethesda,MD, USA).Exon 7 inclusion was calculated using the following formula: exon 7 inclusion ratio (%) = [FL(exon 7 inclusion)/ (FL + ?7(exon 7 exclusion))] × 100(Hua et al., 2007).
Quantitative polymerase chain reaction
Total RNA was extracted from SMA mice or from U87MG cells.The 10-μL qPCR reaction mixture contained 5 μL 2× ChamQ Universal SYBR qPCR Master Mix (Cat# Q711, Vazyme), 0.5 μL forward primer, 0.5 μL reverse primer, 1 μL cDNA, and 3 μL ddH2O.The quantitative polymerase chain reaction (qPCR)conditions were 45 cycles of 95°C for 10 seconds, 60°C for 20 seconds, and 72°C for 20 seconds.The sequences of the primers used for qPCR are shown in Additional Table 2, and the mRNA levels are shown as heat maps.
Western blot assay
Protein samples from the spinal cord at P4 or from U87MG cells were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE) and electroblotted onto polyvinylidene fluoride membranes(Millipore FFP39), followed by blocking with 5% skim milk (Cat# A600669,Sangong, Shanghai, China).Then the membranes were incubated with primary antibodies overnight at 4°C and secondary antibodies for 2 hours at room temperature.The primary antibodies used were as follows: mouse anti-SMN antibody (1:500, BD Biosciences, Franklin Lakes, NJ, USA, Cat# 610647,RRID: AB_397973), rabbit anti-NOVA1 antibody (1:500, Abcam, Cambridge,UK, Cat# ab183024), and mouse anti-β-actin antibody (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA, USA, Cat# sc-47778).Secondary rabbit anti-mouse (1:1000, Cat# D110087) and goat anti-rabbit antibodies (1:1000, Cat#D110058) were purchased from Sangong.Protein signals were detected with the Tanon-5200Multi Gel Imaging System (Tanon Science & Technology,Shanghai, China).The scanned images were imported into ImageJ software 7.0(Schneider et al., 2012).The signals were normalized to actin.
Nissl staining
Nissl staining was carried out with a kit according to the manufacturer’s instructions (Beyotime, Shanghai, China, Cat# C0117).The spinal cord tissue in the control and SMA groups at P1, P4, and P7 was fixed with 4%formaldehyde at 4°C overnight, washed in 0.01 M phosphate-buffered saline (PBS), and embedded in paraffin blocks.Then, 4-μm-thick paraffin sections were cut for Nissl staining.In brief, the paraffin sections were dewaxed, rehydrated, and washed twice with 0.01 M PBS for 2 minutes each.Subsequently, tissue sections were placed in Nissl staining solution for 3-10 minutes at room temperature, washed with pure water twice for a few seconds, dehydrated twice with 95% ethanol for 2 minutes, cleared twice with xylene for 5 minutes, sealed with neutral gum, and observed under a microscope (OLYMPUS BX51, Tokyo, Japan).
Immunofluorescence
Spinal cord tissue from control and SMA mice at P1, P4, and P7 was fixed with 4% formaldehyde at 4°C overnight, dehydrated with 30% sugar solution,embedded in tissue freezing medium (O.C.T), frozen, then cut into 6-μmthick sections using a freezing microtome, rehydrated, and stained for immunofluorescence.In brief, the frozen sections were placed in an oven at 60°C, dried for 30 minutes, washed twice with PBS for 2 minutes, and were incubated with 0.3% Triton X-100 permeabilization solution at room temperature for 30 minutes.Subsequently, these sections were washed with 0.01 M PBS and blocked with 5% bovine serum albumin (BSA) blocking solution for 30 minutes at room temperature.After the blocking solution was discarded, antibodies (rabbit anti-NOVA1 [1:500; Abcam, Cat# ab183024] and chicken anti-choline acetyltransferase [ChAT; 1:50; Abcam, Cat# ab34419])diluted in 0.01 M PBS were added dropwise to the tissue sections.Then,the tissue sections were incubated in a humid box at 4°C overnight.The next day, the tissue sections were washed with 0.01 M PBS for 5 minutes.After dropwise addition of the secondary antibodies Cy3-goat anti-rabbit IgG (1:1000; Sangong, Shanghai, China, Cat# D110062) and fluorescein isothiocyanate-donkey anti-chicken IgY (1:1000; Sangong, Cat# D110201)diluted in 0.01 M PBS, as well as DAPI (1:2000; Sangong, Cat# E607303),the tissue sections were incubated for 2 hours in a humid box at room temperature in the dark.Then, tissue sections were washed three times with 0.01 M PBS for 15 minutes, and the NOVA1 and ChAT co-localization was observed using a fluorescent mounting tablet (OLYMPUS BX51).
RNA pull-down
The biotin-labeled RNA probes WT (5′-AGG UGC UCA CAU UCC U-3′) and Mut(5′-AGG UGC UCA CAU UCC U-3′) were purchased from Shanghai Genepharma for use in the RNA pull-down experiment.After washing streptavidin agarose beads (Sigma, Cat# S1638-1ML) with streptavidin washing buffer three times,biotin-labeled RNA was added to the precleared beads, and the mixture was incubated at 4°C for 1-4 hours on a 100 ×grotator.Then, the beads were precipitated and cleaned using RNA-streptavidin interaction buffer.Next,100 μL of nucleoprotein extracted from U87MG cells was mixed with the beads at a protein concentration of 3-5 μg/μL.The tubes were maintained at 4°C overnight on a 100 ×grotator.After centrifugation, the supernatant was discarded.An appropriate volume of protein loading buffer was added,and the mixture was then placed in a water bath at 90°C for 10 minutes.After cooling on ice, the samples were centrifuged at 10,000 ×gat 4°C for 10 minutes, and the supernatant was removed for western blot analysis.
Statistical analysis
No statistical methods were used to predetermine sample sizes; however,our sample sizes were similar to those reported previously (Mao et al.,2019).The software program SPSS 16.0 (SPSS, Chicago, IL, USA) was used for statistical analysis.Data are presented as mean ± standard deviation (SD).The statistical significance of the differences between groups was analyzed using the Student’st-test.P< 0.05 indicates that the difference was statistically significant.
Results
The inclusion of SMN2 exon 7 is high in neural tissues
To investigate the differences in the inclusion ofSMN2exon 7 in different tissues, we evaluated eight tissues-heart, liver, spleen, lung, kidney, brain,spinal cord, and hindlimb muscle in control and SMA mice at P4 (Figure 1A and B).The results showed thatSMN2splicing resulting in inclusion ofSMN2exon 7 was different between tissues.A high rate of exon 7 inclusion was observed in the brain and spinal cord, and a low rate in muscle in control and SMA mice (P< 0.05).Interestingly, a higher rate of exon 7 was observed in neural tissues than in non-neural tissues in both SMA mice (P< 0.001) and control mice (P< 0.01; Figure 1C and D).
Expression of splicing factors in control and SMA mice
Exon inclusion or exclusion is determined by splicing events that are regulated by splicing factors.Classic splicing factors, such as HNRNP (Martinez-Contreras et al., 2007), SR (Long and Caceres, 2009), and NOVA (Ule et al., 2003, 2006)family members, have been confirmed to regulate a variety of splicing events.To clarify the relationship between the expression of splicing factors in SMA mice and the differential splicing ofSMN2among different tissues, we studied the expression of various splicing factors in neural and non-neural tissues from SMA or control mice at postnatal day 4 using qPCR and heat maps.The results showed that the expression of 20 members of the HNRNP, SR, and NOVA families differed among tissues, whileNovawas highly expressed in nerve tissues in both control mice (Figure 2A) and SMA mice (Figure 2B).
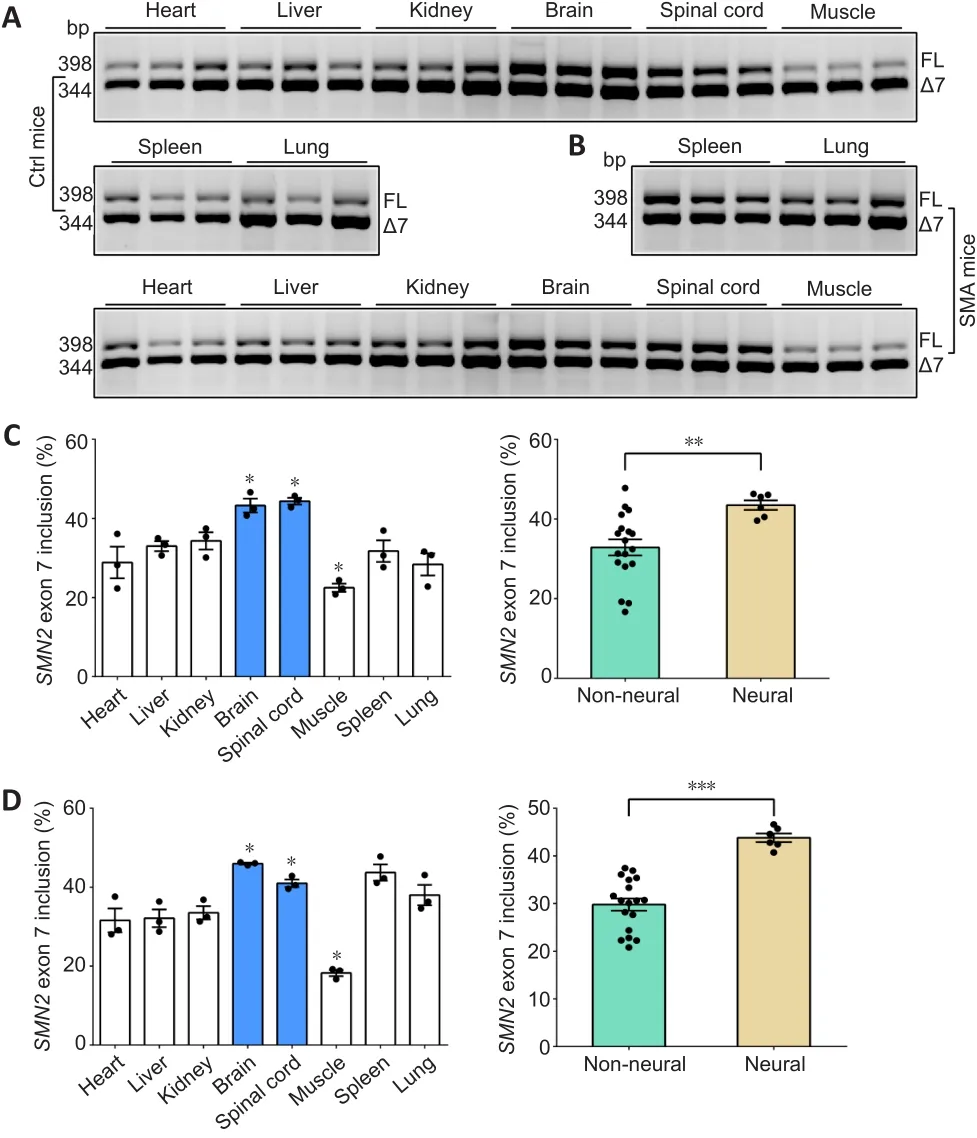
Figure 1 | SMN2 FL expression in neural and non-neural tissues.
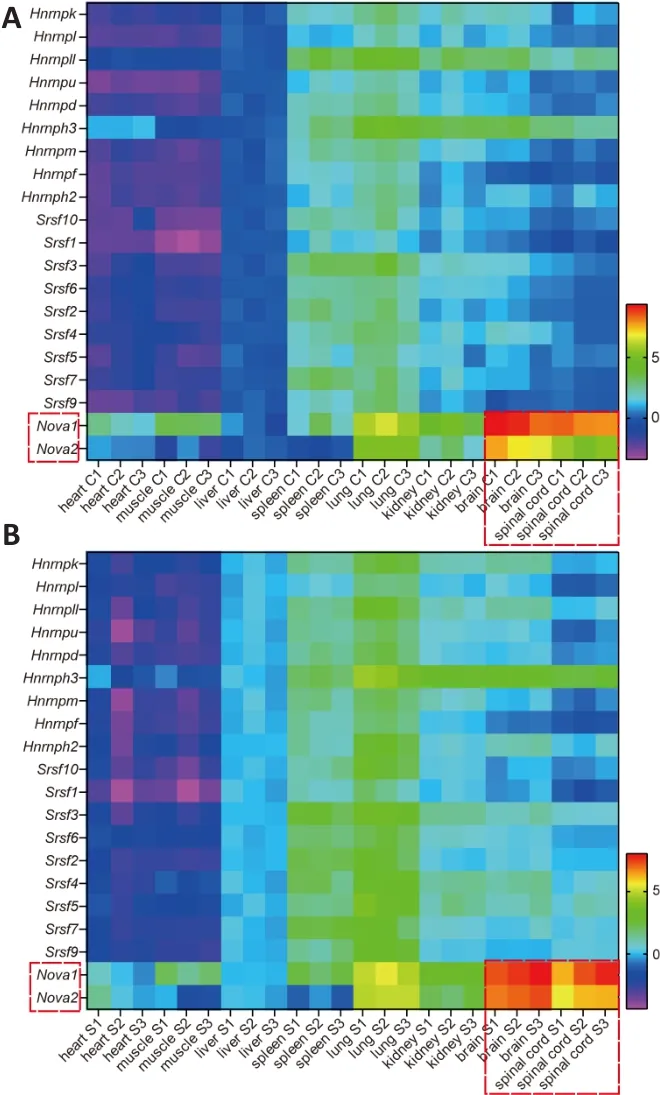
Figure 2|Expression of splicing factors in control and SMA mice.
Nova1 is down-regulated with SMA progression
To investigate the relationship between the neuro-specific expression of NOVA family members and the progression of SMA disease,SMNandNova1mRNA and protein levels were detected in the spinal cord of SMA mice at P1, P4, and P7.In control mice,SMN FLmRNA (P1vs.P7,P< 0.01, P4vs.P7,P< 0.05) and protein (P1vs.P4,P< 0.05, P1vs.P7,P< 0.05) levels were significantly upregulated from P1 to P7 (Figure 3A-C).Meanwhile, NOVA1 mRNA expression levels (P1vs.P4,P< 0.05, P1vs.P7,P< 0.01) were significantly increased on P4 and P7 compared with those on P1, and NOVA1 protein levels (P1 vs.P4,P< 0.01, P1vs.P7,P< 0.05) were significantly higher on P4 and P7 than P1(Figure 3D-F).In SMA mice,SMNFL mRNA (P1vs.P7,P< 0.01, P4vs.P7,P< 0.05) and protein (P1vs.P4,P< 0.05, P1vs.P7,P< 0.05) levels decreased significantly from P1 to P7 (Figure 3G-I).Meanwhile,NOVA1mRNA levels were significantly decreased on P4 and P7 compared with those on P1 (P1vs.P4,P< 0.01, P1vs.P7,P< 0.05), and NOVA1 protein levels were significantly lower on P7 than on P1 and P4 (P1vs.P4,P< 0.01, P1vs.P7,P< 0.001, P4vs.P7,P< 0.05; Figure 3J-L).
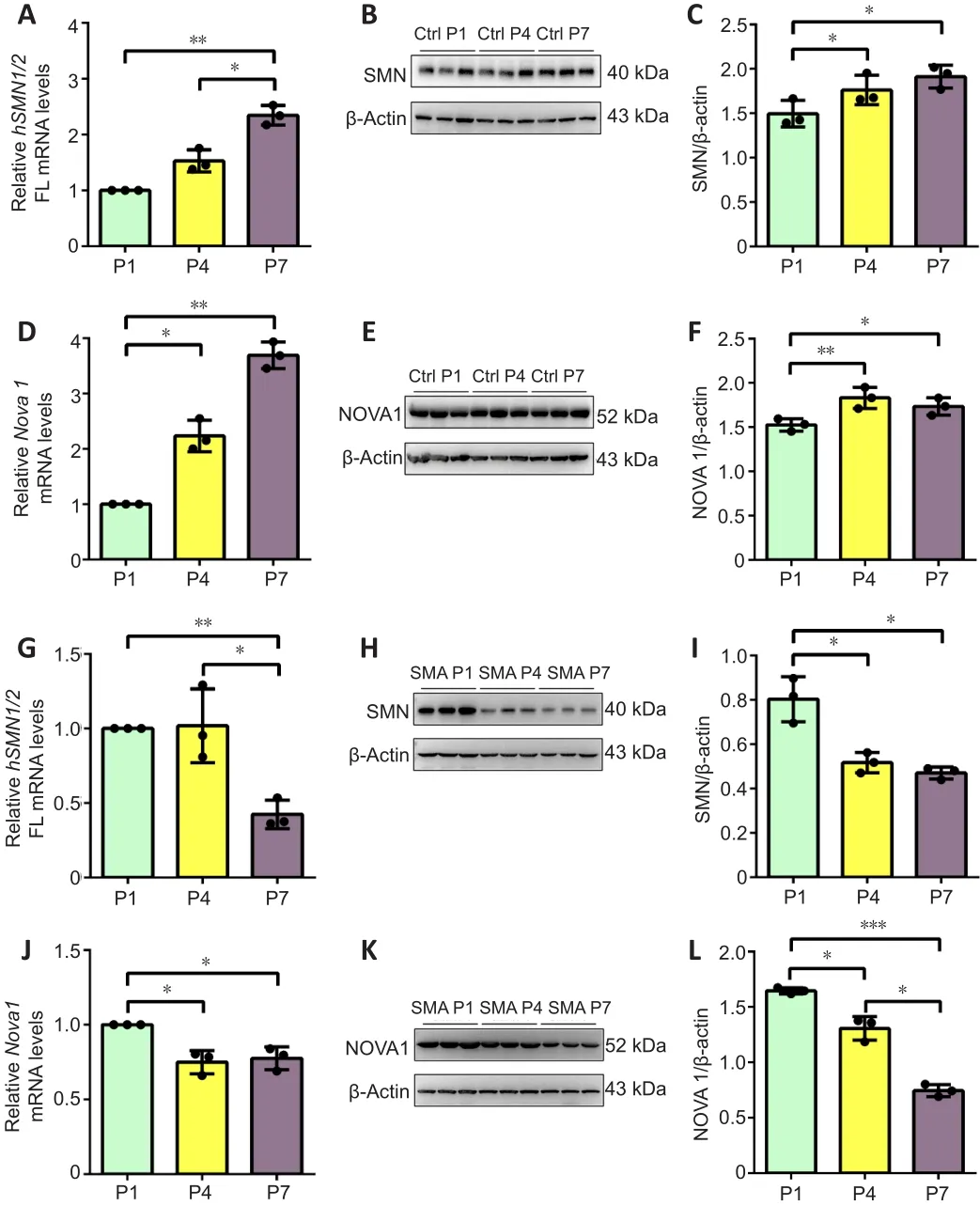
Figure 3|SMN and NOVA1 demonstrated similar expression patterns in spinal cordtissue from control and SMA mice at P1, P4, and P7.
NOVA1 is expressed in motor neurons and decreases with SMA progression in mice
The results described above showed that NOVA1 expression correlated with the expression of the SMA pathogenic protein SMN.Nissl staining showed that the number of neurons in the spinal anterior horn in SMA mice decreased continuously from P1 to P4 to P7 compared with that in control mice (Figure 4A).Bundles of hollow Nissl-positive plaque were found in the SMA anterior horn regions, unlike in control mice.We also found that the motor neuron marker ChAT partially co-localized with NOVA1 in the spinal anterior horn at the time points studied (Figure 4B).Taken together, NOVA1 expression was correlated with splicing of the SMA-related geneSMN2, SMN protein expression, and several motor neurons in the spinal cord anterior horn in SMA mice, indicating that NOVA1 may be involved in the disease progress in SMA mice.
NOVA1 regulates the inclusion of SMN2 exon 7 in vitro
To investigate the mechanism by which NOVA1 regulatesSMN2splicing, that is, inclusion ofSMN2exon 7, we transfected siRNA or over expressed plasmid ofNOVA1into U87MG cells.NOVA1mRNA (P< 0.01; Figure 5A) and protein (P< 0.05; Figure 5B and C) expression levels were significantly decreased after siRNA-mediated knock down.The inclusion ofSMN2exon 7 was significantly reduced whenNOVA1expression was knocked down (P< 0.05; Figure 5D and E).Meanwhile, the SMN protein level was also decreased (P< 0.01; Figure 5F and G).In contrast,NOVA1mRNA (P< 0.001; Figure 5H) and protein (P< 0.01; Figure 5I and J) expression levels were up-regulated after plasmid transfection.NOVA1expression was correlated with the rate ofSMN2exon 7 inclusion (P< 0.05; Figure 5K and L), and SMN protein expression increased(P< 0.05; Figure 5M and N).NOVA1 expression did not change after SMN knockdown (Additional Figure 2).In short, NOVA1 significantly promoted the inclusion ofSMN2exon 7 and increased the expression of SMN protein.
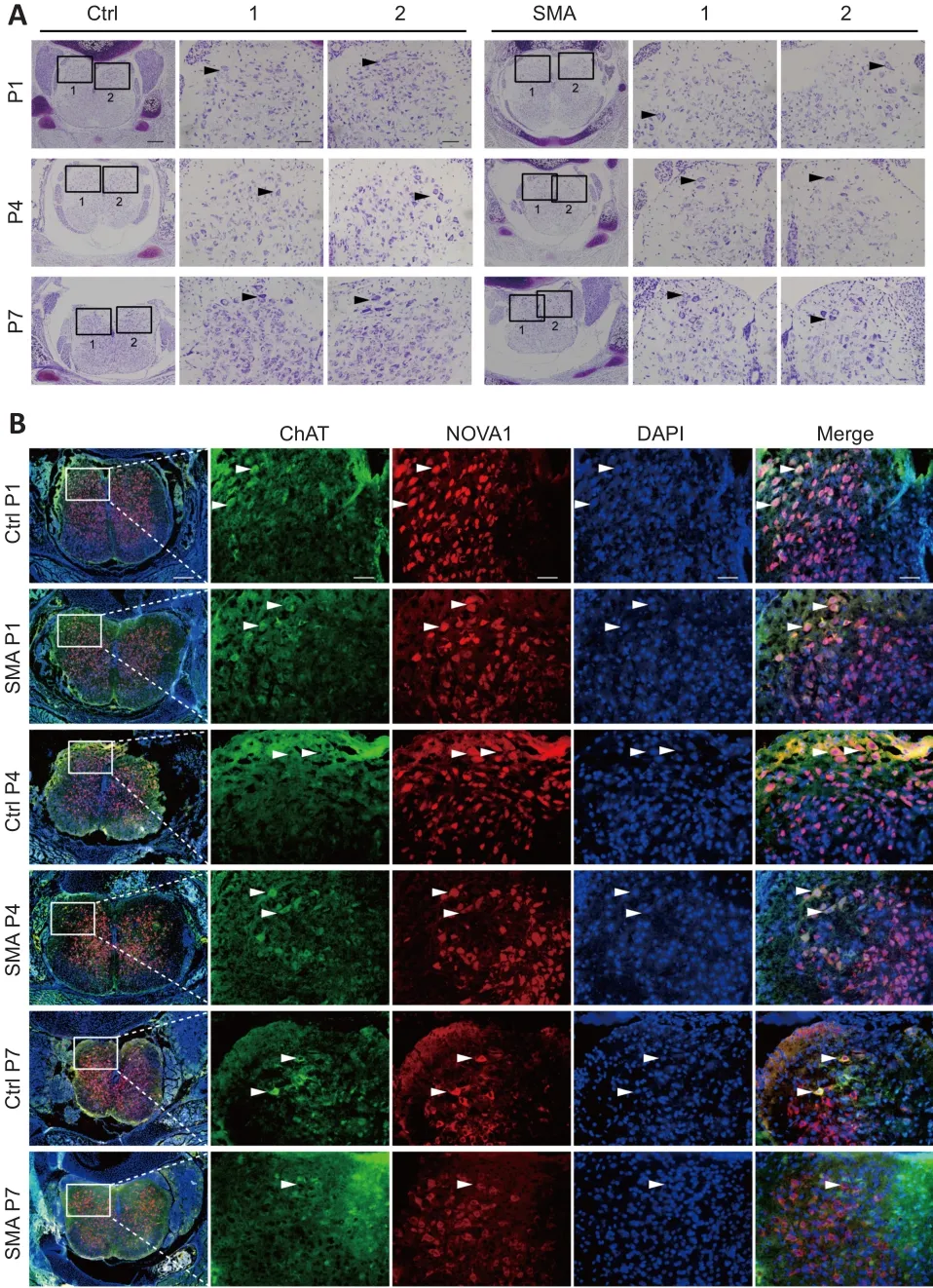
Figure 4|NOVA1 is expressed in motor neurons and decreases with SMA progression in mice.
NOVA1 binds to the UCAC motif in exon 7 of SMN2
Sequence analysis identified the presence of a single UCAC motif, which is the consensus sequence for the YCAY motif, in exon 7 ofSMN2(34 to 37 bp)(Figure 6A).To confirm that NOVA1 targets the UCAC sequence to regulate inclusion ofSMN2exon 7, we constructed anSMN2mini-gene in which the UCAC motif was deleted and a version in which it was mutated to UAAC and investigated the effect of these changes onSMN2splicing (Additional Figure 3).TheSMN2wild-type and mutated mini-genes were then transferred into HEK293T cells.The results showed that deletion of the UCAC sequence significantly reducedSMN2exon 7 inclusion (P< 0.05), and that mutation of the UCAC motif to UAAC reduced its inclusion even more significantly (P<0.001; Figure 6B and C).
To confirm that NOVA1 targeted binding the UCAC sequence to regulateSMN2splicing, RNA pull-down experiments were performed.NOVA1 was detected bound toSMN2WT (UCAC) mRNA, but not toSMN2Mut (UAAC)mRNA (Figure 6D), indicating that the UCAC sequence (34-37 bp) in exon 7 ofSMN2is the binding site for NOVA1.
Next, we tested the effect of mutation in the core “CA” sequence to the following sequences: UGAC, UUAC, UAGC, UGGC, UUGC, and UCGC (Figure 6E).We found that mutation of the UCAC sequence to UCAU (P< 0.001),CCAU (P< 0.05), UGAC (P< 0.01), UUAC (P< 0.001), UAGC (P< 0.001), UGGC (P< 0.001), or UUGC (P< 0.01) reduced the rate ofSMN2exon 7 inclusion (Figure 6F-H).However, mutation to CCAC (P< 0.01) or UCGC (P< 0.01) significantly up-regulated inclusion of exon 7 (Figure 6F-H), indicating that the “CA”sequence has a significant impact on inclusion.
Taken together, these findings show that the UCAC sequence is the key sequence required for NOVA1 to specifically bind toSMN2and regulate splicing.Therefore, more functional, stableSMN2FL is expressed in neuraltissues, which express high levels of NOVA1, than in tissues that express less NOVA1 (Figure 7A).In contrast, in non-neural tissues that express lower levels of NOVA1, more nonfunctionalSMN2?7mRNA is produced, leading to the production of nonfunctional and unstable SMN protein that is quickly degraded (Figure 7B).
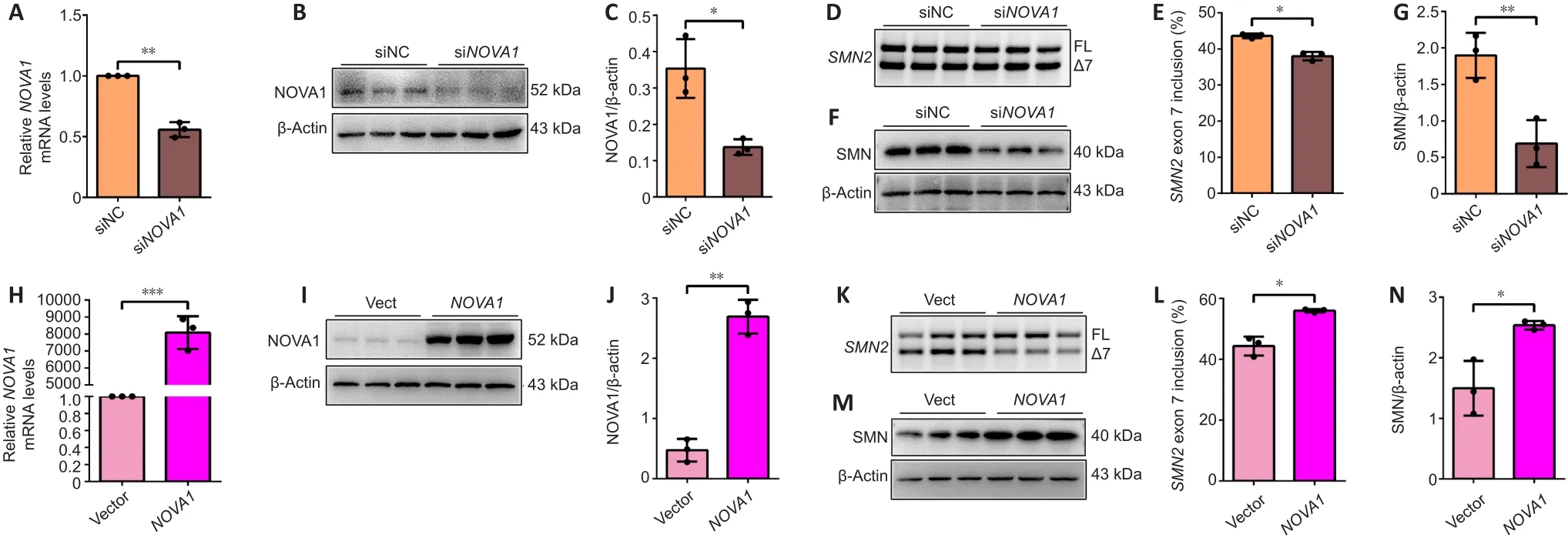
Figure 5|NOVA1 regulates SMN2 exon 7 inclusion in U87MG cells.
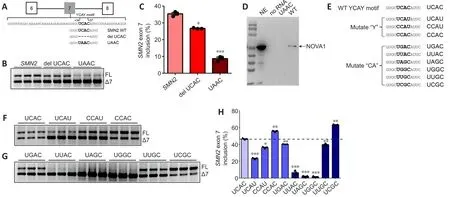
Figure 6|The UCAC motif is the NOVA1 binding site and plays an important role in SMN2 splicing.

Figure 7 | NOVA1 is expressed at high levels in neural tissue and regulates SMN2 splicing by binding the UCAC motif to promote SMN2 FL mRNA and protein expression.
Discussion
SMA, which is caused by a deficit in functional SMN protein synthesis due toSMN1mutation, is a rare disease that seriously patients’ quality of life.Comparison of the splicing ofSMN1and its homologueSMN2has shown that promoting the inclusion ofSMN2exon 7 can effectively reduce the severity of SMA (Son and Yokota, 2018).
In humans, approximately 95% of multi-exon genes are alternatively spliced(Pan et al., 2008).One-third of all disease-causing mutations alter pre-mRNA splicing (Lim et al., 2011), especially in the development of the nervous system, which affects neuronal differentiation and migration, axon growth and guidance, and synapse formation and function.Pre-mRNA splicing has an important impact on the growth and development of nervous tissue and the occurrence of neurological diseases, such as tau protein and Alzheimer’s disease (Love et al., 2015), myelin and demyelinating diseases (Aoyagi et al.,1999), and AMPA receptors and epilepsy (Rogawski and Donevan, 1999).As a neurological disease caused bySMN2alternative splicing, treatment of SMA from the perspective of splicing is currently a highly promising research direction.
There are many limitations to studying the treatment of SMA in human beings.The mouse is a common animal model with highly homologous genome sequences and similar physiological processes to humans and plays an important role in the research and treatment of SMA.A mouse model of SMA with symptoms similar to human SMA was established by the Hung Li laboratory.These mice carry two copies of theSMN2gene (genotypesmn-/-,SMN22tg/0) and develop typical symptoms of SMA 4 days after birth.This indicates that gene polymorphism may play a significant role in regulating the pathological changes that occur during this period (Hsieh-Li et al., 2000).
However, different pathological changes take place in different tissues and organs of SMA mice.The tissue specificity ofSMN2splicing was revealed by a previous study of a mouse model of type III SMA (Chen et al., 2015),suggesting the importance ofSMN2alternative splicing in SMA mice.
In this study, we detected the inclusion ofSMN2exon 7 in eight different tissues from SMA mice at P4.Interestingly, we found thatSMN2exon 7 inclusion differed among tissues, and to a greater degree in the neuraltissues of SMA and control mice.On the basis of the hypothesis that different expression levels of certain splicing factors in neural and non-neural tissues were responsible for the variable inclusion rates, we analyzed the expression of three types of classic splicing factors, HNRNP, SR, and the NOVA family,in different tissues.Enhanced expression of NOVA1 was detected in neural tissues in both control and SMA mice (Graus et al., 1993).Clinical studies have shown that SMA patients develop the disease at 6 months and die of respiratory disease before the age of 2 years.Studies have shown that the development of lung cancer and other respiratory diseases is closely related to NOVA1 (Ludlow et al., 2018; Qin et al., 2018).In addition, to the brain and spinal cord, NOVA1 and NOVA2 were also highly expressed in lung tissues in SMA mice, indicating the important role of NOVA1 in the respiratory system.
NOVA, which is a splicing factor that is specifically expressed in nerves,can regulate the inclusion of γ-aminobutyric acid receptor γ2 (Dredge and Darnell, 2003), deleted in colorectal carcinoma intron 16 (Leggere et al.,2016), and Sept8 exon 10b (Yuan et al., 2018), thereby affecting motor system development and function, as well as motor neuron survival.In addition,NOVA is involved in other neurological diseases, such as familial autonomic dysfunction diseases (Hervé and Ibrahim, 2016).NOVA1 is predominantly expressed in the ventral spinal cord and the midbrain (Graus et al., 1993).NOVA2 is localized in the neocortex and hippocampus (Yang et al., 1998).In this study, we found that NOVA1 expression is correlated with the inclusion ofSMN2exon 7 and is significantly down-regulated as the disease progresses in SMA mice.Immunofluorescence experiments also confirmed that NOVA1 was expressed in the anterior horn motor neurons of the spinal cord of SMA mice,where it may promote the inclusion ofSMN2exon 7.The NOVA-deficient mice created by Jensen and others (Jensen et al., 2000; Ruggiu et al., 2009) using gene-editing technology exhibit motor dysfunction and die within 10 days of birth because of motor neuron apoptosis.The symptoms observed in these mice are similar to those seen in our SMA mice, suggesting an association between NOVA1 expression and SMA severity.Here, we showed for the first time that NOVA1 is highly expressed in neural tissues and enhances SMN translation by promoting the inclusion ofSMN2exon 7.
Decreased SMN synthesis and loss of SMN function dominate the development of SMA.As a ubiquitously expressed protein, SMN protein is involved in small nuclear RNA biosynthesis, pre-mRNA splicing, apoptosis, axon transport, cytoskeleton dynamics, mitochondria, and bioenergy pathways, as well as other activities (Schrank et al., 1997; Paushkin et al., 2002; Fallini et al.,2012).The current study showed that SMN protein expression was notably down-regulated in parallel with SMA progression, which is consistent with the results reported by Groen et al.(2018).These findings confirm that temporal and spatial expression of SMN affects the severity of SMA.However, we also found that, while SMN is translated fromSMN2 FLmRNA, its expression level was remarkably down-regulated with decreased inclusion of exon 7, which may not be able to produce normal levels of SMN.Therefore, we established a positive correlation betweenSMN2exon 7 inclusion and SMN protein expression.We found that both NOVA1 and SMN were simultaneously downregulated in SMA mice.Similarly, NOVA1 up-regulated bothSMN2 FLand SMN protein expressionin vitro.Related studies have shown that SMN can interact with splicing factors, such as SMN and HNRNPR in axons, axon terminals, and the cytoplasm of mouse motor neurons (Dombert et al., 2014).Thus, it was speculated that NOVA1 may directly bind to SMN to regulate its expression,thereby affecting SMA severity; this putative regulation mechanism needs further study (Tadesse et al., 2008; Singh et al., 2017).Interestingly, separate mutation of the outer pyrimidine residues or the core “CA” sequence results in a significant reduction in the rate ofSMN2exon 7 inclusion, and mutation of the core sequence had the greatest impact.Therefore, the “CA” sequence in the YCAY motif is critical for the interaction with NOVA1.In addition,mutation of UCAC to CCAC or UCGC greatly promoted the inclusion of exon 7, suggesting that these mutations may have created new RNA-binding sites.Further research is needed to verify this hypothesis.
Potential treatments for SMA that are currently being investigated include sodium butyrate (Chang et al., 2001) and induced pluripotent stem cells(Corti et al., 2012).Although these methods are somewhat promising, their use is limited by minimal efficacy or by safety concerns.In recent years, gene therapy and antisense oligonucleotides have become popular options for the treatment of SMA.Spinraza is the first antisense oligonucleotide drug,developed by Hua et al.(2011), to treat SMA in children and adults (Hoy,2017).In addition, Zolgensma, the first gene therapy drug used to treat SMA in children younger than 2 years, was also approved by the Food and Drug Administration (FDA) for clinical treatment in May 2019 (Hoy, 2019).However,multiple systems have been reported to be affected in SMA (Farrar et al.,2017), and, while these drugs are effective in one or more systems, their limitations and side effects cannot be ignored.The small nucleic drugs have relatively high efficiency and a short half-life in the circulation.In contrast,overexpression vectors can be persistently expressed, have larger molecular weights, and potentially risk causing mutations in host genes if they cross the blood-brain barrier.At the same time, they cannot completely and effectively cure SMA, creating a greater economic burden on patients.Thus, the development of more effective SMA treatments is urgently needed.
In conclusion, SMA development correlates with down-regulation of NOVA1 expression, which exhibits the same inter-tissue differences asSMN2splicing,and NOVA1 binds to the UCAC sequence located at 34-37 bp ofSMN2exon 7 to regulateSMN2splicing.These findings provide new perspectives on correctingSMN2gene splicing and treating SMA.
Acknowledgments:We thank Professor Yi-Min Hua from Institute of Neuroscience in Soochow University for providing SMA mice and guidance in constructing SMN2 mini-gene.
Author contributions:Study design and manuscript revision: LCW, JJS;experiment implementation: LLD, JJS, LCW, ZHC; data analysis: LCW, JJS;experiment guidance: YXS; manuscript draft: LLD, LCW.All authors read and approved the final manuscript.
Conflicts of interest:We hereby declare that there is no conflict of interest between the authors of this manuscript.
Author statement:This paper has been posted as a preprint on Research Square with doi: https://doi.org/10.21203/rs.3.rs-798194/v1, which is available from: https://assets.researchsquare.com/files/rs-798194/v1/4f3e17b8-317a-4fd5-8894-2e46bf019299.pdf?c=1637245333.
Availability of data and materials:All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Table 1:Primer sequence used to construct the SMN2 mini-gene with UCAC sequence deletion or mutation on SMN2 exon 7.
Additional Table 2:Primer sequence used in this experiment.
Additional Figure 1:PCGT7-NOVA1 sequence by forward CMV primer and reverse primer.
Additional Figure 2:Western blot detection of NOVA1 expressions after SMN knockdown.
Additional Figure 3:Diagrams of sequencing results before and after UCAC sequence deletion or mutation.
 中國(guó)神經(jīng)再生研究(英文版)2022年11期
中國(guó)神經(jīng)再生研究(英文版)2022年11期
- 中國(guó)神經(jīng)再生研究(英文版)的其它文章
- Modeling Alzheimer’s disease:considerations for a better translational and replicable mouse model
- Engineering cerebral folding in brain organoids
- Verapamil, a possible repurposed therapeutic candidate for stroke under hyperglycemia
- Delayed activation of leg somatotopic fibers of an injured corticospinal tract in a patient with cerebral infarction
- Polydopamine-modified chitin conduits with sustained release of bioactive peptides enhance peripheral nerve regeneration in rats
- Obstructive sleep apnea aggravates neuroinflammation and pyroptosis in early brain injury followingsubarachnoid hemorrhage via ASC/HIF-1α pathway
