Molecular mechanisms of intermuscular bone development in fish: a review
Bo Li, Yuan-Wei Zhang, Xiao Liu, Li Ma, Jun-Xing Yang,*
1 Cave Fish Development and Evolution Research Group, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
2 College of Life Sciences, Capital Normal University, Beijing 100048, China
3 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Innovative Academy of Seed Design, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
4 Yunnan Key Laboratory of Plateau Fish Breeding, Yunnan Engineering Research Center for Plateau-Lake Health and Restoration,Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
ABSTRACT Intermuscular bones (IBs) are slender linear bones embedded in muscle, which ossify from tendons through a process of intramembranous ossification,and only exist in basal teleosts. IBs are essential for fish swimming, but they present a choking risk during human consumption, especially in children, which can lead to commercial risks that have a negative impact on the aquaculture of these fish. In this review, we discuss the morphogenesis and functions of IBs, including their underlying molecular mechanisms, as well as the advantages and disadvantages of different methods for IB studies and techniques for breeding and generating IB-free fish lines. This review reveals that the many key genes involved in tendon development, osteoblast differentiation, and bone formation, e.g., scxa, msxC,sost, twist, bmps, and osterix, also play roles in IB development. Thus, this paper provides useful information for the breeding of new fish strains without IBs via genome editing and artificial selection.
Keywords: Intermuscular bones; Molecular mechanisms; Intramembranous ossification; Bone formation; Genome editing; Artificial selection
INTRODUCTION
Vertebrate bones develop from the paraxial mesoderm, lateral plate mesoderm, or cranial neural crest, and their endochondral ossification occurs in six stages: i.e.,commitment, compaction, proliferation, growth, chondrocyte death, and bone cell generation (Gilbert & Barresi, 2016). In teleosts, ribs are formed by endochondral ossification.However, intermuscular bones (IBs), also named intramyoseptal bones or myoseptal spines (Bing, 1962), are late developing bones that appear after axial skeleton and limb bone formation (Bing, 1962) and form from tendons by intramembranous ossification without a cartilage phase. IBs are slender linear structures about one-third to one-half of the length of the ribs (Li et al., 2013) and are inserted into the myosepta between adjacent myomeres (Yang et al., 2019c)from the head to tail in an orderly manner (Li et al., 2013).These bones only occur in teleosts (Patterson & Johnson,1995). In most basal teleosts, the few occurring IBs are relatively simple in morphology. In contrast, IBs have completely disappeared in certain teleost lineages, such as the spiny finfish Perciformes (Li et al., 2013; Patterson &Johnson, 1995), but are found in several ancient species,such asPholidophorus becheiAgassiz andOligopleurus(Lund, 1966). In other teleost lineages, e.g., cyprinids, IBs are extremely complex in morphology and range from 73 to 169 in number (Yang et al., 2019c).
As early as the 1930s, scientists recognized the influence of diet on IB development in carp species (Hirsch, 1938).Although subsequent studies focused on IB number, genetic variability, and breeding in carp (Bing, 1962; Kossmann, 1972;Lieder, 1961; Moav et al., 1975; Sengbusch & Meske, 1967),reducing IBs in such species remains a challenging mission for aquaculturists. In aquaculture, fish species and meat quality are of paramount importance in food production. In 2016, the Food and Agriculture Organization of the United Nations (FAO) estimated that Cypriniformes species account for ~70% of total fish farming production worldwide (Nie et al.,2020). There are four popular domestic species in China,including black carp (Mylopharyngodon piceus), grass carp(Ctenopharyngodon idella), silver carp (Hypophthalmichthys molitrix), and bighead carp (Hypophthalmichthys nobilis). In 2018, these species contributed to 26.4% of total finfish produced from aquaculture worldwide (FAO, 2020). In addition, IB-containing Characiformes species, such as tambaqui (Colossomamacropomum), account for a substantial proportion of consumed fish (142 000 tons in 2016)and are important commercial species in areas such as the Amazon River basin (Woynárovich & Van Anrooy, 2019).
Fish protein is an important part of a healthy human diet and critical for normal physical development in children. However,IBs increase the risk for choking in adults and children and can cause serious and potentially lethal complications if ingested (Knight & Lesser, 1989), thus reducing their commercial value (Nie et al., 2020). Moreover, IBs increase difficulty in food processing, affect meat freshness, and reduce the commercial value of fish products (Li et al., 2013; Yang et al., 2019c). To address these issues, new strains of IB-free fish produced by genome editing and artificial selection would be of enormous significance in both basic research and aquaculture. To the best of our knowledge, however, most current research has focused on the number and morphology of IBs, while the molecular mechanisms underlying the development of IBs remain poorly understood. In this review,we discuss current knowledge on the morphogenesis of IBs,including inducing factors and molecular mechanisms, as well as research methods and practical applications for studies on IBs. This paper should provide a theoretical basis for improving human health and modern aquaculture by the development of new IB-free fish strains.
MORPHOLOGY AND EVOLUTION OF IBs
Early research on IBs primarily focused on bone number and morphology (Nie et al., 2021). In teleosts, IBs can be classified into four categories according to their position and attachment pattern: i.e., epineurals, epicentrals, epipleurals, and myorhabdoi (Li et al., 2017; Nie et al., 2020). Epicentrals are located in the central vertebrae of the horizontal septum,epineurals are located above the horizontal septum and attached to the neural arches in the epaxial myoseptum, and epipleurals are located under the horizontal spetum and attached to the ribs or hemal arches in the hypaxial myoseptum (Gemballa & Britz, 1998; Li et al., 2017; Patterson& Johnson, 1995; Yang et al., 2019c). Furthermore, one or two types of unattached IBs, called myorhabdoi, are also present in a small number of teleost species (Nie et al., 2020).These myorhabdoi are not attached to the axial skeleton but are located in the ventral and dorsal forward flexures of the myoseptum (Patterson & Johnson, 1995). Current data indicate that basal teleosts (Osteoglossomorpha) only have epineurals; Clupeomorpha and Elopomorpha have epineurals,epicentrals, and epipleurals; and Ostariophysi, e.g.,Cypriniformes and Characiformes, lack epicentrals,suggesting that epicentrals appeared later and degenerated earlier during evolution (Nie et al., 2020; Yang et al., 2019c).
In North Atlantic herring (Clupea harengus), IBs are cylindrical, thin, and fiber-like, and the mid-shaft region has a homogenous ellipsoidal cross-sectional area. Although tissue mineral density (TMD) is not dependent on fish size, crystal length is larger and Young’s modulus is lower in the IBs of large fish compared to small fish (Fiedler et al., 2019).Relative to mammalian bones (i.e., mouse tibiae), IBs have similar TMD, lower Young’s modulus, similar strength, and higher ductility (Fiedler et al., 2019). In mature IBs of the blunt snout bream (Wuchang bream,Megalobrama amblycephala),a great number of mature osteoblasts are distributed along the edge of the bone matrix, and osteocytes are apparent in the center area of the bone matrix (Liu et al., 2017).
According to morphological classification, IBs can be divided into seven categories: i.e., non-fork type (I-type), oneend-unequal-bi-fork type (卜-type), one-end-equal-bi-fork type(Y-type), one-end-multi-fork type, two-end-bi-fork type, twoend-multi-fork type, and tree-branch type (Nie et al., 2020).The IBs in cyprinids are more complex than in other species and almost all seven different shaped epineural IBs are represented. Epipleural IBs are simpler than epineurals and most are non-forked (Nie et al., 2020).
The shape of IBs exhibits a pattern from simple to complex during evolution from Osteoglossomorpha to Ostariophysi (Lv et al., 2007), during which time IB number increased gradually, then decreased gradually (Table1). The forked IBs start from the non-forked I-type via two evolutionary paths: 1.I-type — 卜-type — Y-type — one-end-multi-fork type; 2. Itype — 卜-type — Y-type — two-end-bi-fork type — two-endmulti-fork type — tree-branch type (Lv et al., 2007).
ORIGINS OF IBs
The ossification of IBs differs among fish species. In zebrafish(Danio rerio), IBs start to ossify in the post-embryonic stages when standard length reaches 6.52-7.63 mm (Yao et al.,2015). In some cyprinids and characins, such as silver carp,barbel steed (Hemibarbus labeo), Oujiang color common carp(Cyprinus carpio var. color), blunt snout bream, Yellow River carp (Cyprinus carpio haematopterus), and dorado (Salminusbrasiliensis), IBs are first observed at 10-20 days posthatching when body length reaches 10-20 mm (Nie et al.,2020). The process of IB ossification can be divided into four developmental stages: i.e., Stage 1 (S1): IBs have not emerged; Stage 2 (S2): small number of IBs have started to ossify; Stage 3 (S3): IBs ossify and appear rapidly; Stage 4(S4): all IBs show mature morphology (Nie et al., 2019).
There are two pattern types of IB ossification (Nie et al.,2020). One is from the posterior to anterior, which occurs in zebrafish (Yao et al., 2015), silver carp (Ke et al., 2008),barbel steed (Lv et al., 2012), blunt snout bream (Wan et al.,2014), Yellow River carp (Chen et al., 2017b), and Oujiang color common carp (Lv et al., 2014). The other is from the anterior to posterior, as found in Japanese eel (Anguilla japonica) (Yao et al., 2015) and Asian swamp eel (Monopterus albus) (Nie et al., 2018).
The origin of IB ossification is controversial (Nie et al.,2020). Patterson and Johnson (1995) suggested that IBs are ossified in ligaments, through which they join to the proximal axial skeleton, and are often part of an extensive series of ligaments in teleost fish. In contrast, Gemballa and Britz(1998) proposed that IBs are ossified from tendons based on several reasons. First, tendons originate from a bundle of fibers from the vertebral neural arch and extend posterolaterally as a fan-shaped structure toward the integument. Second, IBs can ossify from parts of the intermuscular tendons, which transfer muscular forces from the myomeres to the axial skeleton and can connect to the axial skeleton by a short bundle of fibers. Third, IBs are observed in the proximal portion of each tendon ontogenetically (Gemballa & Britz, 1998). Currently, IBs arising by tendon ossification is the most widely accepted opinion (Nie et al., 2020). The process of IB ossification in zebrafish has been described in epineural or epipleural tendons: i.e., the mesenchymal cells differentiate directly into osteoblasts and begin to ossify, then the collagenous unossified area of the tendon connects the ossified tendon to the vertebra (Danos & Ward, 2012).
Two modes of ossification are reported in vertebrates, i.e.,endochondral ossification and intramembranous ossification.Endochondral ossification includes the differentiation of mesenchymal cells into chondrocytes, cartilage formation, and replacement of cartilage by bone (Gilbert & Barresi, 2016).Intramembranous ossification is the direct differentiation of mesenchymal cells into osteoblasts without forming cartilage(Gilbert & Barresi, 2016). Transcriptome data from the blunt snout bream show that chondrocyte-related genes, includingcol2a1,ihha,mmp13,sox9,timp2a,fgfr3, andctgfa, are expressed at higher levels in stage 1 (S1), but at lower levels in stages 2 (S2) and 3 (S3) during IB development (Chen et al., 2021a).

Table 1 Number of IBs in different species
FACTORS AFFECTING IB DEVELOPMENT
Swimming behavior
The ossification mode of IBs is advantageous for swimming behavior. For example, most cyprinids (e.g., zebrafish) with the posterior to anterior IB ossification pattern exhibit typical carangiform swimming style. In contrast, species that show the anterior to posterior IB ossification pattern (e.g., Japanese eel) exhibit an anguilliform swimming style. In addition, IB morphology varies in fish with different swimming behaviors.For example, zebrafish only contain epineural and epipleural IBs; Japanese eels contain epineural, epicentral, and epipleural IBs, and Asian swamp eels only have epicentral IBs(Nie et al., 2020).
Carangiformes swim by the propulsion of the body and/or caudal fin with undulations in the posterior third or half of the body (Nie et al., 2020). Tail-amputated zebrafish IBs show a shorter posterior area and missing vertebrae (Yao et al.,2015). Anguilliformes swim by locomotion of the body and/or caudal fin, which bends the body to form a backward-moving propulsive wave, which extends to the caudal fin (Nie et al.,2020). The IBs of tail-amputated Japanese eel are shortened beyond the ninth epineural bone (Yao et al., 2015). In these species, tail amputations affect the frequency of the swimming swing, and swimming speed preference may further influence the formation of IBs. In addition, carnivorous fish chase and out swim their prey, which requires faster swimming speeds.Therefore, more IBs may be needed to transmit force and maintain muscle stability and body stiffness during swimming(Yang et al., 2019c).
Ecological factors
Ecological factors, including diet, water depth, and velocity,also affect IB growth in fish. Yang et al (2019c) showed that diet, but not water depth or velocity, is correlated with IB number. Based on diet habits, fish can be divided into carnivorous, omnivorous, and herbivorous. Carnivorous fish mainly eat small invertebrates and vertebrates, herbivorous fish forage on aquatic plants and algae, while omnivores fish consume both plants and animals, but not algae. Certain carnivorous fish species in Danioninae and Cultrinae contain more IBs than non-carnivorous species (Yang et al., 2019c).The number of IBs is different in omnivorous fish, such as the stone moroko (Pseudorasbora parva, average IBs of 89),Chinese lizard gudgeon (Saurogobio dabryi, average IBs of 136), and Chinese false gudgeon (Abbottina rivularis, average IBs of 94) (Yang et al., 2019c). In addition, temperature changes may also influence IB variation during the developmental stages (Moav et al., 1975).
InSinocyclocheilus, IBs generally number from 84 to 120,which is less than that in typical cyprinids (Yang et al., 2015).Most of these fish species live in caves, which lack light and have limited food resources (Yang et al., 2015, 2019c). The reduced number of IBs may be an adaptation to the extreme cave environment, supporting that ecological factors can impact the development of IBs.
FUNCTIONS OF IBs
IBs support lateral muscles during myokinesis in areas without ribs (Bing, 1962; Li et al., 2013). They also promote muscle force (Li et al., 2013) by strengthening the connection between sarcomeres potentially (Wan et al., 2015). For example, in carnivorous fish, more IBs are needed to transmit force and maintain muscle stability and body stiffness during predation(Yang et al., 2019c).
As mentioned above, IBs are likely ossified from tendons,so their functions may exhibit similarity to that of myoseptal tendons, including enhancing body stiffness, constraining myomere deformation during contractions, transmitting force between muscle segments, and storing and releasing energy(Danos & Ward, 2012). An example of the latter is sound production in the ossified myoseptal tendons of the fawn cuskeel (Lepophidium profundorum) (Danos & Ward, 2012; Fine et al., 2007).
MOLECULAR MECHANISMS AND REGULATIONS OF IBs
Research on key genes involved in IB development
Key genes in tendon development:Given that IBs are likely ossified from tendons, key factors involved in tendon development may also participate in IB development. For example,scleraxis(scx), which is a helix-loop-helix (bHLH)transcription factor, is involved in tendon cell differentiation and maturation (Nie et al., 2021). There are twoscxorthologues in zebrafish, includingscxaandscxb(Nie et al.,2021). Notably, zebrafishscxa-1-/-mutants produced by clustered regularly interspaced short palindromic repeats(CRISPR)-Cas9 gene editing show significant reductions in total IB and rib number (70% and 57%, respectively).Furthermore, early IB defects in these mutants include developmental retardation, with fewer IBs found in the tail at 38 days post-fertilization (dpf) compared to normal wild-type controls (Nie et al., 2021). Some phenotypes in CRISPRCas9-knockoutscxazebrafish include cranial tendon and ligament defects, cranial muscle fiber defects in attachment and orientation, reduced body size and muscle volume,abnormal swimming behavior, bone growth, and composition,and defective rib mineralization in tendon-like regions (Kague et al., 2019). However, CRISPR-Cas9-knockoutscxbzebrafish show normal IBs and ribs (Kague et al., 2019; Nie et al., 2021). Thus, these results indicate thatscxalikely plays a role in IB development.
Key genes in bone development:Bone development via intramembranous ossification occurs through mesenchymal cell differentiation into osteoblasts without a cartilage phase(Nie et al., 2020), and is controlled by many different genes and regulatory factors.
Muscle segment homeobox C (MsxC) is an important factor in bone formation and induces epithelial-mesenchymal interaction in vertebrate organogenesis (Bendall & Abate-Shen, 2000; Lv et al., 2015). Studies have indicated thatmsxCis expressed in barbel steed myosepta from 26 to 41 days after hatching (dah), which coincides with the onset of IB development (35 to 62 dah), implying thatmsxCmay induce epithelial-mesenchymal interactions during the formation of IBs (Lv et al., 2015).
Another critical gene involved in bone development issclerostin(sost), which inhibits the Wnt and bone morphogenetic protein (BMP) signaling pathways by binding to co-receptor lipoprotein receptor-related protein 5/6 (LRP5/6)and BMP type Ⅰ and Ⅱ receptors to regulate bone development (Qin et al., 2013). In crucian carp (Carassius auratus),sostmRNA and protein are expressed more strongly in the dorsal muscles than in the caudal muscles; however,IBs are thicker and simpler in the caudal muscles than in the dorsal muscles, suggesting that differential expression ofsostmay influence IB development (Yang et al., 2019d).
The Twist family bHLH transcription factor (Twist) plays a key role in the differentiation of mesenchymal cells into osteoblasts (Marofi et al., 2019) and Twist1 and Twist2 both inhibit osteogenesis (Huang et al., 2014). In barbel steed,twist1andtwist2are significantly changed in transcriptomes during the four stages of IB development.Twist1expression is highest in the S1 stage and decreases significantly in S3 and S4; in contrast,twist2is not significantly changed in stages S1 to S3, but decreases markedly in S4 (Chen et al., 2021b).Thus,twist1andtwist2may be correlated with IB development. Intwist1b-knockdown Japanese rice fish(medaka,Oryzias latipes), neural arches are lacking in some vertebrae located bilaterally at the anterior end of the centra,but the migration of sclerotome-derived cells is not affected.These results indicate thattwist1bfunctions during the conversion of sclerotomal cells to neural arch-forming osteoblasts, but not during the migration of sclerotome-derived cells (Yasutake et al., 2004).
As members of the transforming growth factor-β (TGF-β)superfamily, BMPs are among the best-characterized osteoinductive cytokines (Su & Dong, 2018). In blunt snout bream,bmp3,bmp4,bmp5, andbmp8aincrease significantly during S2, suggesting that they may play important roles in IB formation, whereasbmp7bandbmp16are highly expressed during S3, suggesting that they may participate in osteoblast differentiation and IB maturation; furthermore, mostbmpsare increased during S4, suggesting potential involvement in IB morphogenesis (Zhang et al., 2018). In addition, in the Nile tilapia (Oreochromis niloticus) and blunt snout bream,bmp2bis more highly expressed in IBs than in the fin, brain, liver,muscle, heart, and spleen (Yang et al., 2019a). There is also a positive dose-dependent correlation between the expression ofbmp4and IB distribution in the dorsal and caudal muscles of common carp (Cyprinus carpio)(Su & Dong, 2018).
Osterix (Osx, Sp7) is a specific transcription factor in osteoblasts and can activate genes for pre-osteoblast differentiation into mature osteoblasts and osteocytes. It plays dual roles in intramembranous ossification, i.e., promoting osteoblast differentiation and inhibiting chondrocyte formation(Sinha & Zhou, 2013). In common carp, there are twosp7gene,sp7aandsp7b(Zhong et al., 2016). The CRISPR-Cas9sp7a-knockout mutants are lighter and smaller than wild-type controls, and show severe bone development defects,including insufficient opercula and maxilla, bended backs,deformed centrums, irregular hemal spines, and shorter IBs(Zhong et al., 2016). The bone defects insp7amutant are more obvious than those in the CRISPR-Cas9-sp7bmutant. In bothsp7aandsp7bmutants, craniofacial and centrum bones develop more slowly than in wild-type controls (Zhong et al.,2016). Other defects insp7amutants include irregular shape,small scales, and fewer pharyngeal teeth (Zhong et al., 2016).
Bioinformatics analysis of key genes and molecular regulation involved in IB development
Candidate genes selected by quantitative trait locus and genome-wide association study:Quantitative trait locus(QTL) analysis is used to study the relationship between phenotype and genotype. Candidate genes associated with IBs in common carp, includingcxcr4b,has2,wnt4b,map2k1,rspo3,col5a3a,ucmaa,araf,tmco1,wnt5b,itgb1a,itga2.2,LOC101886891,dot1l,p2rx7,matn1,camk1da,ccn4b,wnt2ba,sp9,nid1a,porcn,lrp2a,igf1ra, andmtmr4, have been identified previously by QTL (Tang et al., 2020).Genome-wide association study (GWAS) has also been widely used to analyze candidate genes. GWAS analysis of tambaqui lacking IBs (Perazza et al., 2017) and wild-type individuals resulted in the identification of various genes, i.e.,actn3b,adamtsl2,atp6v0a1a,atp6v0ca,dchs1b,ebf3a,efnb1,nmu,ntn4,pde4d,plek,wisp1b, andxpr1b, that may be involved in the development of IBs (Nunes et al., 2020).
Candidate single nucleotide polymorphisms (SNPs)involved in IB development:SNPs may also play a role in the development of IBs in blunt snout bream (Wan et al.,2019). Bulk segregant analysis (BSA) is used to identify genes and SNPs and involves determining the number of IBs from whole-genome resequencing data of IB-positive and IBnegative groups (Wan et al., 2019). Five key SNPs related to IB number have been identified: i.e., Chr06-33 022 815 (A, T)in the coding sequence (tryptophan (W)→ arginine (R))involved in Pgap1 (GPI inositol-deacylase), and four others in the intergenic region, including Chr20-18 560 334 (G, A)annotated tobmp3bandFERM and PDZ domain-containing protein 2(frmpd2), Chr11-14 388 171 (T, A) annotated toallantoicase(allc) andXENLA transcription factor sox11-b(sox11b), Chr11-13 516 603 (C, G) annotated tosyntaxin binding protein 5(stxbp5) andSAM and SH3 domaincontaining protein 1(sash1), and Chr11-12 488 304 (G, A)annotated toN-myc proto-oncogene protein(nmyc) (Wan et al., 2019). These results indicate that SNPs may potentially be correlated with IB development and provide a new area for molecular selective breeding of IB-free strains.
Key microRNAs (miRNAs) involved in IB development:MiRNA is known to regulate gene expression in development,and generally represses gene expression at the translational level (Krebs et al., 2018). In blunt snout bream, 13 miRNAs(let-7d-3p,miR-106a,miR-153a,miR-15c,miR-190a,miR-196d,miR-218a,miR-301,miR-454b,miR-457b,miR-460-5p,miR-96, andmiR-9a-3p) are expressed only in IBs and 17 miRNAs (miR-1788-3p,miR-18b-3p,miR-192-3p,miR-196c,miR-19a-3p,miR-203b-5p,miR-430a,miR-733,miR-7553,miR-17b,miR-204b,miR-30d-3p,miR-430b,miR-7b,miR-9-3p,miR-202-3p, andmiR-457a) are found only in the connective tissue. Among abundant miRNAs, both IB and connective tissue sharemiR-1,miR-206,let-7a,let-7b,let-7c,miR-199-3p,miR-21, andlet-7f, whilelet-7dandmiR-199a-3pare IB-specific andlet-7gandmiR-22aare connective tissuespecific (Wan et al., 2015). Understanding the function of these special miRNAs in both IBs and connective tissue will help clarify the formation and development of IBs.
Key signaling pathways involved in IB development:Based on transcriptome-proteome and comparative analysis in the blunt snout bream (Nie et al., 2019), the key signaling pathways involved in the IB developmental stages (S1-S4)include mitogen-activated protein kinase (MAPK), calcium,thyroid hormone, cyclic guanosine monophosphate-dependent protein kinase (cGMP-PKG), cyclic adenosine monophosphate (cAMP), Rap1, Ras, phosphatidylinositol 3’-kinase-Akt (PI3K-Akt), and tumor necrosis factor (TNF)signaling (S1 vs S2, S2 vs S3, and S3 vs S4 showed differential expression); Hedgehog signaling (only S1 vs S2 showed differential expression); TGF-β and nuclear factorkappa B (NF-kappa B) (S1 vs S2 and S2 vs S3 were differentially expressed); osteoclast differentiation and Wnt signaling (S1 vs S2 and S3 vs S4 were differentially expressed); and vascular endothelial growth factor (VEGF)signaling (S2 vs S3 and S3 vs S4 were differentially expressed). Therefore, the developmental time axis of IBs,from beginning of ossification to maturity, is regulated by the differential expression of bone formation signaling pathway proteins (Nie et al., 2019). The identification of these differential proteins is the key to understanding the formation and development of IBs.
Osteoblast differentiation and osteogenesis genes as potential regulators of IB development
During the process of osteoblast lineage commitment,osteoprogenitors (runx2expression in mesenchymal cells)begin to proliferate, then exit mitosis while expressingosxand form osteoblasts, which expressalkaline phosphatase(alp),bone sialoprotein(bsp), andtype Ⅰ collagento produce and mature the osteogenic extracellular matrix (ECM), and finally expressosteocalcin(oc) andosteopontin, which participate in ECM mineralization (Zhang, 2010). For osteoblast formation,runx2andosxare necessary for the commitment of mesenchymal progenitors and are predominantly expressed in osteoblasts (Javed et al., 2010), with their function in IB development described below:
Runt-related transcription factor 2 (Runx2):Runx2 (Cbfa1,Osf2), which belongs to the Runt transcription factor family, is essential for the differentiation of mesenchymal cells into preosteoblasts and plays an important role in the process of intramembranous ossification in mice (Takarada et al., 2016;Zhang, 2010).
Runx2 activity is controlled by many different factors.Vestigial-like family member 4 (VGLL4) can bind to the Yesassociated protein (YAP) binding sites of TEAD/TEF family members TEAD1-4, which produces a competitive relationship with YAP (Suo et al., 2020). When VGLL4 binds to TEADs, the interaction that promotes osteoblast differentiation between TEADs and RUNX2 is prevented (Suo et al., 2020). Epidermal-growth factor receptor (EGFR)signaling attenuates the expression ofOsxand decreasedRunx2in undifferentiated osteoprogenitors, which suppresses osteoblast differentiation (Zhu et al., 2011). Fibroblast growth factor 2 (FGF2) can increase the activity of Runx2/Cbfa1 by phosphorylation of extracellular signal-regulated kinases(ERK1 and ERK2) in the MAPK pathway (Xiao et al., 2002).The cAMP pathway can inhibit Cbfa1 by ubiquitin/proteasomedependent proteolytic degradation (Tintut et al., 1999).Transducin-like enhancer of split 2 (TLE2) interacts with the last 5 amino acid residues (VWRPY motif) ofosf2, which inhibits its transactivation (Thirunavukkarasu et al., 1998).Hairy and enhancer of split 1 (HES1) can bind to Cbfa1 to form a complex, which potentiates Cbfa1 activity (McLarren et al., 2000). The SMAD family members Smad1, Smad2,Smad3, and Smad5 interact with and enhance the transactivation ability of RUNX2 (Zhang et al., 2000). The retinoblastoma protein (pRb) can interact with CBFA1, serving as an associator in osteoblast differentiation (Thomas et al.,2001). Heart and neural crest derivatives-expressed 2(Hand2) binds and inhibits Runx2 as a negative regulator of intramembranous ossification (Funato et al., 2009).YAP/transcriptional co-activator with PDZ-binding motif (Taz)acts upstream ofrunx2in endothelial cells to promoterunx2expression, while the expression ofbmprequires Yap/Taz transcriptional activity and transcriptional activation of Yap/Taz requires appropriate blood flow during intramembranous ossification (Uemura et al., 2016). Nel-like molecule 1 (NELL1)is controlled by Runx2, which is preferentially expressed in osteoblasts, and promotes intramembranous or endochondral ossification of orthotopic bone regeneration by inducing the development of bone marrow stromal cells (Zhang et al.,2010).
Osterix (Osx):Osx inhibits the Wnt signaling pathway by activating the Dickkopf WNT signaling pathway inhibitor 1(Dkk1) and inhibiting the transcriptional activity of β-catenin by disrupting the DNA binding of T-cell factor (Tcf) (Zhang, 2010).Wnt antagonism by Osx inhibits the proliferation of osteoblasts(Zhang, 2010).
The expression ofosxis controlled by many factors. BMP2 and insulin-like growth factor 1 (IGF1) can induce the expression ofosxin human mesenchymal stem cells (Celil &Campbell, 2005). The Bmp2/Smad pathway activatesrunx2,which subsequently activates the expression ofosx(Sinha &Zhou, 2013). Runx2 can directly interact and activate the Runx2-binding element located less than 1 kb from theosxpromoter (Sinha & Zhou, 2013). However, this activation is not entirely dependent on Runx2. For example,runx2-nullcells with BMP2 treatment can promote distal-less 5 (Dlx5)interaction with theosxpromoter by phosphorylation of Dlx5,which, in turn, activatesosxexpression (Lee et al., 2003;Sinha & Zhou, 2013).Dlx5is a positive regulatory target gene in the BMP pathway, and involved osteoblast differentiation,anddlx6plays the similar role asdlx5but works at a lower mRNA level (Miyama et al., 1999; Bendall & Abate-Shen,2000). Whendlx5mRNA is overexpressed in MC3T3-E1 cells,the levels of alkaline phosphatase, mineralization of ECM, and production of osteocalcin are increased (Miyama et al., 1999).Both IGF1 and BMP2 synergistically increaseosxexpression(Sinha & Zhou, 2013). BMP2/IGF1 signaling triggers the MAPK and protein kinase D (PKD) pathways to activateosxexpression, which involves the phosphorylation of p38 and Erk1/2 (Sinha & Zhou, 2013). Additionally, ascorbic acid(vitamin C) (Xing et al., 2007) and 1,25(OH)2vitamin D3(Maehata et al., 2006) can stimulate the expression ofosx.
The p53 tumor suppressor prevents bone development and osteoblast differentiation (Zambetti et al., 2006). Because a p53 binding sequence does not exist in theosxgene, p53 prevents p300 and other activators from being recruited to target chromatin and inhibit the expression ofosx(Sinha &Zhou, 2013). Tumor necrosis factor α (TNFα) can inhibitosxtranscription by 90% through the MAPK/ERK1/2 pathway (Lu et al., 2011). TNFα upregulates thepaired-relatedhomeodomain(prx1) mRNA and protein in cultured primitive mesenchymal cells, and Prx1, in turn, suppresses Osx and Runx2 expression, acting as a molecular mediator with TNFα to inhibit osteoblast differentiation (Lu et al., 2011).
Hormones and physical stress also influenceosxexpression (Sinha & Zhou, 2013). Parathyroid hormone (PTH)targeting of fracture calluses in mice increasesosxexpression and accelerates fracture healing (Kaback et al., 2008). This regulation requires activating transcription factor 4 (ATF4), butosxis inhibited by the cAMP signaling pathway after prolonged exposure to PTH (Hong et al., 2009; Kaback et al.,2008; Yu et al., 2009; Zhang, 2010). Endoplasmic reticulum(ER) stress is caused by mature osteoblasts producing many misfolded proteins in the ECM. During this process, ER stress transducers are activated. Inositol-requiring endonuclease 1α(IRE1α) acts byX-box binding protein 1(XBP1) mRNA processing (Sinha & Zhou, 2013). XBP1 targets unfolded protein response (UPR) elements and activates UPR elements to response to ER stress, andosxpromoter is one of XBP1 binding site (Murakami et al., 2009; Sinha & Zhou,2013).
Osxactivity is regulated by many factors, including positive regulators brahma-related gene 1 (Brg1), basal transcription factor TFII, p300, and nuclear factor of activated T cells(NFTAc) and negative regulator NO66 histone demethylase(Sinha & Zhou, 2013). The interaction between nuclear factor of activated T cells 1 (NFATc1) and Osx activation domain enhances Osx activity by activating thetype Ⅰ collagen(Col1a1) promoter and recruiting NFATc1 (Koga et al., 2005).TFIIB is associated with the transactivation domain and Brg1 with the C-terminal zinc finger domain of Osx (Hatta et al.,2006). P300, a transcriptional activator, interacts with phosphorylated Osx target genes, such asbspandfmod(Sinha & Zhou, 2013). NO66 histone demethylase interacts with Osx to inhibit its activity by regulating histone methylation of Osx-target gene promoters, such asocandbsp(Sinha et al., 2010; Sinha & Zhou, 2013).
Other relevant genes:SRY-box transcription factor 9 (Sox9)has a crucial function in chondrocyte lineage commitment(Nakashima & Crombrugghe, 2003). If Sox9 is inactivated before chondrogenic mesenchymal condensation, therunx2transcripts disappear; however, if Sox9 is inactivated after mesenchymal condensation, therunx2transcripts are not affected and osteoblast differentiation occurs normally(Akiyama et al., 2002). This indicates that Sox9 is required for the formation of chondrocytes and osteoblasts (Akiyama et al.,2002; Nakashima & Crombrugghe, 2003). Bothsox9andrunx1are potentially involved in incipient intramembranous ossification (Yamashiro et al., 2004). SATB homeobox 2(SATB2) is a molecular node in skeletal development and the osteoblast differentiation transcriptional network (Zhang,2010). Schnurri 3 (Shn3) acts as a central regulator in postnatal bone mass development (Zhang, 2010).Muscle segment homeobox 2(msx2) is expressed in skull bones and teeth preceding mouse osteoblast development and plays an important role in craniofacial development (Bendall & Abate-Shen, 2000). Previous research has indicated that a 4 bp deletion in humandlx3results in cranial bone thickening(Bendall & Abate-Shen, 2000). Dermo1 (a bHLH protein)binds to the E-box consensus sequence forming a heterodimer with E12, which blocks its transcriptional activation (Li et al., 1995). Furthermore,dermo1is expressed in mandibular and maxillary processes of craniofacial mesenchyme (Li et al., 1995). Dermo1 plays an important role in mesenchymal cell specification and differentiation (Li et al.,1995). The AP-1 transcription factor subunitfosbencodes a truncated FosB (ΔFosB) in osteoblasts, and its overexpression in mice increases bone formation and mass (Sabatakos et al.,2000). The AP-1 transcription factor subunit Fra1 specifically enhances bone formation, whereby the Fra1 overexpressed transgenic mice showed an increase in bone mass, but unchanged expression level of Runx2/Cbfa1 (Jochum et al.,2000). InSp3-/-mutant mice, some ossification centers are completely absent in E18.5 embryos, the expression level ofosteocalcinis reduced, which causing a delay in ossification,thereby limit differentiation to osteoblasts (G?llner et al.,2001). Regulator of calcineurin 2 (Rcan2) inhibits the calcineurin-nuclear factor pathway during osteoclast and osteoblast differentiation and function (Bassett et al., 2012).Fibroblast growth factor receptors FGFR2 and FGFR3 play important roles in the intramembranous ossification of mandibular bones (Havens et al., 2008). InRNA component of the mitochondrial RNA processing endoribonuclease(Rmrp)mutant zebrafish, the intramembranous ossification of skull bones is inhibited, but Wnt/β-catenin signaling is up-regulated and vertebral ossification is promoted (Sun et al., 2019). SHP2(also known as protein tyrosine phosphatase non-receptor type 11, PTPN11) plays an important role in intramembranous ossification. InSHP2mutants, the formation of calvarial bone is severely defective, and in later stages TGFβ is enhanced and BMP2 signaling is suppressed in mutant mesenchymal progenitors (Wang et al., 2019).Cell division cycle 42(Cdc42)mutants show defects in cranial bone tissue intramembranous ossification and decreased expression ofIndian hedgehog(ihh) andbmps(bmp2,bmp4) (Aizawa et al., 2019). Ihh positively regulates intramembranous ossification by inducingbmp2/4expression (Lenton et al., 2011). Brain and muscle Arnt-like protein 1 (BMAL1) is necessary for bone development, with mutants showing osteoblast inhibition and osteoclast promotion (Chen et al., 2020). Wnt16 suppresses intramembranous ossification and osteoblast differentiation through the Wnt/β-catenin pathway (Jiang et al., 2014). Axin1 and Axin2 promote the degradation of β-catenin in the Wnt pathway (Yu et al., 2005), and hypoxia-inducible factor 2α(HIF2α) targetsTwist2to regulatetumor necrosis factor receptor-associated factor 6(Traf6), which stimulates receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis, causing inhibition of osteoblast differentiation during endochondral and intramembranous ossification (Lee et al., 2019; Rosenblum et al., 2019).
Dentin matrix protein 1 (DMP1) plays important roles in bone ECM and osteoblast differentiation. In the calvaria of mice, overexpression of DMP1 leads to significant differences in the expression ofrunx2(30 and 90-days post-natal),col1a2(15 and 30-days post-natal), andalp(60-days post-natal)compared with the wild-type, indicating that DMP1 may directly or indirectly regulate these genes during intramembranous ossification (Padovano et al., 2014).Solutecarrier family 20 member 2(Slc20a2) participates in tissue mineralization, which is necessary for bone quality and strength (Beck-Cormier et al., 2019). Connexin 43 (Cx43) is a gap junction protein expressed in osteoblasts. In Cx43-null mouse embryos, cranial vault intramembranous and endochondral ossification are delayed (Lecanda et al., 2000).Osteopontin (OPN) has a biphasic expression model and may promote pre-osteoblastic proliferation and migration at an early stage in development and participate in hydroxyapatite crystal formation at later stages (Perrien et al., 2002).
MiRNAs play important roles in the regulation of bone development. Eleven miRNAs (i.e.,miR-133,miR-23,miR-205,miR-135,miR-137,miR-338,miR-218,miR-34,miR-30,miR-217,miR-204) are known to inhibitRunx2expression,whilemiR-125b,miR-138, andmiR-637inhibit Osx expression(Lian et al., 2012). Furthermore,miR-93, via Osx/Sp7 feedback, regulates osteoblast differentiation,miR-23a-27a-24-2inhibits the osteoblast to osteocyte process (Lian et al.,2012), andmiR-181atargetsTgf-beta induced(Tgfbi) andTGF-β type Ⅰ receptor(TβR-Ⅰ/Alk5) to promote osteoblast differentiation (Bhushan et al., 2013).
GENETIC BREEDING FOR IBs REDUCTION
Research on IB-deficient fish by genetic breeding
In the last century, scientists have tried to reduce IBs by changing fish diets (Hirsch, 1938), by selective breeding(Lieder, 1961), and by appropriate strain selection (Sengbusch& Meske, 1967). However, difficulties were encountered with each of these approaches, and Moav et al. (1975) eventually showed that none were likely to be feasible. Following the development of molecular biology and bioinformatics,additional approaches have become available. The genetic parameter estimating of IBs in the blunt snout bream indicates that selective breeding may be a feasible way in which to decrease IB number (Xiong et al., 2019). Selective breeding through family selection targeting ventral IBs may be more effective (Xiong et al., 2019). In common carp, IB (total, Ytype, and I-type) heritability is moderate and significant (Tang et al., 2020), suggesting that aquaculturists should focus on IBs in the posterior region. This could be done by reducing Itype, two-end-bi-fork type, and tree-branch type, and or by converting the Y-type, one-end-multi-fork type, and two-endmulti-fork type to I-type (Cao et al., 2015).
Artificial gynogenetic breeding has been utilized to obtain IB-deficient grass carp, which exhibit large and regular arrayed lateral-line scales, significantly thicker vertebral umbo,and no discernible IBs in muscles (Xu et al., 2015). Xu et al.(2015) also showed that the key genes regulating the development of IBs may also affect the calcification of scales,and that IBs are not necessary for overall growth and development.
Genetic screening is a traditional way to breed new strains.IB-deficient and partially IB-defective zebrafish strains have been successfully bred and do not differ significantly from wild-type strains in terms of fertilization, hatching, and larval abnormality (Yang et al., 2019b). In the development of osteoblasts, the Bmp2/4-Smad pathway targets and activates Runx2, thereby further activating the expression ofosx(Sinha& Zhou, 2013). In IB-deficient zebrafish, the expression of skeletal genesrunx2a,sp7,bmp2a,bmp4a,smad1, andsmad4adoes not significantly change throughout development compared with wild-type embryos (Yang et al.,2020b). In contrast,runx2a,bmp4a,smad4a, andsmad1are significantly higher in wild-type embryos,osx/sp7shows significantly higher expression in the mutant-type, andrunx2adoes not exhibit any significant differences in the postembryonic stages (Yang et al., 2020b). Although the expression levels of these genes changed at different developmental stages, skeletal staining in IB-deficient zebrafish indicates that bone development is the same as in the wild-type, except for IB formation (Yang et al., 2020b). In IB-deficient and partially IB-defective zebrafish mutants, the expression levels of muscle-specific genesmyod,myog,myf5,mef2ca, andsox6are not significantly changed compared with wild-type controls; similarly, no obvious differences occur in density or diameter of muscle fibers between mutant and wildtype controls at these stages (Yang et al., 2019b; 2020a).
Interesting, in a Brazilian fish hatchery, 28 out of 120 tambaquis were missing or had vestige IBs (notably, two females and four males lacked IBs, while 22 had vestige IBs on both sides of their loins). However, the mechanisms underlying these IB-defective phenotypes are still unclear(Perazza et al., 2017).
Reducing IBs by ploidy breeding
In recent years, scientists have used genetic breeding to produce new fish strains with fewer IBs. One way to accomplish this is to breed fish at different ploidies or cross fish of different ploidies and obtain their hybrid offspring. Fish with different ploidies have been bred to improve certain characteristics and increase commercial value (Li et al., 2013).For example, the triploid crucian carp (ITCC, 3n=150,IBs=79.1±2.0), a hybrid strain produced by maternal IRCC(Carassius auratus red var., 2n=100, IBs=70.5±6.4) and paternal G×AT (improved allotetraploids, 4n=200,IBs=82.3±2.5), has significantly fewer IBs than white crucian carp (WCC, 2n=100, IBs=81.2±1.7) orCarassius auratusvar.Pengze (PZCC, 3n=150, IBs=83.8±1.4) (Li et al., 2013). Other features of ITCC include fresher meat, higher protein content,higher tolerance to cold and hypoxia, larger body size, and faster growth (Li et al., 2013).
IB RESEARCH AND APPLICATION
Traditional techniques for IB studies
There are several main techniques used in IB research. Xirradiation is a noninvasive way in which to analyze IB number without damaging the specimen. It is fast and easy to operate,but costly and may cause misreading due to overlapping areas caused by single angle radiographs (Nie et al., 2020;Yang et al., 2019c). This method involves anesthetizing fish using clove oil (2-5 mg/L), imaging via X-ray systems (e.g.,Ajex Meditech 135H/A digital X-ray system, Scan-X ALLPRO Imager), and digitization using specific software (e.g., Metron-DVM v7.07) (Perazza et al., 2017).
Micro-computed tomography (micro-CT) can be used to detect trabecular microstructures and observe small fish, but its resolution for fish IBs is too low, resulting in a relatively large error rate (Nie et al., 2020). In this method, fish are anesthetized (0.03% tricaine) and placed gently into 5 mL centrifuge tubes, then imaged by the micro-CT system (e.g.,SkyScan 1 176 high-resolution micro-CT scanner), followed by image reconstruction (e.g., SkyScan CT Analyser software)and tracing of areas of interest using methods such as“Double Time Cubes” 3D reconstruction (Zhong et al., 2016).
Using R?ntgen-Müller television device to response R?ntgen rays is a method to observe IBs in a large scale. This method improves the efficiency of selecting boneless carp,allowing the examination of several thousand fish a day(Sengbusch, 1967).
As a noninvasive alternative to X-irradiation, ultrasound has been applied to identify IBs (Nie et al., 2020). While specimen pretreatment is similar, a portable MyLabTMOne VET digital ultrasound machine with a multi-frequency rectal linear transducer (6.0/10 MHz) can be used to detect IBs, with the operator moving the ultrasound probe back and forth along both sides of the body. Transverse and longitudinal images are then recorded for further analysis using manufacturerprovided software (Perazza et al., 2017).
Bone staining with Alizarin Red S has also been used to assess mineralization of IBs (Yao et al., 2015). However, the process is complicated and not suitable for precise IB counting in an individual (Nie et al., 2020). In this method, fish are anesthetized using tricaine methanesulfonate solution(200 mg/L), then fixed in 4% paraformaldehyde, washed in phosphate-buffered saline, digested in 1% trypsin, and stained using Alizarin Red in 1% potassium hydroxide (KOH). The stained samples are cleaned in 20% glycerol and 1% KOH and then imaged (Yao et al., 2015).
Direct visualization under a microscope is a good way to count IBs accurately, but it is labor-intensity and destroys the specimen. In this method, fish are wrapped in gauze and boiled, after which the gauze and fish skin is removed carefully. The muscles are then removed from the head to the tail, and IBs are taken out in order and examined (Li et al.,2017; Lv et al., 2007).
Genome editing and RNA interference (RNAi) for generating IB-deficient lines
CRISPR-Cas9:The CRISPR-Cas9 system is a flexible and robust tool for genome editing (Zhang et al., 2014). It utilizes nonspecific Cas9 nuclease and programmable sequence specific CRISPR RNA (crRNA) to edit target sequences precisely. The crRNA recognizes and guides Cas9 to target sequences, and Cas9 recognizes the protospacer adjacent motif and cleaves DNA by generating double-strand breaks as well as desired insertions, deletions, or substitutions to generate cellular DNA repair (Zhang et al., 2014).
CRISPR-Cas9-knockout is highly efficient in fish and has been successfully applied in zebrafish with efficiencies of 59.4%±1.7% (fhl), 24.1%±7.0% (apoea), 35.6%±4.6% (th1),35.8%±4.4% (rgs4), 57.0%±5.0% (tia1l), 36.0%±3.7% (tph1a),27.1%±6.0% (gsk3b), and 28.4%±2.8% (drd3) (Hwang et al.,2013). In 2016, the CRISPR-Cas9 system was successfully applied to common carp, with mutation efficiencies tested under different guide RNA (gRNA) concentrations, resulting in efficiencies of 93.5%, 99.1%, 92.8% (sp7a, 100 pg, 150 pg,200 pg gRNA), 80.7%, 81.9%, 76.8% (mstnba, 25 pg, 50 pg,100 pg gRNA), 55.9%, 65.7% (runx2, 50 pg, 100 pg gRNA),52.8% (sp7b, 100 pg gRNA), 53.1% (opga, 100 pg gRNA),and 76.2% (bmp2a, 100 pg gRNA) (Zhong et al., 2016). In addition, the targeting oftyrosinase(tyr) in Japanese white crucian carp (WCC,Carassius auratus cuvieri) and its hybrid offspring (WR,Carassius auratus cuvieri♀ ×Carassius auratus red var♂) showed efficiencies of 79.38% and 78.13%, respectively (Liu et al., 2019).
CRISPR-Cas9 is also a key tool for studying gene function.For example, disruption ofagouti signaling protein1,2(asip1,asip2) in the Oujiang color common carp results in the disappearance of the black patch and melanophores along the dorsal fin in F0 individuals, suggesting that Asip plays an important role in black patch formation (Chen et al., 2019). In additional, disruption ofdopachrome tautomerase1,2(dct1,dct2) in the same species affects melanocyte morphology and black patch pigment pattens (Si et al., 2020). Disruption oftolllike receptor 22(tlr22) in roho labeo (Labeo rohita) embryos has provided a model carp to help understand the function of Tlr22 in innate immunity (Chakrapani et al., 2016).
CRISPR-Cas9 has also been used for disease prevention in fish. Whenintegrin β-1(itgb1) is disrupted in the rare gudgeon(Gobiocypris rarus), the efficiency of viral entry and expression of apoptosis-related genes are reduced (Chen et al., 2018).CRISPR-Cas9 has been utilized to disruptJunctional Adhesion Molecular-A(gcJAM-A) inC. idellakidney (CIK)cells, which improves their resistance to reovirus (Ma et al.,2018).
TAL effector nucleases (TALENs):TALENs fuse to the FokⅠ cleavage domain and DNA-binding domains of transcription activator-like effector (TALE) proteins (Gaj et al.,2013). TALE recognizes and binds to target DNA and the endonuclease FokⅠ cuts the specific target DNA to produce double-strand breaks after dimerization (Jiang & Shen, 2019).The specific recognition of TALE relies on four repeat variable diresidues: i.e., A-NI (Asn-Ile), T-NG (Asn-Gly), C-HD (His-Asp), and G-NN (Asn-Asn) (Jiang & Shen, 2019).
TALENs have been utilized to generatetnikbanddip2ainheritable zebrafish mutants (Huang et al., 2011). In common carp, targetingrunx2,sp7a,mstnba, andspp1aresulted in mutant efficiencies of 15.2% (runx2), 36.8% (sp7a), 29.1%(mstnba), and 81.5% (spp1a) (Zhong et al., 2016).
Zinc finger nuclease (ZFN):ZFN is a first-generation geneediting technique and consists of a zinc finger protein (ZFP)and FokⅠ restriction endonuclease (Jiang & Shen, 2019).Each ZFP includes three zinc fingers (F1, F2, F3) of nearly 30 amino acid residues, and the -1, +3, and +6 residues bind to three triplet bases to form a 9 bp recognition site. The endonuclease FokⅠ specifically cuts target DNA to produce a double-strand break after dimerization when the two FokⅠsites are located approximately 5-7 bp to each other (Jiang &Shen, 2019).
In zebrafish mutant which targetingno tail/Brachyury(ntl)andslc24a5gene, the efficiencies were dose-dependent and mutants were heritable (Doyon et al., 2008). However, ZFN may be toxic for embryos, there have mild to moderate developmental defects with high ZFN dose (Doyon et al.,2008).
RNAi: RNAi is a technology in which sequence-specific double-stranded RNA (dsRNA) is introduced into cells, which induces mRNA degradation and gene silencing (Li et al.,2000). After its introduction into cells, dsRNA is recognized by Dicer and processed into small interfering RNA (siRNA; 21-23 bp long), which is then incorporated into an RNA-induced silencing complex (RISC) to induce gene silencing by the destruction of target mRNA (Li et al., 2000). RNAi has been successfully applied in zebrafish since 2000 (Li et al., 2000).
RNAi is also a tool for studying gene function. RNAiknockdown of grass carpIL-1R-associated kinase 4(CiIRAK4)in CIK cells indicates thatCiIRAK4has little effect on p65 translocation from the cytoplasm to nucleus in the NF-kappa B pathway (Wu et al., 2019). Following knockdown ofCiSARM1andCiSARM1s(sterile alpha and Toll/IL-1R motif containing 1(SARM1)) in grass carp CIK cells,CiMyD88,CiIPS-1,CiIRF3,CiIRF7,CiIFN-I, andCiMx1expression levels increase significantly, further confirming that SARM1 negatively controls toll/interleukin-1 receptor (TRI) domain containing adapter inducing interferon-β (TRIF)-dependent toll-like receptors (TLR) signaling (Yan et al., 2015).
RNAi can also be used for fish disease prevention. At 20 nM, siRNA can inhibit spring viraemia of carp virusphosphoprotein (SVCV-P) and SVCV-nucleoprotein (SVCV-N)transcripts; in addition, targeting SVCV-P is more efficient for reducing SVCV-glycoprotein (SVCV-G) mRNA (Gotesman et al., 2015).
RNAi has also been applied in fish breeding. The RNAi vector was designed to inhibitmyostatinand inserted to common carp genome, which generated 32.78% successful integration of this an exon into the fish genome. The transgenic fish grew faster, heavier, and have a wider body(Yan et al., 2013).
DISCUSSION
Intermuscular bones first appeared during the evolution of basal teleosts (Patterson & Johnson, 1995), and their development is similar throughout teleost species. Based on data from Osteoglossomorpha, Elopomorpha, Clupeomorpha,and Ostariophysi, the evolutionary pattern of IBs can be considered as simple-to complex-to-simple (Lv et al., 2007). In cyprinids, IBs are extremely complex in morphology and numerically large, but they have completely disappeared in Perciformes (Li et al., 2013; Patterson & Johnson, 1995; Yang et al., 2019c). In addition, IB-deficient species (e.g., zebrafish)show that the absence of IBs does not affect fish growth or development and is heritable. Therefore, the development of IBs may not be a specific requirement in fish development,allowing cyprinids to be produced without IBs for commercial utilization.
Fish are an important source of protein for many humans and have a huge value for the world economy. Fish are rich in high-quality animal proteins, micronutrients, and polyunsaturated fatty acids, which contribute to diversified and healthy diets (FAO, 2020). In 2017, 17% of animal proteins and 7% of all proteins consumed by humans were obtained from fish (FAO, 2020). China is a major fish producer,accounting for 35% of global fish production in 2018,especially of cyprinids (FAO, 2020). However, the existence of IBs can severely impact the economic value of fish as well as human health. In addition, during fish processing, the removal of IBs is labor-intensive and reduces freshness. Thus, it is important to cultivate new strains of cyprinids without IBs.
Early studies in this field used genetic breeding or explored strains without IBs to produce new fish lines. This approach was successful but time and labor-intensive and was not feasible in different species on a large scale. The current genome-editing and molecular cloning techniques described in this review suggest that it is possible to generate IB-free strains by knockout of IB-specific genes.
Previous studies successfully knocked out thesp7andscxagenes, which reduced IB number, but also affected the development of other bones (Nie et al., 2021; Zhong et al.,2016). In Japanese rice fish without IBs,twist1bknockdown resulted in the loss of the neural arch in the vertebrae(Yasutake et al., 2004). These results indicate thatsp7,scxaandtwist1bare not IB specific. Furthermore, compared with mammalian bones, IBs have higher ductility and similar strength and TMD (Fiedler et al., 2019), suggesting that genes involved in development and formation of IBs may be similar to those involved in rib formation. In this review, we explored several genes involved in bone development and intramembranous ossification, providing candidates to highlight IB-specific.
Based on the above literature review, we conclude that IB development may primarily be controlled by gene regulation.Because IBs are a product of connective tissues (tendons),which are ossified similarly to ribs and latter develop than ribs,a certain regulatory element may control the ossification of tendons. IBs are ossified via intramembranous ossification,whereby mesenchymal cells differentiate directly into osteoblasts without undergoing a cartilage phase (Karsenty &Wagner, 2002; Nie et al., 2019). Comparative analysis of the key signaling pathways in the stages (S1-S4) of IB development indicate that these pathways may also play important roles in the development of ribs (Nie et al., 2019).Thus, IB formation may be controlled by genes involved in tendon and bone development (e.g., osteoblast differentiation,osteogenesis, or morphogenesis), related genes, or the regulation of these genes, includingcis-regulation (e.g.,enhancers),trans-regulation (e.g., transcription factors), or other regulatory factors (e.g., miRNAs, epigenetic regulation).
We also summarized current genome-editing techniques,including CRISPR-Cas9, TALEN, ZFN, and RNAi, in cyprinids,thus providing technological support for generating new IBdefected strains. The methods applied in studies on IB morphology, such as X-irradiation, micro-CT, ultrasound, bone staining, and anatomy, were also summarized. Thus, this review should provide a technological reference for future research on IBs in fish. A schematic profile of this review is provided in Figure 1.
COMPETING INTERESTS
The authors declare that they have no competing interests.
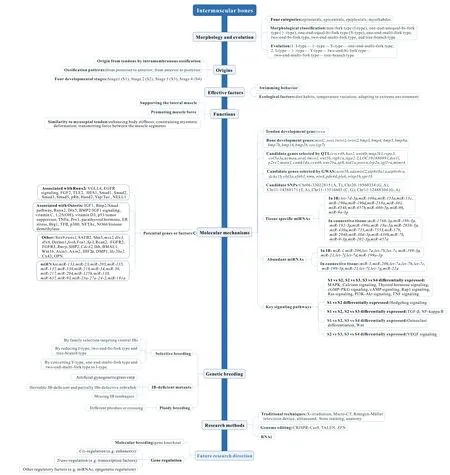
Figure 1 Schematic profile summarizing main points
AUTHOR CONTRIBUTIONS
B.L., L.M., and J.X.Y. conceived the project. B.L. and L.M.wrote the manuscript with input from all authors. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to William R. Jeffery, Zhi-Lian Hu, and You He for comments on a draft manuscript.
- Zoological Research的其它文章
- LIN28A inhibits DUSP family phosphatases and activates MAPK signaling pathway to maintain pluripotency in porcine induced pluripotent stem cells
- Species bias and spillover effects in scientific research on Carnivora in China
- Northern pig-tailed macaques (Macaca leonina)infected with SARS-CoV-2 show rapid viral clearance and persistent immune response
- Potential aquatic environmental risks of trifloxystrobin:Enhancement of virus susceptibility in zebrafish through initiation of autophagy
- Particulate matter exposure exacerbates susceptibility to SARS-CoV-2 infection in humanized ACE2 mice
- A review of the Cypriniform tribe Yunnanilini Prokofiev,2010 from China, with an emphasis on five genera based on morphologies and complete mitochondrial genomes of some species

