Pleiotropy within gene variants associated with nonalcoholic fatty liver disease and traits of the hematopoietic system
Carlos Jose Pirola, Adrian Salatino, Silvia Sookoian
Abstract Genome-wide association studies of complex diseases, including nonalcoholic fatty liver disease (NAFLD), have demonstrated that a large number of variants are implicated in the susceptibility of multiple traits — a phenomenon known as pleiotropy that is increasingly being explored through phenome-wide association studies. We focused on the analysis of pleiotropy within variants associated with hematologic traits and NAFLD. We used information retrieved from large public National Health and Nutrition Examination Surveys, Genome-wide association studies, and phenome-wide association studies based on the general population and explored whether variants associated with NAFLD also present associations with blood cell-related traits. Next, we applied systems biology approaches to assess the potential biological connection/s between genes that predispose affected individuals to NAFLD and nonalcoholic steatohepatitis, and genes that modulate hematological-related traits—specifically platelet count. We reasoned that this analysis would allow the identification of potential molecular mediators that link NAFLD with platelets. Genes associated with platelet count are most highly expressed in the liver, followed by the pancreas, heart, and muscle. Conversely, genes associated with NAFLD presented high expression levels in the brain, lung, spleen, and colon. Functional mapping, gene prioritization, and functional analysis of the most significant loci (P < 1 × 10-8) revealed that loci involved in the genetic modulation of platelet count presented significant enrichment in metabolic and energy balance pathways. In conclusion, variants in genes influencing NAFLD exhibit pleiotropic associations with hematologic traits, particularly platelet count. Likewise, significant enrichment of related genes with variants influencing platelet traits was noted in metabolic-related pathways. Hence, this approach yields novel mechanistic insights into NAFLD pathogenesis.
Key Words: Nonalcoholic fatty liver disease; Nonalcoholic steatohepatitis; Platelets; Leukocytes; Hematologic traits; Genetics
INTRODUCTION
Nonalcoholic fatty liver disease and pleiotropic associations
Nonalcoholic fatty liver disease (NAFLD) is regarded as the most prevalent chronic liver disease[1]. A worldwide increase in NAFLD prevalence is acknowledged not only in the adult but also the pediatric population[1]. Consequently, NAFLD has become a serious health issue on a global scale.Like many other complex diseases, NAFLD develops due to the combined effect of environmental and genetic factors[2-4]. NAFLD presents phenotypic complexity and inter-individual variability, implying that its natural course is characterized by different histological stages-from simple fat accumulation to steatohepatitis (NASH), cirrhosis, and eventually to hepatocellular carcinoma[1]—and considerable variability in disease progression exists among affected patients. One of the proposed factors contributing to the observed inter-patient differences in the disease prognosis and severity is genetic susceptibility[2-4], which might explain up to approximately 20% of the disease variance[5].
Furthermore, NAFLD has not only a high degree of comorbidity with disorders of the metabolic syndrome, including type 2 diabetes, obesity, and cardiovascular disease, but also shared disease mechanisms and disease pathways[6]. These comorbidities have a strong negative impact on the course of NAFLD and vice versa, whereby the presence of NAFLD substantially modifies the course and prognosis of metabolic syndrome-associated diseases[6]. More importantly, the impact of long-term consequences of these comorbidities on cardiovascular health is, at least in part, independent of the presence of general obesity. In fact, it was shown that lean patients with NAFLD have an altered metabolic profile mostly related to increased visceral adiposity that predispose them to cardiovascular risks[7].
Notably, knowledge gained from genome-wide and exon-wide association studies [Genome-wide association studies (GWAS) and EWAS, respectively] of complex diseases, including NAFLD, shows that a large number of single nucleotide polymorphisms are implicated in the susceptibility of multiple traits, which is known as pleiotropy. Phenome-wide association studies (PheWAS) that exploit a significant amount of clinical characteristics gathered mostly from electronic clinical records have thus become a powerful strategy for uncovering pleiotropy.
Among the many traits explored to date in large GWAS/EWAS and PheWAS, including disease traits and biochemical parameters, pleiotropy within gene variants associated with NAFLD and biochemical traits of the hematopoietic system is remarkably consistent across different datasets, specifically those concerning platelet count and platelet volume[8-10]. However, the mechanisms behind the biological connection between NAFLD and platelet-related traits remain poorly understood.
Therefore, in this review, we focused on the analysis of pleiotropy within variants associated with biochemical hematologic traits and NAFLD. We took advantage of information retrieved from large public GWAS and PheWAS based on the general population and examined whether variants known to be associated with NAFLD also exhibit associations with blood cell-related traits, specifically platelet-related phenotypes. After collecting that information, we adopted systems biology approaches to assess the potential biological connection/s between genes that predispose to NAFLD and NASH, and genes that modulate related hematological traits, including platelet count and platelet crit. We reasoned that focusing on clinically meaningful traits for which associations with liver disorders would be plausible from the biological perspective may help elucidate NAFLD biology. Likewise, these analyses would conceptually allow the identification of potential molecular mediators that link NAFLD with platelets.
NAFLD AND HEMATOLOGICAL TRAITS
Recent observations shed light on the concept that NAFLD not only co-exists with cardiovascular disease, including functional and structural myocardial abnormalities[11], but is also associated with other diseases, including cancer, kidney disease, and hematological disorders, among many others[5]. To illustrate the relevance of the association between NAFLD and hematological traits, we analyzed data from the National Health and Nutrition Examination Surveys (NHANES) 2017-2018 database. The dataset and further information are freely available online https://www.cdc.gov/nchs/nhanes/index.htm). The NHANES are population-based surveys conducted by the National Center for Health Statistics of the Centers for Disease Control and Prevention of the United States. They are frequently used to study liver disease. The National Center for Health Statistics Research Ethics Review Board approved the NHANES protocol, and informed consent was obtained from all participants. Liver steatosis was defined by the controlled attenuation parameter (CAP) obtainedviatransient elastography (FibroScan?). Liver steatosis was diagnosed when CAP >268 dB/m was obtained, which is the threshold for significant steatosis based on a large study[12]. We modeled the relationship between NAFLD and relevant factors by linear logistic regression with an interaction term for the specific hematologic trait and gender while adjusting for subjects' demographic and clinical characteristics. Figure 1 shows the effect of white blood cells (WBC), including lymphocytes (Figure 1A), eosinophils (Figure 1B), neutrophils (Figure 1C), and platelet (Figure 1D) count number by gender. Myeloid and lymphoid lineages, as well as platelets show interaction effects with gender even after adjusting for log-transformed confounding factors such as age, waist circumference, diabetes, glycohemoglobin, total cholesterol, and systolic blood pressure. More importantly, NAFLD risk increased steadily with the number of different blood cell types, particularly in men.
These results are reproducibly observed in the Asian population. Wanget al[13]reported that WBC count is a significant factor associated with incident NAFLD in Han Chinese[13]. The authors observed that the association between WBC count and NAFLD remained valid after adjusting for confounding factors including age, gender, smoking, regular exercise, BMI, hypertension, hyperglycemia, and lipid traits[13]. Similarly, WBC count was found to be independently associated with NAFLD regardless of components of the metabolic syndrome in the Korean population[14,15].
PLATELETS AND LIVER DISORDERS—A CONCISE APPRAISAL OF THE RELATIONSHIP
For many biological reasons, the liver and platelets are biologically connected. For example: (1) The fetal liver is a privileged organ of megakaryocyte progenitor differentiation[16]; (2) Thrombocytopenia is a major and debilitating complication of liver cirrhosis[17]; (3) Platelets are involved in the process of fibrogenesis by remodeling the extracellular matrix[18]and secretion of cytokines, including platelet-derived growth factors[19]; and (4) Platelets functioning in the liver exhibit a collaborative effect with endothelial and Kupffer cells in liver regeneration[20]. A more comprehensive update on the role of platelets in liver disorders has been recently published[21].

Figure 1 Steatosis and hematological traits. Panels show the interaction effects between hepatic steatosis and hematological traits (A: Lymphocyte number; B: Eosinophil number; C: Neutrophil number; D: Platelet number) on the probability of having hepatic steatosis according to gender. Interaction analyses were performed by linear logistic regression for the presence of steatosis (no = 0, yes = 1) as the dependent variable adding an interaction term among gender (women = 0, men = 1) and the specific continuous variable. Diabetes and log-transformed age, waist, HgbA1c, total cholesterol, systolic blood pressure, and triglycerides were also included as cofactors. The probability of liver fibrosis was estimated by margins as implemented in the STATA software. Data analysis was based on National Health and Nutrition Examination Surveys 2017-2018; the National Center for Health Statistics Research Ethics Review Board approved the National Health and Nutrition Examination Surveys protocol, and participants gave informed consent. Datasets and further information are available online (https://www.cdc. gov/nchs/nhanes/index.htm). Liver steatosis was defined by the controlled attenuation parameter[12]. Only participants that consumed less than 30 g and 20 g of alcohol for men and women, respectively, were included in the present analysis. Those participants with positive tests for viral hepatitis were excluded.
Earlier evidence also demonstrates that platelets are involved in NASH-related complications. For example, we have observed that patients with NASH have high expression ofTGFB1(transforming grown factor β1)-mRNA in circulating platelets[22]. Platelets are also involved in the process of atherogenesis by upregulating many molecules[23], includingTGFB1[24], which is also related to NAFLD and the risk of cardiovascular disease. Results yielded by a recent study suggest that plateletmediated inflammation in NAFLD drives hepatocellular carcinogenesis[25].
PLEIOTROPIC EFFECTS OF THE NONSYNONYMOUS PNPLA3-RS738409 (P.ILE148MET) VARIANT
The nonsynonymous rs738409 C/G variant inPNPLA3(patatin-like phospholipase domain containing protein 3, also known as adiponutrin or calcium-independent phospholipase A2-epsilon), which encodes the amino acid substitution I148M, is regarded as the major genetic variant associated with the susceptibility to NAFLD and NASH[26,27].
The rs738409 has also been associated with alcoholic liver disease[28-30], hepatitis C[31], hepatitis B[32], and hepatocellular carcinoma[33],[34].
Results of a large PheWAS performed in subjects of European ancestry (816903 participants) with genome-wide genotyped data linked to phenotypic information, including the United Kingdom Biobank cohort, 23andMe cohort, FINRISK (workingage population of Finland), and Children’s Hospital of Philadelphia, confirmed that rs738409 presents pleiotropic effects beyond the liver. For instance, rs738409-G was associated with increased risk of type 2 diabetes and decreased risk of high total cholesterol, acne, gout, and gallstones; all these associations remained significant after adjusting for elevated transaminases[35].
In addition, it was suggested that rs738409 might be used to predict race-related hepatotoxicity in pediatric patients with acute lymphoblastic leukemia[9].
Another study revealed the association of rs738409 with mean corpuscular hemoglobin (P= 6 × 10-9) based on the analysis of individuals of 116666 British ancestry[36]. Consistently, Kichaevet al[37]established the association of rs738409 with mean corpuscular hemoglobin (P= 4 × 10-25) by using genome-wide genotyping array in a sample of 443000 individuals of European ancestry[37]. Variants in or nearPNPLA3have also been associated with the aspartate transaminase-to-platelet ratio index[38].
A summary picture illustrating the genetic associations withPNPLA3locus, including its pleiotropic effects on diverse laboratory measurements, is shown in Figure 2. It reveals that the strength of the association scores of rs738409 and levels of liver enzymes, particularly ALT levels (association score = 1), is shared with the effect of this variant on many blood-related traits, for instance platelet count (association score = 0.8) (Figure 2).
PHEWAS: THE EFFECT OF VARIANTS ASSOCIATED WITH NAFLD ON BLOOD-RELATED TRAITS
We next used information sourced from electronic health records and GWAS data computed from United Kingdom Biobank entries pertaining to 452264 individuals, which was retrieved from the Gene ATLAS (http://geneatlas.roslin.ed.ac.uk) and Neale's database (http://www.nealelab.is/uk-biobank/).
We specifically searched for PheWAS associations of variants influencing NAFLD, includingPNPLA3-rs738409,TM6SF2-rs58542916,MBOAT7-TMC4rs641738,GCKRrs780094, andHSD17B13-rs72613567. As expected, rs738409, rs58542916, and rs72613567 were associated with liver-related traits in the United Kingdom-Biobank GWAS (Table 1).
Of note, all aforementioned variants showed GWAS-significant associations (P< 5 × 10?8) with blood-related traits. The strongest associations pertained to platelet traits, including platelet crit (which represents the proportion of blood volume that is occupied by platelets, expressed as a percentage), platelet volume, and platelet count (Table 1). Specifically,PNPLA3-rs738409 was associated with platelet count (P= 2.9 × 10-45) and platelet crit (P= 3.6 × 10-29) with thePvalues for association exceeding those for association with liver diseases (Table 1). A similar pattern was obtained forTM6SF2-rs58542926 andHSD17B13-rs72613567 (Table 1) variants and their associations with platelet traits.
GENES ASSOCIATED WITH NAFLD AND PLATELET COUNT SHARE PATHWAYS INVOLVED IN EXTRACELLULAR MATRIX REMODELING AND CYTOSKELETAL SIGNALING PROTEINS
To explore shared pathways between NAFLD and platelet-related traits, we retrieved from the Open Target Genetics platform (https://genetics.opentargets.org) the list of genes (human protein-coding genes) associated with phenotypes of interest. Specifically, we focused on platelet count, which expresses the number of platelets per unit volume in a sample of venous blood.
The genetic associations in the Open Target platform are derived from GWAS Catalog (https://www.ebi.ac.uk/gwas) and PheWAS (https://phewascatalog.org/), whereby the former contains publications indexed in PubMed and the latter is a repository of electronic medical records with links to the Vanderbilt DNA biobank. The list of genes associated with platelet count (n= 305) and NAFLD (n= 161) is shown in Supplementary Table 1 and Supplementary Table 2, respectively.
To analyze and interpret the pathways shared between genes associated with NAFLD and those associated with platelet count, we used the FUMA platform available at https://fuma.ctglab.nl/. FUMA utilizes positional expression quantitative trait loci and chromatin interaction mappings to build gene-based pathways and tissueenrichment heatmaps[39]. Hence, we first tested the tissue specificity of the list of genes/proteins associated with each phenotype, namely NAFLD and platelet count. Specifically, we explored tissues in which those genes/proteins present higher expression levels (differentially expressed genes are defined for each label of each expression dataset). Genes showing aP≤ 0.05 after Bonferroni correction and absolute log fold change ≥ 0.58 were defined as differentially expressed. Interestingly, genes associated with platelet count were most highly expressed in the liver, followed by the pancreas, heart, and muscle (Figure 3A). Conversely, genes associated with NAFLD presented high expression levels in the brain, lung, spleen, and colon (Figure 3B).
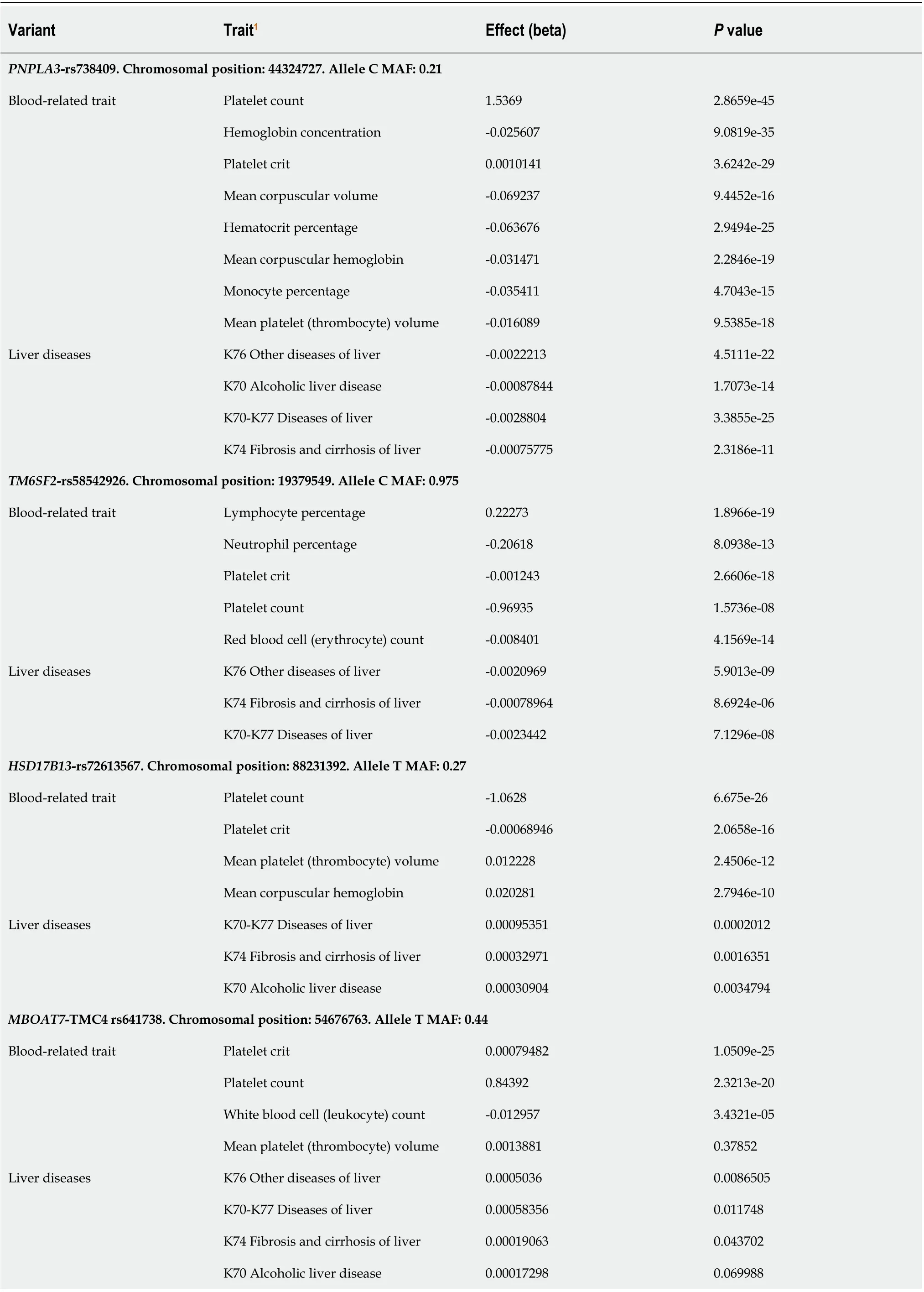
Table 1 Phenome-wide association studies associations of variants influencing nonalcoholic fatty liver disease and their effect on blood-related traits

GCKR-rs780094. Chromosomal position: 27741237. Allele C MAF: 0.38 Blood-related trait Mean reticulocyte volume 0.16064 5.9197e-33 Neutrophil percentage -0.14035 2.7743e-19 Neutrophil count -0.037139 6.3804e-47 Monocyte percentage 0.073076 7.914e-79 Mean platelet (thrombocyte) volume 0.014342 1.3297e-18 Platelet crit -0.0013887 1.3581e-71 Platelet count -1.9052 6.2404e-92 Hematocrit percentage 0.049511 4.3172e-21 Liver diseases K76 Other diseases of liver -0.00058708 0.0026207 K70-K77 Diseases of liver -0.0004415 0.060918 K74 Fibrosis and cirrhosis of liver -0.00011872 0.2169 K75 Other inflammatory liver diseases -0.00012854 0.13954 K70 Alcoholic liver disease 9.7922e-05 0.31331 1Letters and numbers such as K70, K76, K74 and K75 represent the codes for diseases (ICD10) in the United Kingdom Biobank. Approximately 30 million variants in the United Kingdom Biobank from the Gene ATLAS (http://geneatlas.roslin.ed.ac.uk) and Neale's database (http://www.nealelab.is/ukbiobank/) resources were comprehensively tested for their association with liver (ICD10 codes: K70, K76, K74 and K75) and blood cell-associated traits, including platelet, leukocyte and neutrophil counts. PNPLA3: Patatin-like Phospholipase Domain Containing protein 3; TM6SF2: Transmembrane 6 Superfamily Member 2; HSD17B13: Hydroxysteroid 17-Beta Dehydrogenase 13; MBOAT7: Membrane Bound O-Acyltransferase Domain Containing 7; TMC4: Transmembrane channel like 4; GCKR: Glucokinase regulator.
The Venn diagram provided in Figure 3C shows the genes shared between NAFLD and platelet count according to the information retrieved from GWAS and PheWAS catalogs, as explained earlier, among which we found eleven shared loci that includedPNPLA3. Next, we performed pathway analysis on the list of shared genes, which revealed an enrichment of genes (ACTN1andTNFRSF13B) belonging to the predicted pathway “Syndecan 4 pathway” (PID_SYNDECAN_4_PATHWAY) (Figure 3D). The Syndecan 4 pathway is involved in cell growth, differentiation, and adhesion, and in the modulation of extracellular matrix proteins[40]. Syndecans are type I transmembrane proteins with an N-terminal ectodomain that contains several consensus sequences for attachment to glycosaminoglycan, heparan sulfate, and to a lesser extent chondroitin sulfate chains, and a short C-terminal cytoplasmic domain. Syndecans may act as integrin co-receptors. Interactions between fibronectin and syndecans are modulated by tenascin-C. Syndecans bind a wide variety of soluble and insoluble ligands, including extracellular matrix components, cell adhesion molecules, and growth factors, including VEGFs, cytokines, and proteinases[41]. It is worth noting that parvin beta (PARVB), which has been significantly associated with NAFLD[42,43]and hematological traits[44], encodes a member of the parvin family of actin-binding proteins that play a role in cytoskeleton organization and cell adhesion. This family member binds to alphaPIX and alpha-actinin, and can inhibit the activity of integrinlinked kinase. This protein also functions as a tumor suppressor. As thePARVBlocus is located nearPNPLA3, further studies are needed to establish whether the association of the locus with NAFLD and hematological traits merely reflects a linkage between the two genetic loci.
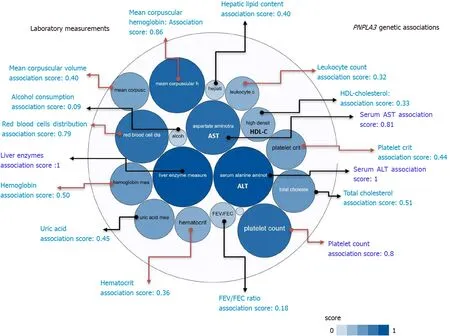
Figure 2 PNPLA3 and genetic associations with laboratory measurements in genome-wide association studies and phenome-wide association studies. The score for the associations ranges from 0 to 1, with higher scores indicating stronger evidence for an association. Bubbles in the figure represent the different scores with varying shades of blue: the darker the blue, the stronger the association. Red arrows highlight the association of the rs738409 variant in PNPLA3 and laboratory measurements related with hematological traits. HDL: High density lipoprotein cholesterol; FEV: The ratio of forced expiratory volume to forced vital capacity, used as a measure of pulmonary function; uric: Uric acid; AST: Aspartate aminotransferase level; ALT: Alanine aminotransferase level; total chol: Total cholesterol. Source: Open Target Database (https://genetics.opentargets.org/gene/ENSG00000100344). Score summaries: Data sources and factors that affect the relative strength of the evidence scores can be found at: https://docs.targetvalidation.org/getting-started/scoring.
We further explored the Gene Ontology (GO) biological processes in which the list of genes associated with platelet count was specifically enriched. We found that, in addition to expected pathways that included hemostasis, response to wound healing, and platelet degranulation, there were pathways that support the plausibility of sharing genes with NAFLD, for example the triglyceride catalytic process. The GO biological processes associated with the top 50 genes on the list of loci associated with platelet count is shown in Figure 4. Remarkably,PNPLA3andFABP6(Fatty Acid Binding Protein 6) are the two overlapping loci that would be responsible for the enrichment of triglyceride metabolism (Figure 4).
BIOLOGICAL PATHWAYS OF GENES ASSOCIATED WITH PLATELET COUNT VS GENES ASSOCIATED WITH NAFLD
It is also noteworthy that, despite the shared genes, there are differences in the biological processes in which the sets of genes associated with platelet count and with NAFLD are involved. To explore these pathways, we performed overrepresentation analysis based on a more comprehensive list of genes associated with each phenotype.
The input list of platelet count-associated variants (P< 1 × 10-6) was generated by searching the United Kingdom Biobank GWAS database as provided by Neale’s lab resource (http://www.nealelab.is/uk-biobank/), which shows variants in about 2424 loci with genome-wide significance (P< 5 × 10-8) for an association with platelet count as a continuous trait (109cells/L; mean = 252.023 ± Std.dev = 60.0604). We used the United Kingdom Biobank GWAS database because it provides one of the largest publically available sources of genetic associations with laboratory traits in the general population involving 479367 individuals of both sexes (http://biobank. ctsu.ox.ac.uk/crystal/field.cgi?id=30080).
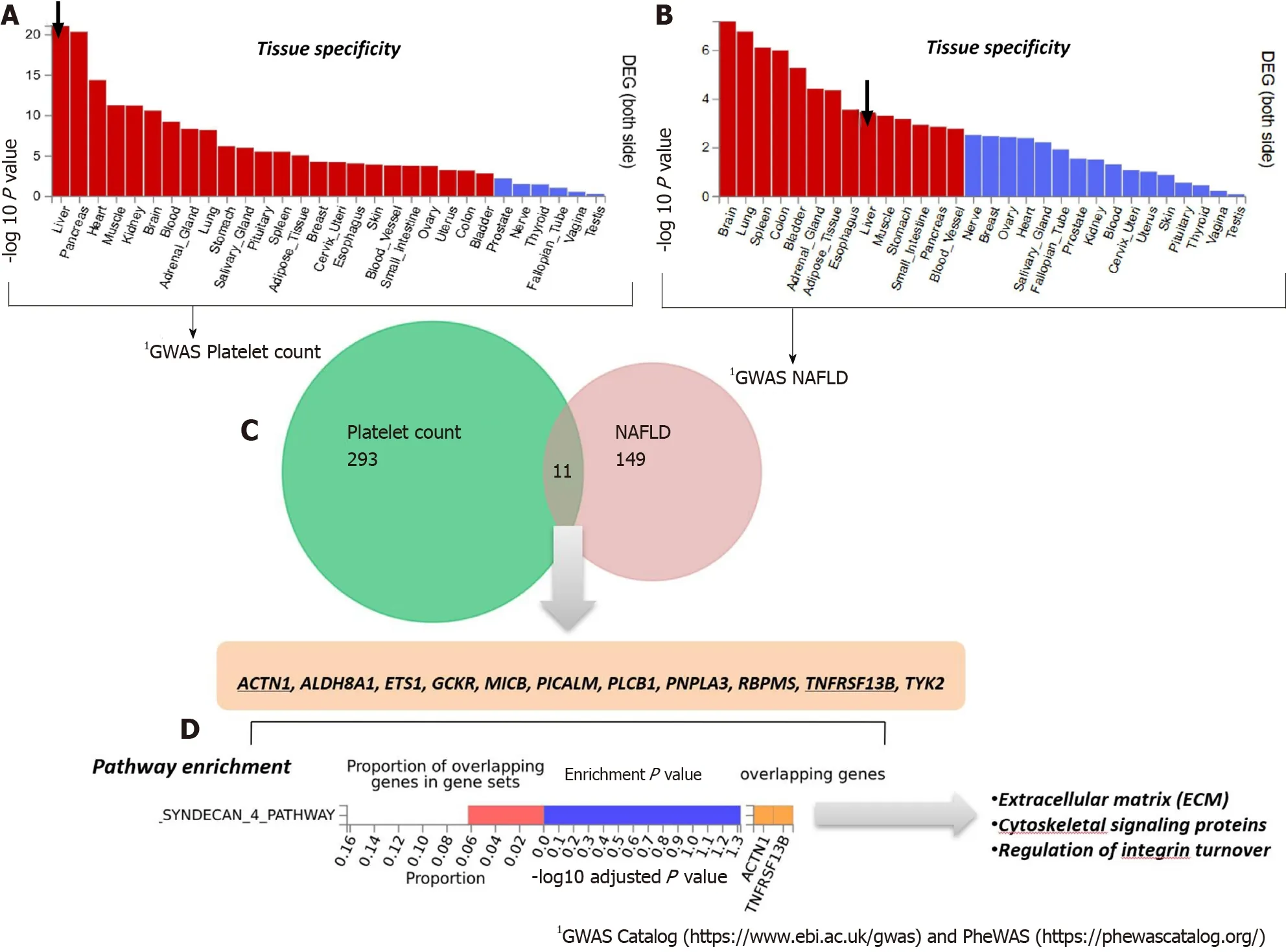
Figure 3 Genes that are shared between nonalcoholic fatty liver disease and platelet count. A and B: Significantly enriched Differentially Expressed Gene (DEG) Sets (Pbon < 0.05) are highlighted in red (FUMA). DEG sets were pre-calculated by performing two-sided t-test for any one of the labels against all others. For this purpose, expression values were normalized (zero-mean) to obtain a log2 transformation of expression value (EPKM or TPM). Genes with P ≤ 0.05 after Bonferroni correction and absolute log fold change ≥ 0.58 were defined as differentially expressed genes in a given label compared to others. In addition to DEG, upregulated DEG and down-regulated DEG were also pre-calculated by taking the sign of t-statistics into account. Input genes were tested against each of the DEG sets using the hypergeometric test. The background genes are genes that have average expression value > 1 in at least one of the labels and exist in the userselected background genes. Significant enrichment at Bonferroni-corrected P ≤ 0.05 is colored in red. C: Venn diagram showing the number of genes that are common (overlapping areas) and dissimilar (non-overlapping areas) in nonalcoholic fatty liver disease (NAFLD) and platelet count gene lists. D: Pathway analysis using all Canonical Pathways (MsigDB c2) in the web-based FUMA platform available at http://fuma.ctglab.nl. Overlapping genes (underlined): ACTN1: Actinin Alpha 1; TNFRSF13B: TNF Receptor Superfamily Member 13B. The input lists of platelet count- and NAFLD-associated variants were generated by searching the Open Target platform, which contains data retrieved from GWAS Catalog (https://www.ebi.ac.uk/gwas) and phenome-wide association studies (https://phewascatalog.org/). The list of genes associated with platelet count contains 305 loci and that associated with NAFLD contains 149 loci, as shown in Supplementary Figures 1 and 2, respectively.
In the case of NAFLD, and to avoid issues arising from the paucity of GWAS/ EWAS discovered genes, we included a more comprehensive list of 928 loci obtained by data mining[5]that represents genetic and molecular associations with the disease.
We chose to conduct the overrepresentation analysis on the list of loci obtained from data mining because NAFLD as the disease trait (K76.0 Fatty change of liver: http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=41202) is underrepresented in the United Kingdom Biobank GWAS database, affecting only 460 of 410332 individuals whose data are stored in this repository.
Differences in biological pathway enrichment between the two datasets (NAFLD and platelet count) are summarized in Figure 5, which shows specificity in the function of genes in each of the gene lists. For instance, the NAFLD list of associated genes/proteins is enriched in expected pathways, such as metabolism of lipids and amino acids, purine metabolism, and circadian rhythm, among others (Figure 5). The platelet count list is enriched with genes involved in DNA repair, telomere maintenance, and the Hedgehog pathway, among many others, as shown in Figure 5[45]. Interestingly, some pathways related to platelet-derived growth factors, such as the platelet-derived growth factors receptor-alpha signaling pathway, are over-represented in the list of genes associated with NAFLD. The many types of integrin cell surface interaction pathways seem to play a more important role in NAFLD pathophysiology than in regulating platelet count. Notably, the gene set associated with platelet count presents enrichment in the leukotriene synthesis pathways, which have been linked to the progression of NAFLD[46-49].

Figure 4 Gene Ontology biological processes of the top 50 genes associated with platelet count. The chart shows fold enrichment in biological processes of genes associated with platelet count with respect to those present in the whole genome. The input list of platelet count-associated variants was generated by conducting a search via the Open Target platform, which contains data retrieved from genome-wide association studies Catalog (https://www.ebi.ac.uk/gwas) and phenome-wide association studies (https://phewascatalog.org/). The whole list of genes associated with platelet count contains 305 loci, as shown in Supplementary Figure 2. Gene mapping and analysis was conducted using the FUMA genome-wide association studies tool (https://fuma.ctglab.nl/). Genes belonging to the pathways as overlapping genes are shown as a heatplot to the right.
FUNCTIONAL ASSESSMENT OF VARIANTS ASSOCIATED WITH LABORATORY HEMATOLOGICAL-RELATED TRAITS
To understand the potential involvement of genes associated with laboratory hematological-related traits in NAFLD biology, we performed functional analysis. Based on the reverse biology premise, we reasoned that genes involved in hematological-related traits might be associated with some metabolic function/s that would presumably affect NAFLD pathogenesis. Thus, the top blood-related associated traits in terms of their statistical significance were further searched for genetic associations in the entire United Kingdom Biobank dataset. Specifically, approximately 30 million variants in the United Kingdom Biobank from the Gene ATLAS (http://geneatlas.roslin.ed.ac.uk) and Neale's database (http://www.nealelab.is/ukbiobank/) resources were comprehensively tested for associations with blood cellassociated traits, including platelet, leukocyte and neutrophil counts. We further explored the pathways in which the lists of genes associated with these traits are involved. For this purpose, we used the FUMA resource that allows using functional and biological information to prioritize genes based on GWAS outcomes.
Interestingly, functional mapping, gene prioritization, and functional analysis using FUMA of the most significant genetic variants (P< 1 × 10-6) revealed 85 mapped genes in the full list of loci associated with hematological traits that are also associated with liver traits, including chronic liver diseases, fibrosis and liver cirrhosis, NAFLD, and other inflammatory liver diseases (Figure 6A).PNPLA3, SAMM50, PARVBandHSD17B13are among the ten genes shared by liver traits and platelet count.TM6SF2is shared by liver traits, platelet count, and neutrophil count, andGCKRis shared by liver and all hematological traits (Figure 6A).

Figure 5 Overrepresentation analysis of biological pathways of genes associated with platelet count vs. genes associated with nonalcoholic fatty liver disease. Overrepresentation analysis of biological pathways of platelet count-associated genes in comparison with that pertaining to nonalcoholic fatty liver disease-associated genes. The input list of platelet count-associated variants (P < 1 × 10-6) was generated by searching the United Kingdom Biobank genome-wide association studies database as provided by Neale’s lab resource (http://www.nealelab.is/uk-biobank/). The input list associated with nonalcoholic fatty liver disease includes 928 genes/proteins obtained by data mining[5]. The analysis was performed by applying the functional enrichment and interaction network analysis (FunRich) tool[46].
In addition, Figure 6B shows the consistent association ofPNPLA3, TM6SF2, SAMM50, andPARVBwith liver and hematological traits identifiedviaFUMA analysis in reported GWAS. Finally, we performed functional enrichment analysis on genes significantly associated with platelet count, leukocyte count, and neutrophil count by applying the FunRich tool. Notably, loci involved in the genetic modulation of platelet, leukocyte, and neutrophil counts presented significant enrichment in metabolic, energy balance, xenobiotics, and CYP-450-related pathways (Figure 7A?C). However, platelet-related loci are particularly involved in regulating key aspects of metabolism, while leukocyte and neutrophil counts are related to more general homeostasis regulation processes (Figure 7D?F).
CONCLUSION
PheWas revealed that variants in genes influencing NAFLD present pleiotropic associations with laboratory-related hematologic traits and are relevant to the hematopoietic liver function. Similarly, related genes with variants influencing hematological traits, platelet count in particular, presented significant enrichment in metabolic and energy balance-related pathways.
By using different resources and datasets of variants associated with the genetics of platelet count and NAFLD, we found consistency in the results, suggesting that there are shared mechanisms and pathways between the two phenotypes. In particular, we found metabolic and lipid pathways shared by NAFLD and platelet traits. It is anticipated that potential therapeutic targets, including novel ligands of peroxisome proliferator-activated receptors may also play a role in modulating platelet-related phenotypes such as platelet activation and the cascade of events associated with inflammation and cardiovascular risk.
In summary, our approach provides novel mechanistic insights into NAFLD pathogenesis. Further research is nonetheless necessary to ascertain whether genes associated with liver diseases present ample pleiotropy and, therefore, modify functions of diverse organs simultaneously. If, conversely, some phenotypes are found to act as intermediaries between genes and disease, a Mendelian Randomization approach can be used to study the relationship between, in this case, hematological and liver traits or vice versa.
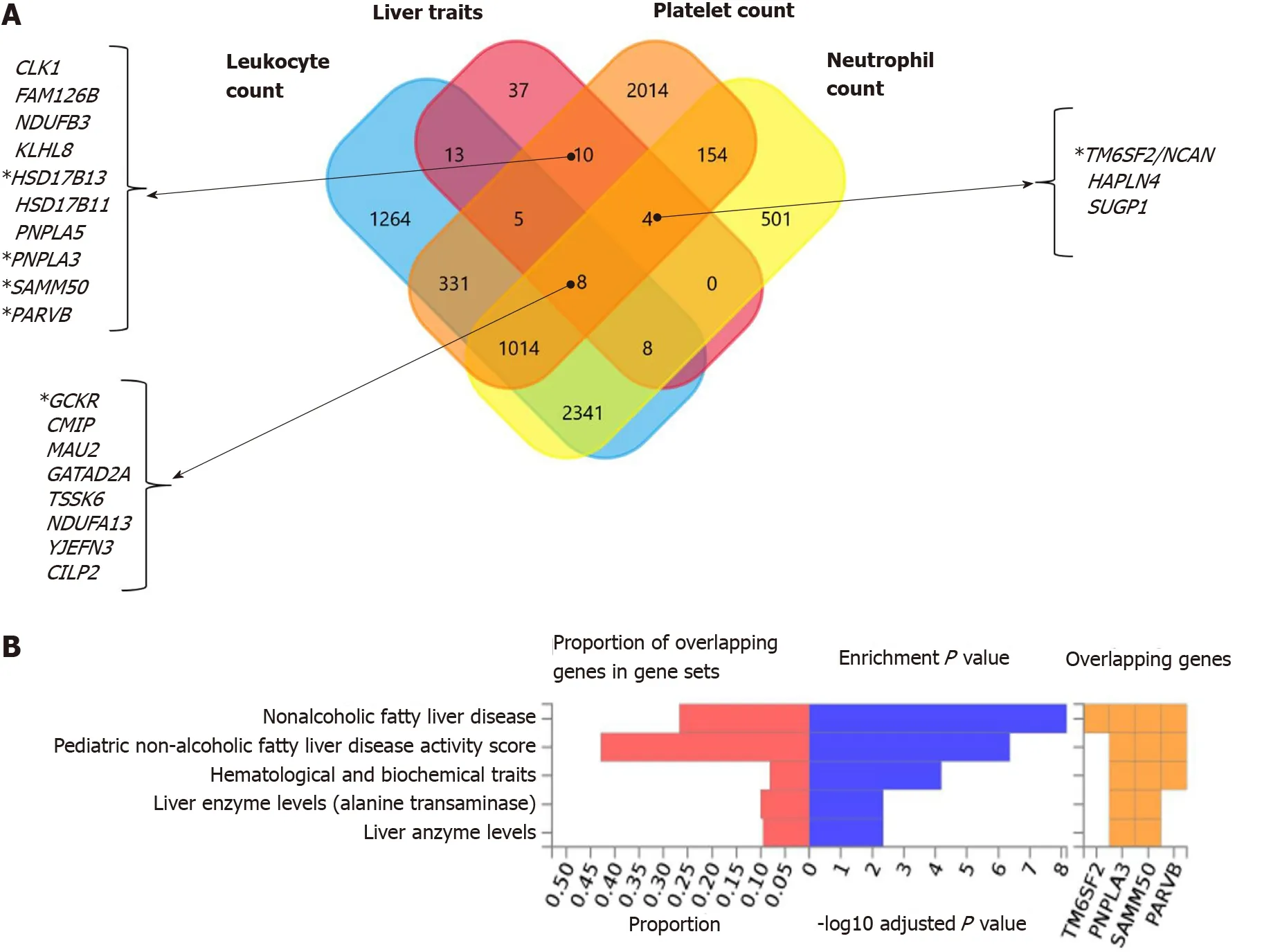
Figure 6 Nonalcoholic fatty liver disease and hematological traits associated genes. A: Venn diagram showing the set of genes associated with liver traits (ICD10 codes: K70, K76, K74 and K75), platelet count, leukocyte count, and neutrophil count, as well as their overlapping genes in the United Kingdom Biobank. Approximately 30 million variants in the United Kingdom Biobank dataset sourced from the Neale's database (http://www.nealelab.is/uk-biobank/) were comprehensively tested for association with liver and blood cell traits, including platelet, leukocyte and neutrophil counts. B: Overlap between nonalcoholic fatty liver disease-associated genes and those associated with hematological traits. The chart shows information on previously known single nucleotide polymorphisms-trait associations reported in the genome-wide association studies (GWAS) catalog for all single nucleotide polymorphisms associated with nonalcoholic fatty liver disease in the United Kingdom Biobank GWAS database; the analysis was conducted using the FUMA GWAS tool (https://fuma.ctglab.nl/).
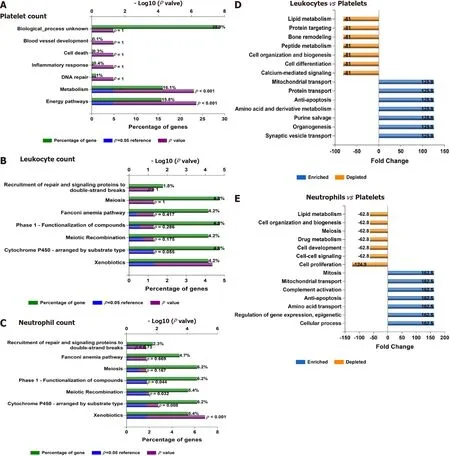
Figure 7 Functional assessment of variants associated with laboratory hematological-related traits. A-C: Functional analysis of genes significantly associated with platelet, leukocyte, and neutrophil count in the whole genome-wide association studies dataset (452264 individuals whose data are included in the United Kingdom Biobank). Functional enrichment analysis was performed using the FunRich tool, while Bonferroni and Benjamini-Hochberg methods were used to correct for multiple testing. D-E: Charts show fold changes in biological processes of genes associated with leukocyte or neutrophil counts vs biological processes of genes associated with platelet count. The input list of genes associated with platelet count, leukocyte count, and neutrophil count (P < 1 × 10-8) was generated by searching the United Kingdom Biobank genome-wide association studies database.
 World Journal of Gastroenterology2021年4期
World Journal of Gastroenterology2021年4期
- World Journal of Gastroenterology的其它文章
- Duplication of the common bile duct manifesting as recurrent pyogenic cholangitis: A case report
- Could saline irrigation clear all residual common bile duct stones after lithotripsy? A self-controlled prospective cohort study
- Effect of remote ischemic preconditioning among donors and recipients following pediatric liver transplantation: A randomized clinical trial
- Clinic-pathological features of metabolic associated fatty liver disease with hepatitis B virus infection
- Preoperative maximal voluntary ventilation, hemoglobin, albumin, lymphocytes and platelets predict postoperative survival in esophageal squamous cell carcinoma
