Polymorphism in the Ras and β-Glucosidase Genes and Their Association with Growth Traits in the Pacific Oyster Crassostrea gigas
HAN Ziqiang, CONG Rihao, and LI Qi, 2), *
Polymorphism in theandGenes and Their Association with Growth Traits in the Pacific Oyster
HAN Ziqiang1), CONG Rihao1), and LI Qi1), 2), *
1),,,266003,2),,266237,
Thegene, a conserved member of the insulin pathway, andgene, an important cellulase, are two im- portant growth-related genes.However, there is no study on the association between mutations of these two genes and growth traits in bivalves. Here, the polymorphism of these two genes inwere revealed.Their association with growth traits was evaluated in 290 oysters from five families, and was further confirmed in another 186 oysters from three fast-growing strains. Seven- teen and twelve SNPs were identified in thegene andgene, respectively. Among these SNPs, four SNPs in each gene (: C.86C>A, C.90T>C, C.112A>G and C.118G>A;: C.247G>A, C.284C>T, C.1260C>T and C.1293T>C) were significantly (<0.05) associated with the growth ofthese oysters. Furthermore, eight and nine haplotypes were constructed in thegene andgene, respectively. Oysters with both haplotypes R-Hap5 (CCAA) and β-Hap7 (ACCT), or with both R-Hap 6 (ATGG) and β-Hap 6 (ACTC), or with both R-Hap 6 and β-Hap 9 (ACTT), or with both R-Hap 7 (ATAA) and β-Hap 7, show- ed the highest growth performances. These results provide candidate markers for selectingwith fast growth.
; growth; SNP;;
1 Introduction
The Pacific oyster (), which was once endemic to China, Japan and Korean, is now one of the most widely farmed aquaculture species worldwide due to its strong adaptability (Miossec., 2009). However, the production and profitability ofhas been limited, as world production is mainly based on un-improved po- pulations or stocks (Gjedrem., 2012; Gjedrem and Rye, 2018). With the development of efficient hatchery technique, many breeding programs ofhave been es- tablished in many countries (Ward., 2000; Langdon., 2003; Dégremont., 2007; Li., 2011). Fast growth, as one of the traits that directly affects the produc- tion and profitability, is the preferred target trait in almost all breeding programs (Ward., 2000; Langdon., 2003; Li., 2011). Compared with the selection only relying on phenotype, incorporating genome information in- to selective breeding has become an important aspect to enhance the efficiency of selection (Hollenbeck and Johns- ton, 2018).
Recently, a lot of candidate genes associated with growthtraits have been revealed in scallop (Sun., 2020), mus-sel (Prieto., 2019) and clam (Saavedra., 2017) with the rapid development of genome resources.Many single nucleotide polymorphisms (SNPs) in these candi- date genes, such asgene (Huang., 2016),gene (Niu, 2015; Fan., 2017), long- chain fatty acid-CoA ligases gene (Dai., 2015), zinc finger transcription factor (Yang., 2020) and insulin- like growth factor (Feng., 2014; Ning., 2018) have been identified associated with growth traits. For, however, onlygene (Huvet., 2008),gene (Chen., 2020) and a few genes from in- sulin family (Cong., 2013, 2014; Moon and Choi, 2019) have been reported to have growth-related muta- tions. Considering the complexity of the genetic mecha- nism underling growth traits and the growth performances are affected by a series of processes, including the food acquisition and absorption, energy conversion and distri- bution,., it is necessary to further clarify the relation- ship between more genes and growth and their role in growth, which will be helpful to selectwith fast growth.
The insulin family is widely distributed and highly con- served in vertebrates and invertebrates, and plays critical roles in regulating development, reproduction, growth and carbohydrate metabolism (Gricourt., 2006; Schlueter., 2007; Zhang and He, 2020). In, several genes from the insulin-like family, such as insulin recep-tor-related receptor (Cong., 2014), insulin-related pep- tide (Cong., 2013; Shi., 2013), insulin growth factor binding protein (Zhang., 2017; Choi., 2018),have been identified as being associated with growth traits.gene, as a conserved member of the insulin family,plays an important role in regulating cell growth, differ- entiation and apoptosis (Ciocan., 2006). It has been cloned and identified to be involved in carcinogenesis in(Ciocan., 2006) and(Lima., 2008). In,gene is mainly expressed in mantle edges and visceral ganglia, and is overexpressed during the tissue rebuilding stage in the storage tissue, which indicates that this gene may be related to the regulation of growth (Jouaux., 2012).
is a filter-feeder whose natural diet is mainly phytoplankton. An important component of phytoplank- ton is cellulose, which is a macromolecular polysaccha- ride composed of glucose (Watanabe and Tokuda, 2001). Cellulose needs to be degraded into monosaccharide under the catalysis of cellulase, before it can be absorbed by organisms (Beguin and Aubert, 1994). Cellulase is not only widely distributed in protozoa, bacteria, fungi, and plants, but also in many aquatic invertebrates (Tanimura., 2013). The, an important cellulase, plays a critical role in the cellulose degradation (Lynd., 2002). In bivalves, endogenoushas been identified in(Sakamoto., 2009)and(Liu, 2012). It is expressed only in the digestive glands, suggesting that it might be related to the digestive and absorption functions (Liu, 2012).
In order to determine whetherandgenes can be used as indicators in breeding program for, this study explored the polymorphisms of these two genes and analyzed their association with growth traits in different families and fast-growing strains of.
2 Materials and Methods
2.1 Animals and Traits
In June 2009, 54 full-sib families were established with each male mating to three females, using 80 wildcollected from Rushan Bay, Shandong province, China as parents (Cong., 2013, 2014). In March 2011, five fa- milies (55 to 60 oysters per family) were randomly select- ed from these 54 families, and 290 oysters were randomly selected from these five families for preliminary association analysis.
Three selected strains of, strain C, strain J and strain K, were initiated in 2007 in our breeding programtargeting at fast growth. The oysters collected from three stocks in Rushan in Shandong province, China (36.4?N, 121.3?E), Onagawa Bay in Miyagi Prefecture, Japan (38.3?N,141.3?E), and Pusan, South Korea (35.1?N, 129.1?E) as pa- rents, respectively (Li., 2011). In 2008, the second- generation of these three strains were established with oysters collected from the first-generation of strain C (30×31 cross, selection intensity=1.872), strain J (34×36 cross, selection intensity=1.870) and strain K (30×35cross, selection intensity=1.728) as parents, respectively (Wang., 2012). In July 2009, ten females and ten males were selected from each strain as parents to generate the third generation of strain C, strain J and strain K. After four- teen months, 62 oysters were randomly selected from each strain for verifying analysis.
The adductor muscles of 290 oysters from these five fa- milies and 186 oysters from these three strains were ob- tained and stored in ?20℃ for extracting DNA. Shell height, shell length and shell width of 476 oysters were measured using an electronic vernier caliper (0.01mm), and body weight and soft-tissue weight were weighed using an elec- tronic balance (0.01g).
2.2 DNA Extraction and Primer Design
Genomic DNA of each sample was extracted by using the phenol-chloroform method (Li., 2002). PCR pri- mers were designed by Primer Premier 5.0 according to the cDNA sequence ofgene andgene (Table 1). The cDNA sequence ofgene (1145bp, see Fig.1) was amplified by primers 5’-CCCGTCCTCATGT ACTGGTC-3’ and 5’-ATCTTGGATACGGCAGGTCA-3’ reported by Jouaux. (2012), while the cDNA sequence ofgene has been reported by Liu (2012).
2.3 SNP Genotyping
A total of 476 oysters were genotyped using the single- strand conformation polymorphism (SSCP) technique and confirmed by random sequencing as described by Cong. (2014). Briefly, 5μL of each PCR product was add- ed to 10μL denaturing buffer (98% formamide, 0.09% xy- lene cyanole FF, and 0.09% bromophenol blue). These sam- ples were denatured at 94℃ for 5min and then immedi- ately placed on ice for 10min. Electrophoresis of the de- natured DNA was performed in 8%–12% nondenaturing polyacrylamide gel with 120V for 12–14h at 4℃. Final- ly, SSCP patterns on the gels were visualized by silver stain- ing (Ou., 2005). In order to confirm these genotypes obtained by SSCP, the ABI 3730 sequencer (Applied Bio- systems) was used to sequence more than three individual PCR products with the same SSCP pattern in both directions.
2.4 Association Analysis
The association analysis included the preliminary asso- ciation analysis between SNPs and growth traits using oys- ters from families, and verification of growth-related SNPs using oysters from three strains. First, the polymorphisms of bothgene andgene were analyzed with the parents of five oyster families. According to the polymorphisms of these two genes in the parents, the fa- milies that need to be genotyped were determined, and a preliminary association analysis was preformed to screen out potential SNPs associated with growth traits. Then the association between these SNPs and growth traits were ve- rified in three fast-growing strains. Finally, haplotypes based on verified growth-related SNPs were constructed to fur- ther verify their correlation with growth traits of.

Table 1 Primers for analysis of SNPs in the Ras and β-glucosidase genes in C. gigas

Fig.1 Distribution of 17 SNPs in the Ras gene of Crassostrea gigas.
2.5 Statistical Analysis
The association between SNPs and growth traits were analyzed using the general linear model (GLM) procedure of SAS v8.2 (SAS Institute Inc.). The significance of the differences between genotypes or between haplotypes wasanalyzed by Bonferroni’s multiple comparison, and the sig- nificance of the differences was<0.05.
The model of ANOVA for the genotype or haplotype of each growth trait isy=GorH+e, whereyis the observed value ofth individual of genotype or haplotype;is the mean of observed values;Gis the fixed effects of the genotype;His the fixed effects of the haplotype; andeis the random residual effect corresponding to the observed values. No other effects such as generation and sitewere taken in these analyses, because all the oysters of preli- minary association analysis or verified analysis were cul- tured under the same condition and sampled at the same age.
The allelic frequency, heterozygosity and polymorphism information content (PIC) were calculated using an online software PowerMarker v3.25 (http://statgen.ncsu.edu/power marker/).
3 Results
3.1 SNP Identification
Forgene, a total of seventeen SNPs, including C.63C>T, C.86C>A, C.87A>G, C.90T>C, C.92C>A, C.112A>G, C.118G>A, C.625T>A, C.642C>T, C.672G>A, C.675G>A, C.747T>A, C.775T>A, C.786G>A, C.918G>A, C.949G>A and C.953A>G, were detected in the 669bp coding sequence amplified by these four pairs of pri- mers (Table 1). In addition, twelve SNPs, including C.247G>A, C.270C>A, C.276C>T, C.284C>T, C.288C>T,C.301C>T, C.352G>C, C.1260C>T, C.1272G>A, C.1287G>A, C.1293T>C and C.1301C>T were identi- fied in the 716bp coding sequence ofgene amplified by these five pairs of primers (Table 1).
3.2 Preliminary Association Analysis Between SNPs and Growth Traits
Among the seventeen SNPs locating ingene, four SNPs were determined to be significantly (<0.05) related to at least one of the five growth traits by preliminary as- sociation analysis in five families (Table 2). At C.86C>A, oysters with genotypes AA or AC were significantly (<0.05) larger than those with genotype CC in shell height, shell length, body weight and soft-tissue weight. Similarly, significant (<0.05) differences in these four growth traits were also observed at C.112A>G and C.118G>A. At C.112A>G, oysters with genotypes AA were significant- ly (<0.05) larger than those with genotype GA, and oys- ters with genotypes GG or GA at C.118G>A were signi- ficantly (<0.05) larger than those with genotype AA. In addition, at C.90T>C, significant (<0.05) difference was only observed in shell height, in which the oysters with genotype TC were significantly larger than those with genotype TT. As the result, all four growth-related SNPs belong to the same intron.

Table 2 Association between SNPs in Ras and β-glucosidase genes and growth traits in five families of C. gigas
Notes: Means with different superscripts within the same column differ significantly at<0.05. Bold values indicate-value<0.05.
Forgene, six SNPs were determined to be significantly (<0.05) related to at least one of the five growth traits (Table 2). At C.247G>A, oysters with geno- type GG were significantly (<0.05) larger than those with genotype GA in shell height, shell length, body weight and soft-tissue weight. Similarly, for these four growth traits, oysters with genotype AA at C.257A>T were significant- ly (<0.05) larger than those with genotype AT, and oys- ters with genotype CC in C.270C>A were significantly (<0.05) larger than those with genotype CA. In both C.284C>T and C.1260C>T, oysters with genotype CC showed significantly (<0.05) higher performances in shell height, body weight and soft-tissue weight. In addi-tion, in C.1293T>C, oysters with genotypes TT and CC were significantly (<0.05) larger than those with geno- type TC in shell length, total weight and soft-tissue weight. C.247G>A and C.284C>T were nonsynonymous muta- tions (p.Asp63Asn and Thr75Ile), while C.1260C>T and C.1293T>C were synonymous mutations.
3.3 Verification of Growth-Related SNPs
Similar trends as the abovementioned results were ob- served in these verified individuals, although there were slight differences in specific traits (Table 3). Forgene, at C.86C>A, oysters with genotype AA or AC were sig- nificantly (<0.05) larger than those with genotype CC only in shell height and soft-tissue weight, and oysters with genotype AA were significantly (<0.05) larger thanthose with genotype CC in shell weight and body weight, but no significant (>0.05) differences were observed in shelllength. At C.112A>G, oysters with genotype AA were significantly (<0.05) larger than those with genotype GA not only in shell height, shell length, body weight and soft- tissue weight, but also in shell width. Similarly, at C.118G>A, oysters with genotypes GG and GA were significantly (<0.05) larger than those with genotype AA in all five growth traits. Moreover, at C.90T>C, no significant (> 0.05) difference was observed in shell height, but the oys- ters with genotype TC were significantly (<0.05) lar-ger than those with genotype TT in soft-tissue weight.
Forgene, only four of the six growth-re- lated SNPs were observed significantly related to growth traits in verified individuals (Table 3). At C.247G>A, oys- ters with genotype AA were significantly (<0.05) larger than those with genotype GA in all five growth traits, while the performances of oysters with genotype GG were be- tween AA and GA. At C.284C>T, oysters with genotype CC were significantly (<0.05) larger than those with geno- type CT in shell length, body weight and soft-tissue weight, while no significant (>0.05) difference was observed in shell height. Notably, at C.1260C>T, the greatest perfor- mances were observed in oysters with genotype TT not in those with genotype CC. In addition, at C.1293T>C, oys- ters with genotypes TT and CC were significantly (<0.05)larger than those with genotype TC not only in shell length, total weight and soft-tissue weight, but also in shell width.

Table 3 Association between SNPs in Ras and β-glucosidase genes and growth traits in three strains of C. gigas
Notes: Means with different superscripts within the same column differ significantly at<0.05.Bold values indicate-value<0.05.
3.4 Construction of Haplotypes and Their Association with Growth Traits
Eight haplotypes were constructed based on four growth-related SNPs (C.86C>A, C.90T>C, C.112A>G and C.118G>A) ofgene, and these eight haplotypes were signi- ficantly (<0.05) associated with all five growth traits (Table 4). For all five growth traits, oysters with the haplotype R-Hap8 (ACAA) had the highest performances, and were significantly (<0.05) larger than those with haplotype R-Hap1 (CTAA) and R-Hap2 (CCAG).

Table 4 Associations between haplotypes in Ras gene and growth traits in three strains of C. gigas
For, nine haplotypes constructed basedon four growth-related SNPs (C.247G>A, C.284C>T,C.1260C>T and C.1293T>C) were significantly (<0.05) associated with all five growth traits (Table 5). Further- more, oysters with the haplotypeβ-Hap9 (ACTT) had the highest performances, and were significantly (<0.05) lar- ger than those with other haplotypes.
From the above eight growth-related SNPs fromandgenes, 27 haplotypes significantly (<0.05) associated with four growth traits were constructed (Table 6). Oysters with haplotypes R5_β7, R6_β6, R6_β9 and R7_β7 were significantly (<0.05) higher than those of haplotype R1_β2 in shell height, body weight and soft-tis- sue weight.
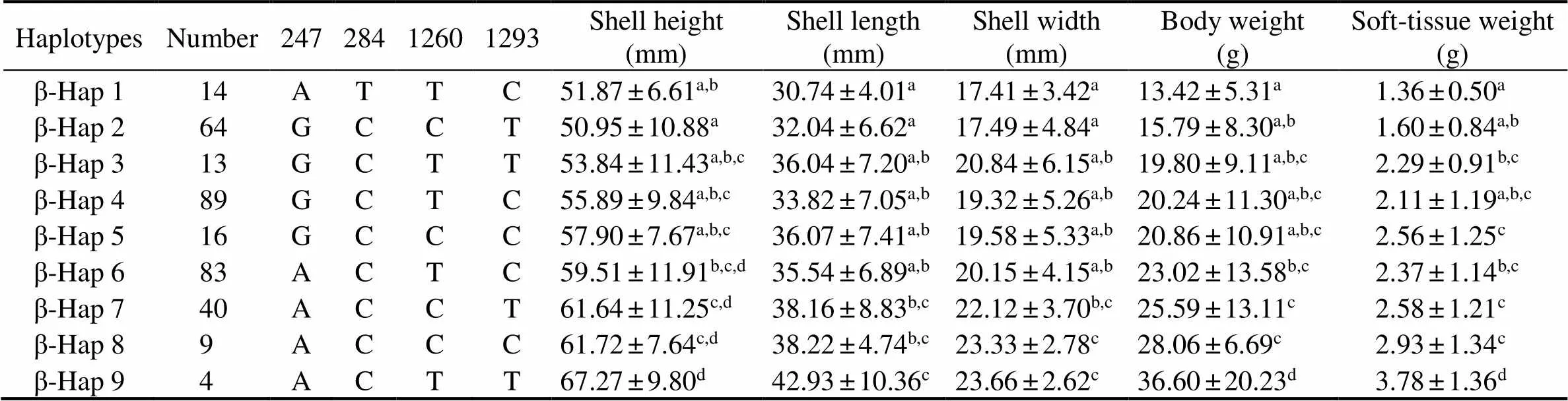
Table 5 Associations between haplotypes in β-glucosidase gene and growth traits in three strains of C. gigas

Table 6 Associations between haplotypes in Ras and β-glucosidase genes and growth traits in three strains of C. gigas
()
()

HaplotypesNumberShell height(mm)Shell length(mm)Shell width(mm)Body weight(g)Soft-tissue weight(g) R5_β51058.15±6.92a,b39.91±8.60a,b20.78±6.38a26.83±13.06a,b3.27±1.55b R5_β6563.6±11.20b37.93±10.12a,b22.19±2.57a30.68±20.83a,b3.12±1.27a,b R5_β71064.55±13.18b41.08±9.61b21.86±3.82a29.62±16.82b3.11±1.43b R6_β2347.03±22.23a,b30.89±9.92a,b15.57±7.65a13.93±10.82a,b1.60±1.45a,b R6_β31257.05±9.58a,b36.6±6.58a,b22.32±5.77a19.82±8.28a,b2.44±0.78a,b R6_β44455.79±8.69b34.27±7.76a,b19.86±5.53a21.42±12.02a,b2.28±1.31a,b R6_β65260.29±12.31b35.74±7.77a,b20.82±5.31a23.52±13.10b2.38±1.17b R6_β7559.92±9.30a,b40.09±7.51a,b23.22±5.18a23.72±13.03a,b2.48±1.26a,b R6_β9567.29±17.73b43.39±13.28a,b23.58±2.92a37.28±22.55b3.68±2.05b R7_β2350.36±22.72a,b31.21±9.80a,b19.16±5.11a17.17±16.35a,b1.47±1.07a,b R7_β4372.83±2.65b39.35±7.08a,b21.61±6.69a33.33±14.77a,b3.57±1.84a,b R7_β7473.61±8.78b42.75±7.84a,b24.73±3.81a37.95±5.88b3.58±0.56b
Note: Haplotype R1_β2 means haplotype R-Hap1 in Table 4 is grouped with β-Hap2 in Table 5, and so on.
3.5 Allelic Frequencies of Eight Growth-Related SNPs in Three Strains
Allelic frequencies of these eight growth-related SNPs were shown in Table 7. The PIC of C.284C>T were from 0.02 to 0.11, while the rest seven SNPs were from 0.26 to 0.37. For C.86C>A and C.284C>T, Hardy-Weinberg equi- librium was observed in all three strains of China, Japan and Korea, while C.90T>C, C.112A>G and C.247G>A did not meet Hardy-Weinberg equilibrium in any of these three strains. In addition, for C.118G>A, Hardy-Wein- berg equilibrium was observed in China and Korea strains, but not in Japan strain. For C.1260C>T and C.1293T>C, Hardy-Weinberg equilibrium was observed in Japan strain, but not in China and Korea strains.
4 Discussion
The SNP detection of the candidate genes to obtain po- tential markers for specific traits can be used to improve the efficiency of selective breeding (Yang., 2020). Thegene andgene are two important growth- related genes (Lynd., 2002; Jouaux., 2012), but there is no study on the association between mutations of these two genes and growth traits in bivalves so far. In thisstudy, seventeen SNPs were revealed in 669bp of the cod- ing region of thegene, and twelve SNPs were reveal- ed in 716bp of the coding region of thegene.The SNP densities of these two genes were 2.54% and 1.68respectively, which is consistent with the average polymor- phism rate 2.3% in(Zhang., 2012).
Forgene, four SNPs significantly (<0.05) associat- ed with growth traits have been identified through preli- minary and validation analysis. Consideringgene iden- tified by this study, as well as the insulin receptor-related receptor (Cong., 2014), insulin-related peptide (Cong., 2013; Shi., 2013), insulin growth factor bind- ing protein (Zhang., 2017; Choi., 2018) report- ed in previous studies, now a total of four genes from the insulin pathway have been identified to be associated with growth traits in. The regulatory effect of insulin pathway on growth traits has also been well documented in other animals (Gricourt., 2006; Schlueter., 2007;Zhang and He, 2020). Moreover, thegene is abun-dantly expressed in the mantle that plays an important rolein the shell formation and soft body growth during the rapid growth stage (Jouaux., 2012). However, the mole- cular mechanism under the association between these four SNPs and growth ofremain unclear. These four SNPs may affect the efficiency of transcription, because introns can increase transcript levels and increase the ef- ficiency of mRNA translation by affecting the rate of trans-cription, nuclear export, and transcript stability (Shaul, 2017). In addition, the haplotype R-Hap8 was found to show the best growth performance. Haplotype, as a combination of alleles of multiple loci inherited on the same chromosome, can provide more information than SNP (Daly., 2001).
Similarly, forgene, four SNPs significant- ly (<0.05) associated with growth traits have been iden- tified through preliminary and validation analysis. The, as an important cellulase, plays a critical role in the cellulose degradation (Lynd., 2002).that is expressed in the mammalian liver, kidney, intestine and spleen might be involved in the degradation and absorption of flavonoid glycosides (Berrin., 2003).In,gene is expressed only in the di- gestive gland, indicating that it might be involved in the digestion and absorption (Liu, 2012). Among the four growth-related SNPs ofgene, C.247G>A and C.284C>T were nonsynonymous mutations (p.Asp63Asnand Thr75Ile), which may cause changes in the spatial structure and function of the encoded protein, thereby af- fecting its physiological functions. C.1260C>T and C.1293T>C were synonymous mutations, which may affect tran- scription efficiency as a positive regulator, or may closely relate to the causal mutation (Beuzen., 2000). The haplotype β-Hap9 showed the highest performances. How- ever, if it is used as a selective marker, some smaller oys- ters might be selected as a big variance of body weight was observed in this haplotype.
The accuracy of selection can be improved by consider- ing both the haplotypes ofandgenes. Haplotypes R-Hap5and β-Hap7, R-Hap 6 and β-Hap 6, R-Hap 6 and β-Hap 9, as well as R-Hap 7 and β-Hap 7 can be used to assist the selection of fast-growing individuals. In addition, the effect of both haplotypes R-Hap 8 and β-Hap 9 is not included here, because the oysters with this com- bination were too few to be analyzed.

Table 7 Allelic frequencies of growth-related SNPs in three C. gigas strains
Note: PIC, polymorphism information content.
The polymorphism level can be classified into three types according to the PIC value, including low polymorphism (PIC value<0.25), intermediate polymorphism (0.25
5 Conclusions
This study explored the polymorphisms ofandgenes and analyzed their association with growth traits in. Four SNPs withingene and four SNPs withingene were significantly as- sociated with growth traits. Grouping differentandgenes together, haplotypes R-Hap5 and β-Hap7, R-Hap 6 and β-Hap 6, R-Hap 6 and β-Hap 9, and R-Hap 7 and β-Hap 7 can be used to assist the selection of fast- growing individuals. These results provide candidate mar- kers for the selective breeding ofwith fast growth.
Acknowledgements
This work was supported by the National Natural Sci- ence Foundation of China (No. 31972789), the Industrial Development Project of Qingdao City (No. 20-3-4-16- nsh), and the Science and Technology Development Pro- ject of Weihai City (No. 2018NS01).
Beguin, P., and Aubert, J. P., 1994. The biological degradation of cellulose., 13: 25-58, DOI: 10.1016/0168-6445(94)90099-x.
Berrin, J. G., Czjzek, M., Kroon, P. A., McLauchlan, W. R., Puigserver, A., Williamson, G.,., 2003. Substrate (agly- cone) specificity of human cytosolic beta-glucosidase., 373: 41-48, DOI: 10.1042/bj20021876.
Beuzen, N. D., Stear, M. J., and Chang, K. C., 2000. Molecular markers and their use in animal breeding., 160: 42-52, DOI: 10.1053/tvjl.2000.0468.
Chen, N., Li, L., Li, C. H., Lin, Z. H., Meng, J., Liu, S.,., 2020. bHLH genes polymorphisms and their association with growth traits in the Pacific oyster., 38: 862-868, DOI: 10.1007/s00343-019-9070-4.
Choi, Y. H., Kim, E. Y., and Nam, T. J., 2018. Involvement of insulin-like growth factor in intraspecific variation in growth of Pacific oysterduring winter., 84: 1017-1024, DOI: 10.1007/s12562-018-1232-3.
Ciocan, C. M., Moore, J. D., and Rotchell, J. M., 2006. The role of Ras gene in the development of haemic neoplasia in., 62: S147-S150, DOI: 10.1016/j.marenvres.2006.04.020.
Cong, R. H., Kong, L. F., Yu, H., and Li, Q., 2014. Association between polymorphism in the insulin receptor-related receptor gene and growth traits in the Pacific oyster., 54: 144-149, DOI: 10.1016/j.bse.2014.02.003.
Cong, R. H., Li, Q., and Kong, L. F., 2013. Polymorphism in the insulin-related peptide gene and its association with growth traits in the Pacific oyster., 46: 36-43, DOI: 10.1016/j.bse.2012.09.008.
Dai, P., Huan, P., Wang, H. X., Lu, X., and Liu, B. Z., 2015. Cha- racterization of a long-chain fatty acid-CoA ligase 1 gene and association between its SNPs and growth traits in the clam., 566: 194-200, DOI: 10.1016/j.gene.2015.04.047.
Daly, M. J., Rioux, J. D., Schaffner, S. E., Hudson, T. J., and Lan- der, E. S., 2001. High-resolution haplotype structure in the hu- man genome., 29: 229-232, DOI: 10.1038/ng1001-229.
Dégremont, L., Ernande, B., Bedier, E., and Boudry, P., 2007. Summer mortality of hatchery-produced Pacific oyster spat(). I. Estimation of genetic parameters for sur-vival and growth., 262: 41-53, DOI: 10.1016/j.aqua-culture.2006.10.025.
Fan, S. G., Xu, Y. H., Liu, B. S., He, W. Y., Zhang, B., Su, J. Q.,., 2017. Molecular characterization and expression ana- lysis of the myostatin gene and its association with growth traits in Noble scallop ()., 212: 24-31, DOI: 10.1016/j.cbpb.2017.07.004.
Feng, L. Y., Li, X., Yu, Q., Ning, X. H., Dou, J. Z., Zou, J. J.,., 2014. A scallop IGF binding protein gene: Molecular charac- terization and association of variants with growth traits., 9: 7, DOI: 10.1371/journal.pone.0089039.
Gjedrem, T., 2012. Genetic improvement for the development of efficient global aquaculture: A personal opinion review., 344: 12-22, DOI: 10.1016/j.aquaculture.2012.03.003.
Gjedrem, T., and Rye, M., 2018. Selection response in fish and shellfish: A review., 10: 168-179, DOI: 10.1111/raq.12154.
Gricourt, L., Mathieu, M., and Kellner, K., 2006. An insulin-like system involved in the control of Pacific oysterreproduction: hrIGF-1 effect on germinal cell prolifera- tion and maturation associated with expression of an homolo- gous insulin receptor-related receptor., 251: 85-98, DOI: 10.1016/j.aquaculture.2005.05.015.
Hollenbeck, C. M., and Johnston, I. A., 2018. Genomic tools and selective breeding in molluscs., 9: 253, DOI: 10.3389/fgene.2018.00253.
Huang, G. J., Guo, Y. H., Li, L., Fan, S. G., Yu, Z. N., and Yu, D. H., 2016. Genomic structure of the alpha-amylase gene in the pearl oysterand its expression in response to salinity and food concentration., 587: 98-105, DOI: 10.1016/j.gene.2016.04.044.
Huvet, A., Jeffroy, F., Fabioux, C., Daniel, J. Y., Quillien, V., Van Wormhoudt, A.,., 2008. Association among growth, food consumption-related traits and amylase gene polymorphism in the Pacific oyster., 39: 662-665, DOI: 10.1111/j.1365-2052.2008.01776.x.
Jouaux, A., Franco, A., Heude-Berthelin, C., Sourdaine, P., Blin, J. L., Mathieu, M.,., 2012. Identification of Ras, Pten and p70S6K homologs in the Pacific oysterand diet control of insulin pathway., 176: 28-38, DOI: 10.1016/j.ygcen.2011.12.008.
Langdon, C., Evans, F., Jacobson, D., and Blouin, M., 2003. Yields of cultured Pacific oystersThunberg improved after one generation of selection., 220: 227-244, DOI: 10.1016/s0044-8486(02)00621-x.
Li, Q., Wang, Q. Z., Liu, S. K., and Kong, L. F., 2011. Selection response and realized heritability for growth in three stocks of the Pacific oyster., 77: 643-648, DOI: 10.1007/s12562-011-0369-0.
Lima, I., Peck, M. R., Rendon-Von Osten, J., Soares, A. M. M., Guilhermino, L., and Rotchell, J. M., 2008. Ras gene in marine mussels: A molecular level response to petrochemical exposure., 56: 633-640, DOI: 10.1016/j.marpolbul.2008.01.018.
Liu, C. H., 2012. Cloning, characterization and expression ana- lysis of cellulose gene in. Master thesis. Ocean University of China (in Chinese with English abstract).
Lynd, L. R., Weimer, P. J., van Zyl, W. H., and Pretorius, I. S., 2002. Microbial cellulose utilization: Fundamentals and bio- technology., 66: 506-577, DOI: 10.1128/mmbr.66.3.506-577.2002.
Ma, L., Qu, Y. J., Huai, Y. T., Li, Z. J., Wang, J., Lan, X. Y.,., 2011. Polymorphisms identification and associations ofgene with cattle growth traits., 135: 1-7, DOI: 10.1016/j.livsci.2010.04.014.
Miossec, L., Le Deuff, R.M., and Goulletquer, P., 2009. Alien species alert:(Pacific oyster).. 42pp.
Moon, J. S., and Choi, Y. H., 2019. Multiplex PCR for the rapid detection of insulin-like growth factor in the Pacific oyster,: A useful indicator for growth assessment., 46: 1023-1031, DOI: 10.1007/s11033-018-4559-z.
Ning, X. H., Feng, L. Y., Li, X., Wang, S. Y., Zhang, M. R., Wang, S.,., 2018. The scallop IGF2 mRNA-binding pro- tein geneand association of a synonymous mutation with growth traits., 93: 91-100, DOI: 10.1266/ggs.17-00028.
Niu, D. H., Wang, L., Bai, Z. Y., Xie, S. M., Zhao, H. G., and Li, J. L., 2015. Identification and expression characterization of the myostatin () gene and association analysis with growth traits in the razor clam., 555: 297-304, DOI: 10.1016/j.gene.2014.11.020.
Ou, L. J., Li, X. Y., Wu, G. Q., and Yang, N., 2005. Efficient and sensitive method of DNA silver staining in polyacrylamide gels., 26: 99-101, DOI: 10.1002/elps.200406177.
Prieto, D., Markaide, P., Urrutxurtu, I., Navarro, E., Artigaud, S., Fleury, E.,., 2019. Gill transcriptomic analysis in fast- and slow-growing individuals of., 511: 734242, DOI: 10.1016/j.aquaculture.2019.734242.
Saavedra, C., Milan, M., Leite, R. B., Cordero, D., Patarnello, T., Cancela, M. L.,., 2017. A microarray study of carpet-shell clam () shows common and organ-spe- cific growth-related gene expression differences in gills and digestive gland., 8: 943, DOI: 10.3389/fphys.2017.00943.
Sakamoto, K., Uji, S., Kurokawa, T., and Toyohara, H., 2009. Mole-cular cloning of endogenous beta-glucosidase from common Japanese brackish water clam., 435: 72-79, DOI: 10.1016/j.gene.2009.01.011.
Schlueter, P. J., Peng, G., Westerfield, M., and Duan, C., 2007. In-sulin-like growth factor signaling regulates zebrafish embryo- nic growth and development by promoting cell survival and cell cycle progression., 14: 1095-1105, DOI: 10.1038/sj.cdd.4402109.
Shaul, O., 2017. How introns enhance gene expression., 91: 145-155, DOI: 10.1016/j.biocel.2017.06.016.
Shi, Y., Guan, Y. Y., and He, M. X., 2013. Molecular identifi- cation of insulin-related peptide receptor and its potential role in regulating development in., 408: 118-127, DOI: 10.1016/j.aquaculture.2013.05.038.
Sun, X. J., Li, L., Liu, Z. H., Zhao, D., Yang, A. G., Zhou, L. Q.,., 2020. Molecular characterization of the myostatin gene and its regulation on muscle growth in Yesso scallop., 520: 734982, DOI: 10.1016/j.aquaculture.2020.734982.
Tanimura, A., Liu, W., Yamada, K., Kishida, T., and Toyohara, H., 2013. Animal cellulases with a focus on aquatic inverte- brates., 79: 1-13, DOI: 10.1007/s12562-012-0559-4.
Wang, Q. Z., Li, Q., Kong, L. F., and Yu, R. H., 2012. Response to selection for fast growth in the second generation of Pacific oyster ()., 11: 413-418, DOI: 10.1007/s11802-012-1909-7.
Ward, R. D., English, L. J., McGoldrick, D. J., Maguire, G. B., Nell, J. A., and Thompson, P. A., 2000. Genetic improvement of the Pacific oyster(Thunberg) in Australia., 31: 35-44, DOI: 10.1046/j.1365-2109.2000.00388.x.
Watanabe, H., and Tokuda, G., 2001. Animal cellulases., 58: 1167-1178, DOI: 10.1007/pl00000931.
Yang, C. Y., Yang, J. M., Hao, R. J., Du, X. D., and Deng, Y. W., 2020. Molecular characterization ofand association of allelic variants with growth traits., 516: 734617, DOI: 10.1016/j.aquaculture.2019.734617.
Zhang, G. F., Fang, X. D., Guo, X. M., Li, L., Luo, R. B., Xu, F.,., 2012. The oyster genome reveals stress adaptation and complexity of shell formation., 490: 49-54, DOI: 10.1038/nature11413.
Zhang, H., and He, M. X., 2020. The role of a new insulin-like peptide in the pearl oyster., 10: 433, DOI: 10.1038/s41598-019-57329-3.
Zhang, H., Shi, Y., and He, M. X., 2017. Molecular identification of an insulin growth factor binding protein (IGFBP) and its potential role in an insulin-like peptide system of the pearl oyster,., 214: 27-35, DOI: 10.1016/j.cbpb.2017.09.003.
September 16, 2020;
December 1, 2020;
March 2, 2021
? Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
. E-mail: qili66@ouc.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2021年6期
Journal of Ocean University of China2021年6期
- Journal of Ocean University of China的其它文章
- Molecular Cloning, Characterization and Expression Profile of Myf5 and Myf6 During Growth and Development in the Seriola lalandi
- Hydrodynamic Coefficient Investigation on a Partial Permeable Stepped Breakwater Under Regular Waves
- Covariability of Subantarctic Mode Water and the Southern Branch of the Subtropical Indian Ocean Countercurrent in Argo Observations
- Heat Insulation and Dissipation Processes in Nordic Seas in the Summer
- Effects of Climate Variability on Habitat Range and Distribution of Chub Mackerel in the East China Sea
- Interaction of Irregular Waves with Vertical Breakwater and Characteristics of Secondary Wave Generated by Overtopping
