Visualizing Meiotic Chromosome Pairing and Segregation in Interspecific Hybrids of Rice by Genomic in situHybridization
Liu Mao-Sen, Tseng Shih-Hsuan, Chen Ting-Chu, Chung Mei-Chu
Research Paper
Visualizing Meiotic Chromosome Pairing and Segregation in Interspecific Hybrids of Rice by GenomicHybridization
Liu Mao-Sen, Tseng Shih-Hsuan, Chen Ting-Chu, Chung Mei-Chu
()
Meiotic disturbances in F1hybrids and their progenies are still major problems in wide hybridization. To investigate the genome affinity reflected in chromosome pairing and segregation, we studied chromosome behaviors during meiosis in two interspecific F1hybrids [×(×, BCE genome) and×(, EHJ genome)] by using both traditional staining methods and genomichybridization (GISH). GISH analysis has been successfully performed on mitotic chromosomes to distinguish differentgenomes, but relatively fewer systematic analyses of meiotic chromosomes of interspecific hybrids have been reported. In the hybrids, highly irregular chromosome behaviors through meiosis resulted in producing microspores with unbalanced genome. At diakinesis of these two hybrids, most chromosomes present as univalent, with low frequency as bivalents and occasionally as trivalents. In a pollen mother cell, 2 to 8 bivalents and 0 to 4 trivalents were observed in the hybrid, and 1 to 8 bivalents and 0 to 5 trivalents were observed in the hybrid. GISH results indicated that 51.52% bivalents inand 79.65% bivalents ininvolved allosyndetic association, which indicates that recombination and introgression should be possible if viable backcrosses can be recovered even from triploid hybrids. In this study, we revealed that the meiotic disturbance due to low affinities between parental genomes is the major reason for the sterility of these two triploid interspecific hybrids. The two hybrids showing vigor in reproductive growth are potential genetic resources in future breeding programs. A better understanding of genomic affinities between these distantspecies can facilitate planning an effective breeding program by using wide hybridization, and efficient and routine GISH analysis is helpful to monitor alien introgression in the process.
; wide hybridization;genomic affinity; allosyndesis; autosyndesis; pollen viability
Developing new crops to produce enough food in a sustainable and environmental-friendly way is feasible for meeting the challenges of climate change and increasing human population. One efficient strategy is to introduce useful agronomic traits from wild species into cultivars, which has been successful in several crops including rice (Sanchez et al, 2014). Some important agronomic traits, including disease resistance and cytoplasmic male sterility, have been transferred from wild species to cultivars by wide hybridization (Sanchez et al, 2014).
The genusconsists of 27 species including two cultivated rice species,and,and other wild species (Stein et al, 2018). From taxonomic and cytogenetic studies,species were classified into 11 genome types: six diploids (2=24; AA, BB, CC, EE, FF and GG) and five tetraploids (2=48; BBCC, CCDD, HHJJ, HHKK and KKLL) (Stein et al, 2018). Two cultivated rice species (and) are the AA genome type.
Numerous hybrids between cultivated rice and wildspecies have been developed (Jena and Khush, 1984; Sitch et al, 1989; Brar et al, 1991; Brar and Khush, 1995). However, the sterility of hybrids and their progeny is still the main problem of adopting wide hybridization for improving rice (Piegu et al, 2006). In the genus, the seed-setting of interspecific hybrids is commonly < 10% (Sitch et al, 1989). Most F1hybrids are sterile because of failure of male and/or female gametogenesis by several kinds of meiotic disturbances in chromosome pairing and segregation. Although numerous genetic and environmental factors may also interfere in chromosome pairing, low affinity between parental genomes is considered a general cause of meiotic aberrations in interspecific hybrids (Jauhar and Joppa, 1996; Lee et al, 2011). After wide hybridization, the immature embryo could becultured to rescue the embryos of F1and backcross progenies (Mariam et al, 1996). Through embryo rescue technologies and repeated backcrossing, a series of interspecific hybrids ofspecies, monosomic alien addition lines (MAALs) and introgression lines have been produced (Brar and Khush, 1997; Jena, 2010). Several MAALs with alien chromosomes from tetraploid were also developed after producing allotriploid hybrids by crossingand tetraploid wild species (Jena and Khush, 1989; Jena et al, 1991). In general, hybrid progenies that resemble the recurrent parent and show restoration of fertility and stability could be obtained by repeated backcrossing to the cultivated rice up to BC5or BC6generations (Kalloo, 1992). In rice wide crosses, such diploid progenies that resemble the recurrent parent with alien characters could be obtained after two backcrosses (Amante-Bordeous et al, 1992; Multani et al, 1994). The rapid recovery of the recurrent parent phenotype suggested that rare recombination occurred between the parental genomes of F1hybrids. However, restriction fragment length polymorphism analysis indicated the occurrence of recombination events that allow genes to transfer from the alien chromosomes into cultivated rice (Jena et al, 1992; Brar and Khush, 1997; Angeles-Shim et al, 2014). No cytogenetic and molecular evidence has shown alien chromosome substitution in introgression lines derived fromand distantly relatedspecies.
Meiosis is a successive event including one replication and two segregations of chromosomes. Normally, the first segregation occurs between paired homologous chromosomes at anaphase I and the second occurs between replicated chromatids at anaphase II, which reduces the genetic material from diploid (2) to haploid (). At the end of meiosis, each pollen mother cell (PMC) produces a tetrad with four haploid microspores () that further develop into pollen grains by two mitosis processes (Dawe, 1998). However, low affinity between parental genomes of an interspecific hybrid would disturb the meiosis process, which would result in failure to form functional gametes and cause low fertility in most interspecific hybrids (Jauhar and Joppa, 1996; Lee et al, 2011). Traditionally, genomic affinity between two parental species of an interspecific hybrid is explained as the number of univalent, bivalent and multivalent chromosomes observed at diakinesis or metaphase I in F1hybrids. Molecular cytogenetic techniques such as fluorescencehybridization (FISH) and genomichybridization (GISH) have been successfully used to discriminate different genomes, elucidate the extent of genetic recombination and identify the genomic constitution of hybrids (Le et al, 1989; Schwarzacher et al, 1989). Furthermore, GISH can distinguish autosyndetic from allosyndetic pairing, which is considered difficult by conventional meiotic analysis (Lee et al, 2011). In an interspecific hybrid, autosyndesis represents pairing between non-homologous chromosomes from the same genome, whereas allosyndesis represents pairing between chromosomes from different but related genomes. Allosyndesis, which facilitates genetic recombination, is desirable for wide hybridization (Mason et al, 2010). Therefore, molecular cytogenetic analysis of the meiosis of F1hybrids will assist in precisely understanding the process of introgression for planning effective breeding programs by using wide hybridization. GISH is able to discriminate differentgenomes and has been successfully used to determine genome composition in allopolyploid species and interspecific hybrids, identify alien chromosome in MAALs and investigate the relationships amonggenomes (Iwata et al, 2014). However, these studies mainly involved GISH of somatic chromosomes, rarely meiotic chromosomes for dissecting chromosome behavior during meiosis.
To study the genome affinity betweengenomes reflected on the meiosis irregularities of wide hybrids, we examined pollen viability by Alexander staining and investigated chromosome behaviors during meiosis by conventional staining and GISH. Two investigated hybrids,()×() and×(), are the most vigorous in reproductive growth among the 14 interspecific hybrids obtained from the International Rice Research Institute (IRRI), the Philippines. These hybrids represent the combination of distant wild species as well as the combination of diploid and tetraploid chromosomes.(EE type) is a native species of tropical Australia with the largest genome size among diploid rice (Ammiraju et al, 2006) and it features several important agronomic traits including resistance to brown planthopper, bacterial blight and rice blast, drought avoidance and tolerance to heat and drought (Sanchez et al, 2014).may have some potential value as a crop because it contains desirable starch properties in grains. In comparison with otherspecies, the starch of its grains contains more short chain amyloses and shows low pasting viscosity and low retrogradation (Tikapunya et al, 2016, 2017).(BBCC type) is distributed in tropical Asia including the Philippines and Papua New Guinea, and it is an important resource of resistance genes against rice blight, bacterial blight, white backed planthopper and brown planthopper (Vaughan, 1994).(HHJJ type) grows across south Asia and is known as a genetic resource of resistance to rice blast, bacterial blight, tungro virus, stem borer and whorl maggot (Sanchez et al, 2014).
One interspecific hybrid, IR64 (ssp.) ×(, AA type), was used as a check in the pollen viability test and in the demonstration of a normal meiosis process. Both IR64andare diploid and belong to the AA genome type. The variety IR64, developed at IRRI, features high yield and medium growth duration but is highly prone to drought (Baghyalakshmi et al, 2018).is native to sub-Saharan Africa and Madagascar and is the origin of the genewith broad resistance to bacterial blight (Khush et al, 1990).
In this study, we provided cytogenetic information to evaluate the genomic affinity and the possibility of genetic recombination in these hybrids.
Results
Pollen viability
Normally, a tetrad with four haploid microspores is formed at the end of meiosis in a PMC. The hybrid IR64 ×produced predominantly normal tetrads (Fig. 1-A), whereas×and×produced mainly tetrads; however, triads, dyads and monads were frequent (Fig. 1-B). For IR64 ×, 57.8% of pollen was solid and stained magenta-red and so were considered to be viable (Fig. 1-C and -D). All pollen observed in×(Fig. 1-E) and×(Fig. 1-F) were empty, shriveled and non-viable. No seed was obtained from×and×; both were full sterile.
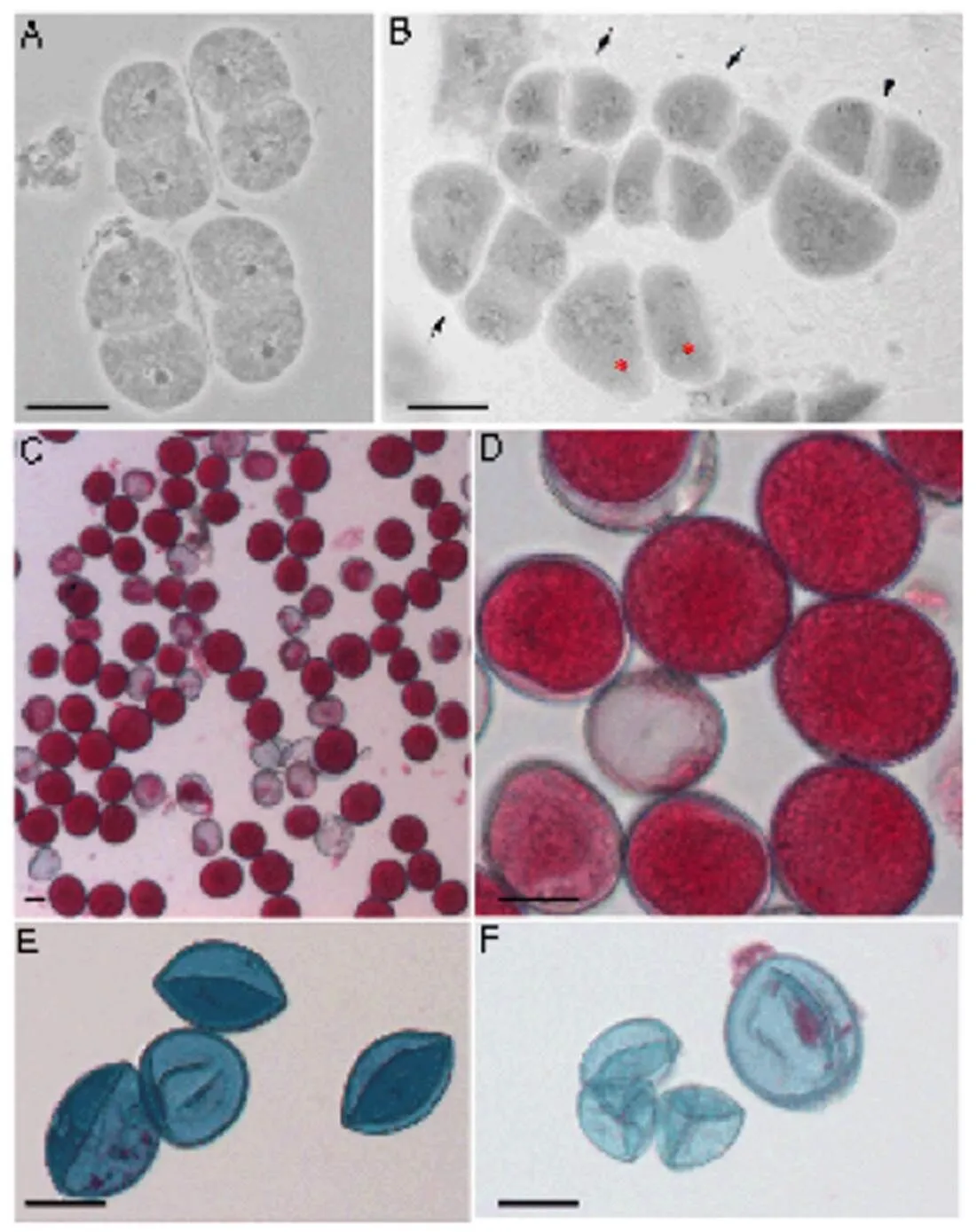
Fig. 1. Viability of pollen grains from interspecific hybrids.
A, Tetrad of hybrid IR64 ×(). B, Meiotic end-products of the hybrid() ×() including tetrad (arrow), triad (arrowhead) and dyad (*). C–F, Pollen viability test by Alexander staining. Viable pollens were solid and stained magenta-red, and non-viable pollens with empty and shriveled features were stained blue. C and D, Pollens of the hybrid IR64 ×; E, Pollens of the hybrid×; F, Pollens of the hybrid×.Scale bars, 20 μm.
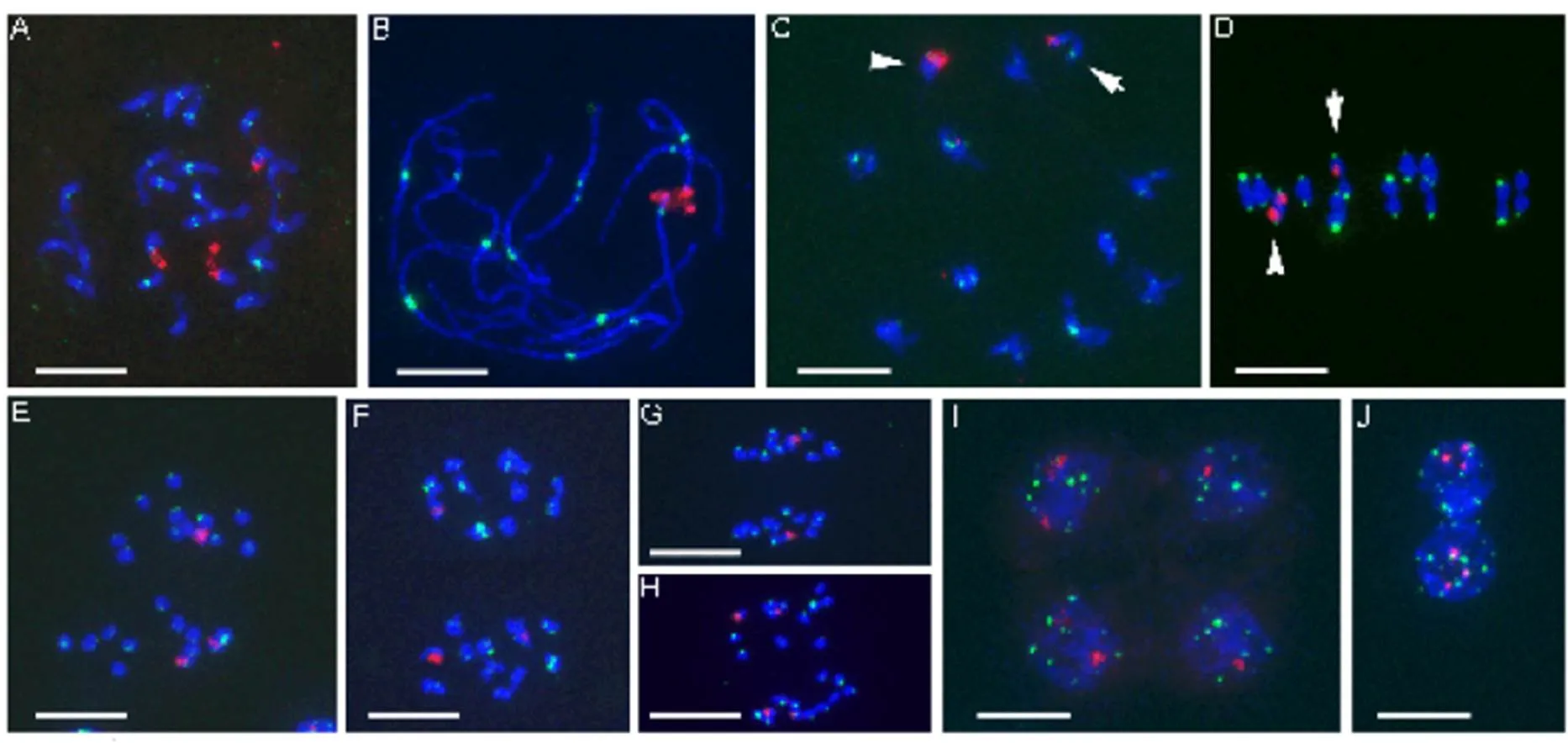
Fig. 2. Mitotic chromosomes and meiotic chromosome behaviors of IR64 ×(2= 24, AA genome).
A, Mitotic metaphase. B, Pachytene. C, Diakinesis. D, Metaphase I. E, Anaphase I. F, Metaphase II. G and H, Anaphase II. I, Tetrad. J, Somatic interphase nuclei. In C and D, arrow indicates the bivalent with a single 45S rDNA signal (heteromorphic 45S rDNA site). Arrowhead indicates the bivalent with two 45S rDNA signals. 45S rDNA and centromeric repeats are indicated in red and green, respectively. Scale bars, 10 μm.and
Meiotic chromosome configuration in IR64 ×Ol
The course of meiosis in IR64 ×is well organized, with no apparent irregularities in chromosome pairing and segregation (Fig. 2). The hybrid IR64 ×has 24 chromosomes; three have 45S rDNA sites (Fig. 2-A). At pachytene stage, 24 chromosomes fully synapsed to form 12 bivalents (Fig. 2-B). The bivalent configuration in the pachytene stage was held through diakinesis (Fig. 2-C) to metaphase I (Fig. 2-D). Three 45S rDNA sites were detected on two bivalents: one has two 45S rDNA-FISH signals and the other is heteromorphic with only one signal (Fig. 2-C and -D). At anaphase I, two homologous chromosomes in each bivalent segregated and moved to opposite poles; chromosomes were equally pulled to two daughter cells, one having two 45S rDNA sites and one having only one 45S rDNA site (Fig. 2-E and -F). Chromosome movement and segregation at metaphase II (Fig. 2-F) and anaphase II (Fig. 2-G and -H) were regular to produce tetrads at the end of meiosis. In most tetrads, two of the microspores had two 45S rDNA signals and the other two had one 45S rDNA site (Fig. 2-I), as compared with the somatic nuclei of anther, which have three 45S rDNA sites (Fig. 2-J).
Meiotic chromosome configuration in Oa × Or
The hybridis an allotriploid with 36 chromosomes and showed irregular meiosis (Fig. 3). At diakinesis, chromosomes presented mostly as univalents, with a few as bivalents and trivalents (Fig. 3-A). In the survey of 30 nuclei at diakinesis, the number of univalents, bivalents and trivalents in a PMC were in ranges of 17 to 30, 2 to 8 and 0 to 4, respectively. Across all observed PMCs, the mean chromosome configuration was (24.30 ± 3.88) univalents, (4.40 ±1.63) bivalents, and (0.97 ±1.20) trivalents, respectively. At metaphase I, most univalent chromosomes were scattered irregularly throughout the cell, whereas bivalent and trivalent chromosomes were oriented on the metaphase I plate. However, the chromosomes in some bivalents were precociously separated to opposite poles (Fig. 3-B). Typical anaphase showing the separation and opposite movement of paired chromosomes was rarely observed inDuring metaphase I to anaphase I, some chromosomes were aggregated at the metaphase plate and some were pulled in opposite directions (Fig. 3-C and -D). At the end of meiosis I, most of the chromosomes reached the opposite poles to form dyad nuclei (Fig. 3-E). From metaphase II to anaphase II, the chromosome numbers in individual daughter cells were highly irregular (Fig. 3-F). At the end of meiosis, a single PMC produced four microspores (tetrad) accompanied by several micronuclei. The nuclei of each microspore varied in size (Fig. 3-G and -J).
GISH with the gDNA probe derived fromandallowed for discriminating the origin of chromosomes in×(Fig. 4). At metaphase, 45S rDNA sites were detected on four chromosomes, one from the E genome and three others from the HJ genome (Fig. 4-A). The meiotic disturbance found in×is attributed to asynapsis and desynapsis. At pachytene stage, most of the chromosomes seemed to be univalent; a few chromosomes were closely paired along their total length or with an unpaired region in some cases (Fig. 4-B and -C). GISH signals on pachytene chromosomes indicated that those bivalents may contain chromosomes derived from different genomes (allosyndesis) (Fig. 4-C). According to the GISH signals on chromosomes at diakinesis (Fig. 4-D) and metaphase I (Fig. 4-E), three types of chromosomal associations could be identified:,and. Among 297 bivalents, 23 (7.74%) wereassociation, 153 (51.52%) wereassociation and 121 (40.7%) wereassociation. The number of chromosomes allocated to each pole was highly irregular (Fig. 4-F). In some cases, GISH signals also indicated that two chromatids of some univalents precociously separated at metaphase I (Fig. 4-G). For example, the signals of 45S rDNA indicated four chromosomes with 45S rDNA sites present as univalents. Two moved to one pole and one to the other pole, whereas chromatids of the last one at the middle plate were precociously separated (Fig. 4-G). Laggards and chromosome bridges were commonly observed at telophase I (Fig. 4-H and -I). As indicated by GISH signals, irregular segregation resulted in uneven distribution of chromosomes at each pole at metaphase II (Fig. 4-J), anaphase II (Fig. 4-K) and telophase II (Fig. 4-L). GISH results also showed recombination chromosomes (Fig. 4-J to -L).
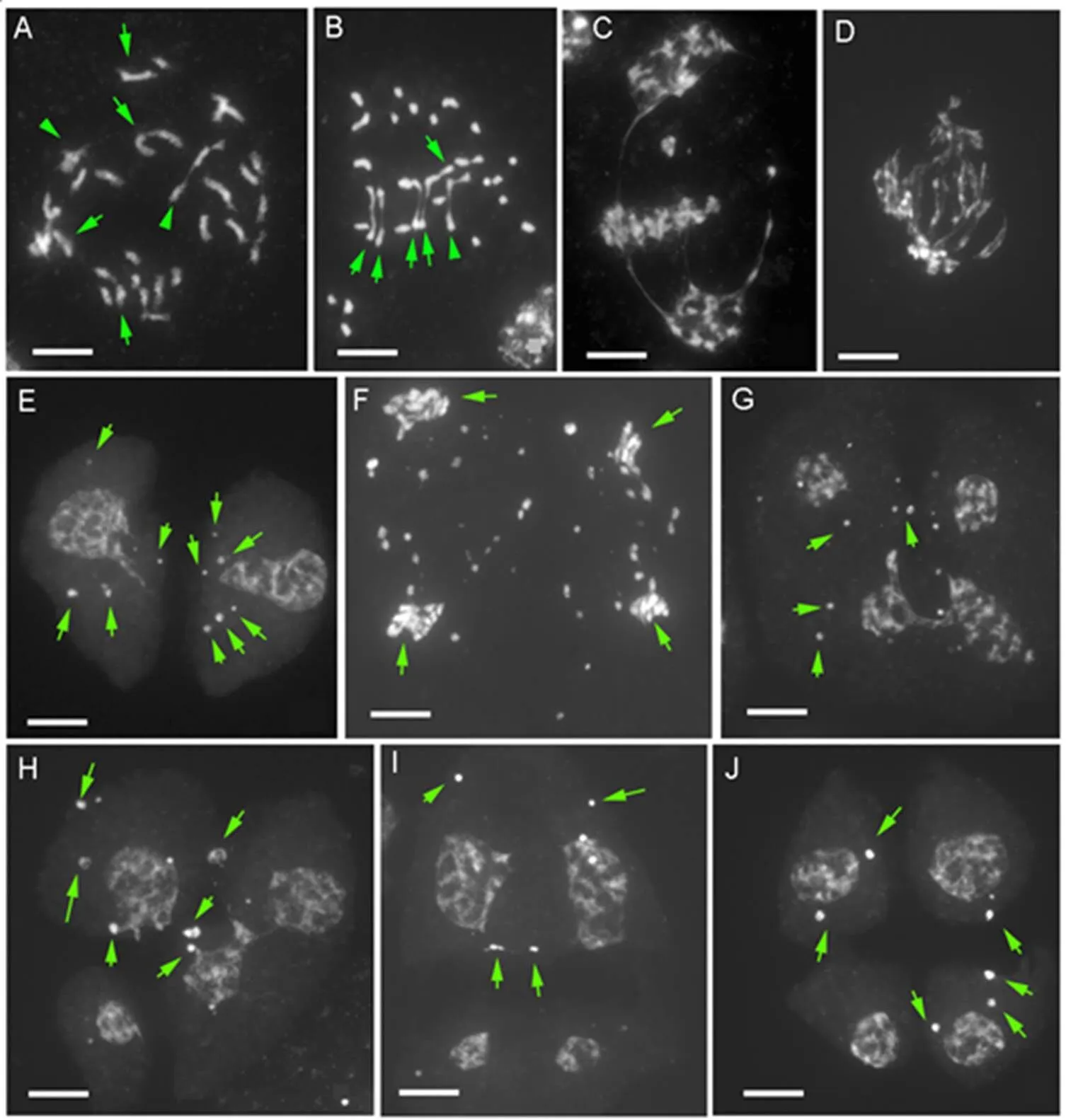
Fig. 3. Meiotic irregularities in pollen mother cells of×(×).
A, Diakinesis showing mostly univalents and a low frequency of bivalents (Arrow) and trivalents (Arrowhead).B, Metaphase I showing 23 univalents, 5 bivalents (Arrow) and trivalent (Arrowhead). C and D, Metaphase I to anaphase I showing irregular separation of chromosomes with some chromosomes aggregated at the metaphase plate and some pulled in opposite directions. E, Dyad nuclei with several chromosomes not being included (Arrow). F, Anaphase II showing a high variable number of chromosomes in each pole (Arrow) and many lagging chromosomes. G–J, Tetrad with four microspores having nuclei in varied sizes and several micronuclei (Arrow). Scale bars, 10 μm.
Meiotic chromosome configuration in Om × Oa
The hybrid×is an allotriploid with 36 chromosomes. Dual-color GISH allowed for discriminating three different genomes (Fig. 5). GISH with two genomic DNA probes ofand(CC type) clearly indicated 12 chromosomes from the EE genome and 12 chromosomes belonging to CC genomes, and the rest without GISH signals belonging to the BB genome (Fig. 5-A). Four univalents presented 45S rDNA-FISH signals, one from the E genome and three from the BBCC genome (Fig. 5-B). Similar meiotic disturbances in×were also observed in×. In the survey of 30 nuclei at diakinesis, the number of univalents, bivalents and trivalents in a PMC were in the ranges of 14 to 27, 1 to 8 and 0 to 5, respectively (Table 2). Across all cells, the mean chromosome configuration was (20.67 ± 3.75) univalents, (4.37 ±1.75) bivalents and (2.20 ±1.19)trivalents. At metaphase I, most chromosomes were scattered irregularly throughout the cell, and some associated chromosomes were ready to separate and head to polar opposites (Fig. 5-C to -F). Among these bivalents/trivalents, chromosomal associations included allosyndesis (BC, BE, CE and CBE) and autosyndesis (BB, CC and EE) (Fig. 5-D to -F). Among 226 bivalents, 180 (79.65%) were allosyndetic and 46 (20.35%)were autosyndetic. Allosyndetic associations included 62 BC, 64 BE and 54 CE. Autosyndetic associations were 14 BB, 16 CC and 16 EE. Highly meiotic irregularities, including laggards, bridges and uneven distribution, were observed throughout meiosis (Fig. 5-G to -L).
Fig. 4. GISH and 45S rDNA-FISH analysis of meiotic chromosome pairings in×(×).
A, Mitotic metaphase: GISH distinguishing chromosomes of the EE genome (Red) from those of the HHJJ genome (Blue). Four 45S rDNA sites (Green) were detected, one from the EE genome and three from the HHJJ genome. B, Pachytene: Showing mostly univalents (Arrow) and a few closely paired bivalents (Arrowhead). Red arrow indicates an unpaired region of a bivalent. C, Pachytene: GISH indicating allosyndetic pairing (Arrowhead) between the EE genome (Red) and HHJJ genome (Blue). Arrows indicate some univalent. D, Diakinesis: GISH showing univalents, bivalents and trivalents. Arrows indicate autosyndetic bivalents, and arrowheads indicate allosyndetic bivalents and trivalents. EE genome and HHJJ genome are indicated in red and blue, respectively.E, Metaphase I: GISH revealing three types of chromosomal associations [i.e.,-(Green-Green),-(Green-Red), and-(Red-Red)]. EE genome and HHJJ genome are indicatd in red and blue, while trivalent is indicated by arrow, respectively.F, Anaphase I: GISH showing irregular and unequal allocation of chromosomes at opposite poles. EE genome and HHJJ genome are indicated in red and blue, respectively. G, Metaphase-anaphase I: GISH signals and 45S rDNA FISH signals showing most chromosomes aggregated at a metaphase plate, a few chromosomes moved to opposite poles (Arrowheads), and precocious separation of chromatids (Arrows). EE genome and 45S rDNA are indicated in red and green, respectively. H, Telophase I: GISH showing two daughter nuclei with different chromosome composition and different origin of laggards (Arrow). EE genome and HHJJ genome are indicated in red and green, respectively.I, Telophase I: GISH and FISH showing irregular and unequal allocation of chromosomes at opposite poles. Arrow indicates the chromosome bridge. EE genome and 45S rDNA are indicated in red and green, respectively.J, Metaphase II: GISH and FISH showing different chromosome number and composition at each metaphase plate. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively.K, Anaphase II: GISH and FISH indicating chromatid segregation and different chromosome number and composition at each pole. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively.L, Telophase II: GISH and FISH indicating different number and composition of chromosomes in each pole and all with peripheral micronuclei. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively. FISH, Fluorescencehybridization; GISH, Genomichybridization. Scale bars, 10 μm.

Fig. 5. GISH and 45S rDNA-FISH analysis of meiotic chromosome pairings in×(×; 2= 36, BCE genome).
A, Mitotic metaphase: GISH signals showing 12 chromosomes from the EE genome (Red) and 12 from the CC genome (Green), and the remaining chromosomes from the BB genome (Blue). B, Diakinesis: GISH and 45S rDNA-FISH signals indicating mostly univalents, four with 45S rDNA sites (Green) including one from the EE genome (Red) and the others from the BBCC genome (Blue). Arrow indicates bivalent.C, Metaphase I: GISH and 45S rDNA-FISH signals indicating mostly univalents scattered irregularly throughout the cell. Arrow indicates bivalent and arrowheads indicate trivalent. EE genome and 45S rDNA are indicated in red and green, respectively. D–F, Metaphase I: GISH signals demonstrating allosyndetic (Arrows) and autosyndetic (Arrowhead) pairing. Green signals indicate chromosomes of CC genome; red signals indicate BB genome in D and EE genome in E and F.G–I, Metaphase I to anaphase I: GISH and 45S rDNA- FISH showing uneven allocation, lagging chromosomes, bridge (Arrow) and precociously separated chromatids (Arrowhead). Red signals indicate chromosomes from the EE genome and green signals from the BBCC genome in G and H and 45S rDNA sites in I. J–L, GISH and 45S rDNA-FISH showing highly meiotic irregularities, including laggards and uneven allocation of chromosomes. J, Telophase I; K, Metaphase II; L, Telophase II. Red signals indicate chromosomes from the EE genome, green signals from 45S rDNA sites in J and K, and CC genome in L. FISH, Fluorescencehybridization; GISH, Genomichybridization. Scale bars, 10 μm.
Discussion
In this study, we used GISH to visualize chromosome behaviors through meiosis in two triploid interspecific F1hybrids. We provided cytogenetic information to evaluate the genomic affinity and the possibility of genetic recombination in these hybrids. Extremely low fertility or even full sterility is the major problem in adopting wide hybridization for rice breeding programs (Khush and Brar, 1988). The diverse genetic backgrounds of the parental accessions may also give various seed setting (0–26.5%) in combinations of interspecific hybrid ofspecies (Sitch et al, 1989; Sitch, 1990). For the hybrid IR64 ×, meiosis was normal without apparent irregularities, producing mainly normal tetrads (Fig. 2) and finally forming a considerable number of viable pollen (57.8%) (Fig. 1-A and -B); however, the hybrid was almost sterile, with a seed setting of 0.8%. In the case of IR64 ×, high sterility seemed to be regulated by a complex genetic model. Similar results were reported in other interspecific hybrids ofspecies with the AA genome (Lu et al, 1998). Two genetic models, ‘double recessive gamete lethal’ (Oka, 1974) and ‘a(chǎn)llelic interaction’ (Ikehashi and Araki, 2008; Zhao et al, 2011), are considered the underlying mechanisms for sterility in interspecific hybrids ofspecies with the AA genome. More than 50 loci have been identified for controlling female gamete sterility or male gamete sterility (Ouyang et al, 2010).
In comparison to IR64 ×, two other interspecific hybrids were fully sterile, which absolutely resulted from meiotic irregularities due to the triploid nature and low affinities between parental genomes (Figs. 3, 4 and 5). Our cytogenetic evidence confirmed that these two hybrids were successful interspecific hybrids betweenand(Fig. 4-C to -F, -H), and betweenand(Fig. 5-A, -D to -H), and these two hybrids were allotriploid with 2=36. Several MAALs with alien chromosomes of tetraploid species in a cultivated rice background were developed after producing allotriploid hybrids betweenand tetraploid species (Jena and Khush, 1989; Jena et al, 1991). The two hybrids,×and×, showing vigor in reproductive growth, are potential genetic resources in future breeding programs to develop MAALs or introgression lines with alien chromosomes in the background ofif viable backcrosses can be recovered even from triploid hybrids.

Table 1. Chromosome associations in pollen mother cells (PMCs) at diakinesis in hybrids O. australiensis (Oa)× O. ridleyi (Or) and O. minuta (Om).
Based on a survey of 30 PMCs at diakinesis. Data are Mean ± SD (= 30). Values in the parentheses are the range of chromosome association in observed PMCs.
The 45S rDNA-FISH experiments provide interesting cytological evidence of genome diversity amongspecies. In the hybrid, four chromosomes had 45S rDNA sites, one from the EE genome and the others from the HJ genome (Fig. 3-A). In the hybrid, the four chromosomes with 45S rDNA sites presented as univalents (Fig. 4-Band -C), which suggested less homology between these chromosomes despite having 45S rDNAs in common. The 45S rDNA sites have been reported to be variable in number and location amongspecies (Chung et al, 2008). For example,has five pairs of 45S rDNA sites, including one on chromosome 4 (BB genome), two on chromosome 9 (BB and CC genome) and the other two on chromosome 10 (BB and CC genome), whereashas only one pair of 45S rDNA sites on chromosome 9 (Chung et al, 2008). Therefore, six 45S rDNA sites are expected in the hybrid×; however, two 45S rDNA sites were undetected in this hybrid due to FISH efficiency or genomic modification following interspecific hybridization (Chung et al, 2008). The number and distribution of 45S rDNA signal are helpful markers for identifying univalent from bivalent. As shown in Fig. 4-G, fluorescent signals indicated that four chromosomes with 45S rDNA sites presented as univalent, three of them at different poles and the one left at middle plate showing precocious separation of chromatids.
During meiosis of these two hybrids, more than half of the chromosomes present as univalents (Table 1), which suggests low affinity between parental genomes in these two hybrids. Despite less homoeology between the parental genomes of individual hybrids, bivalents and trivalents at low frequency were occasionally observed (Table 1). Homologous chromosome pairing (synopsis) at prophase ensures the sequence of normal meiotic events. Conventionally, the numbers of bivalent, univalent and multivalent chromosomes are the direct index of the genomic affinities between parental genomes in a hybrid. The diversities in chromosome number, structure and homology of parents of interspecific hybrids may cause irregular chromosome behaviors during meiosis, which results in unbalanced gametes and low fertility or even sterility. In wide hybrids, chromosome association can be autosyndetic and allosyndetic. Allosyndetic recombination following wide hybridization is an important process to obtain alien DNA introgression into cultivars. However, distinguishing allosyndesis from autosyndesis by traditional meiotic analysis is difficult. GISH is able to discriminate the parental origin of chromosomes in hybrids, which provides a powerful method to visualize different patterns of chromosomal associations among genomes in wide hybrids. GISH has played an excellent role in studying the mechanism for genome evolution within thecomplex and in assisting thebreeding program (Kopecky et al, 2008).
GISH has been applied for genome discrimination, genome evolution, genome phylogeny, micronuclei origin in MAALs and genome recombination in rice. However, these studies mainly used GISH with somatic chromosomes; relatively fewer reports have concerned GISH analyses of meiotic chromosomes ofspecies. GISH results identified that the univalents in MAALs (2+1) and in double MAALs (2+1+1) originated from(Hue et al, 2003). The autosyndetic and allosyndetic pairing could be differentiated in the F1hybrids×(Abbasi et al, 1999, 2009),×(FF)(Abbasi et al, 2010),×(CC) (Yan et al, 1999),×(HHJJ) and×(GG) (Xiong et al, 2006).
PMCs at meiotic stages are advantageous to investigate chromosome behaviors of interspecific hybrids by GISH. A single anther contains numerous PMCs at almost synchronized division stage, but a low mitotic index is always a problem in using somatic tissue for preparing chromosome slides. As we show here, GISH analysis of chromosome samples prepared by using an enzymatic maceration and flame-drying method provided reproductive and informative images. The differentiation efficiency of GISH mainly depends on the components and organization of dispersed repetitive sequences on chromosomes of individual genomes (Anamthawat-Jonsson and Reader, 1995). For better differentiation, excess unlabeled DNA from another species is usually used as a blocking agent in the hybridization mixture (Anamthawat-Jonsson et al, 1990). As shown in this study, GISH allowed for clearly distinguishing the parental genomes inandwithout using blocking DNA (Figs. 3 and 4), which suggests a high divergence among thesespecies. The karyotypes ofspecies, including diploids (2= 24) and tetraploids (2= 48), show high similarity, which implies that few large-scale chromosomal rearrangements occurred during speciation in thegenus (Chung et al, 2008). Instead, selective amplification and elimination of transposable elements (TEs) play main roles in the evolutionary stories ofspecies (Uozu et al, 1997). The chromosome-level reference assemblies for 13 reference genomes spanning thespecies tree confirmed that the genome assemblies contain 27% to 50% repeats, including TEs; both amplification of lineage-specific TEs and deletion of TE-related sequences drove genome dynamics with a very high rate of turnover during the evolution ofspecies (Stein et al, 2018).
GISH results clearly indicated the origin of each chromosome involved in bivalents and trivalents (Figs. 4 and 5), which showed the occurrence of both autosyndetic pairing (BB, CC, EE and HJHJ) and allosyndetic pairing (BC, BE, CE, EHJ and BCE) amonggenomes. Among these bivalents, more than half involved allosyndetic association, which suggested the occurrence of recombination events and the possibility of gene transfer. The most important GISH results indicated the occurrence of recombination between chromosomes of allosyndetic pairing bivalents in the hybrid×(Fig. 4-J to -L). GISH analysis showed that chromosomes of respective parental genomes were unequally allocated to each pole after anaphase I (Figs. 3-I to -K,and 4-G to -K) and showed unequal distribution of parental genomes in the meiotic end-products (Figs. 4-L and 5-L). Therefore, the end-products of meiosis in these two hybrids predictably contained an unbalanced and incomplete complement of chromosomes. Noteworthily, GISH results indicated that each daughter nucleus of the end-products of meiosis in these two hybrids retrieved more or less genetic materials from both parental genomes. Generally, univalents are scattered irregularly throughout the cell, and they may be randomly transmitted to daughter nuclei or lag behind and finally eliminated (Kalinka et al, 2010; Xie et al, 2013). Therefore, the existence of univalents may lead to producing microspores with unbalanced chromosome composition, which ultimately reduces functional pollen and seed setting (Reddi and Rao, 2000). Multivalent and chromosomal bridge findings (Figs. 3, 4 and 5) suggested the existence of structural changes among these chromosomes of different parental genomes. Together with the elimination of laggards and micronuclei and the unequal distribution, haploid gametes could be formed that contain parts of both parental genomes for a chance to produce progenies with introgression by sequential backcrossing. GISH analysis of meiotic chromosomes can be an efficient and convenient tool in rice breeding programs to assist in detecting alien chromosomes or chromosome fragment transfers in interspecific hybrids and introgression progenies.
Our GISH results detailed the chromosome behaviors during meiosis in PMCs of triploid interspecific hybrids, which indicates that irregular chromosome association and segregation could be the main reasons for sterility in wide hybrids. The results also showedthe genomic affinities and revealed frequent allosyndetic pairing betweengenomes, which indicates possible recombination and introgression. Together with GISH, FISH-mapping of more chromosome-specific markers such as 45S rDNA would be helpful to reveal meiotic disturbance of interspecific hybrids. A better understanding of genomic affinities of the distant species can assist in planning an effective breeding program for introducing agronomically important genes from alien species to cultivars by wide hybridization. However, GISH is still a challenge in rice breeding programs for reaching a similar performance as inbecause rice has small chromosomes and lacks available markers for chromosome identification. Precision of the introgressive hybridization programs in rice can be further enhanced by incorporating marker-assisted selection and backcross breeding with physical mapping of the alien introgression with minimum linkage drag by using molecular cytogenetic tools such as FISH and GISH.
Methods
Plant materials
Two F1hybrids were×and×. Table 2 shows the parental species and the chromosome number of interspecific hybrids investigated in this study.species used for genomic DNA probes in GISH are also listed in Table 2. These F1plants, although sterile, are perennial and vigorous, having been maintained in the greenhouse of Academia Sinica, Nankang, Taipei, China, for some years.
Pollen viability test by Alexander staining
Non-dehiscent anthers in flowers about to open were collected and fixed in Farmer’s solution (95% ethanol: acetic acid=3:1) overnight. Anthers were placed on a microscope slide with a drop of water for 5 min; the liquid was removed and replaced with 2 to 4 drops of Alexander staining solution (Dupláková et al, 2016). Anthers were gently chopped by using fine forceps to release pollen. A coverslip was placed over the sample for incubation in the dark for 60 min. Slides were examined under a microscope (Axioplan, Carl Zeiss AG, Germany), and micrographs were taken by using a CCD camera (Axiocam ICc 1, Carl Zeiss) and edited with Adobe Photoshop CS5 (Adobe Systems Inc., CA, USA).
Chromosome preparation
For preparing meiotic chromosome samples, immature panicles, with the flag leaf collar extending 10 cm above the last leaf collar, were harvested and fixed in freshly prepared Farmer’s solution at room temperature overnight. Fixed samples were replaced in fresh fixative and stored at -20 oC before use. For preparing mitotic chromosome samples, newly emerged root tips were excised and ?xed as the meiotic sample after pretreatment with 2 mmol/L 8-hydroxyquinoline at 18 oC for 2 to 3 h. Chromosome samples were prepared by using an enzymatic maceration and flame-drying method (Chung, 2015). The maceration enzyme mixture contained 2% cellulose (Onozuka R-10; Yakult Honsha, Tokyo, Japan) and 2% pectinase (Sigma Chemical Co., St. Louis, MO, USA) in 10 mmol/L citrate buffer (pH 4.5). Slides with good chromosome spreads, which retained all chromosomes with less overlap, were selected for FISH and GISH experiments.
Fluorescence in situ hybridization (FISH) and genome in situ hybridization (GISH)
FISH and GISH were performed as described by Chung (2015). For detecting the 45S rDNA sites, a fragment of partial 18S rDNA region (1.6 kb) from total genomic DNA (gDNA) of IR64was amplified and labeled with digoxigenin-11-dUTP (Roche Diagnostics, GmbH, Mannheim, Germany) by PCR (Primer 1: 5-CGAACTGTGAAACTGCGAATGGC-3; and Primer 2: 5-TAGGAGCGACGGGCGGTGTG-3) (Chang et al, 2010). Plasmid pRCS2 containing centromeric repeats (CentO, 155 bp) ofspecies (Dong et al, 1998) was used to indicate the centromere position on chromosomes. Total genomic DNA ofspecies was isolated with the CTAB extraction method. Before labeling for probes, genomic DNA was broken into short fragments (2–10 kb) by boiling for 30 to 60 min. Plasmid DNA and genomic DNA were labeled with biotin-16-dUTP (Roche Diagnostics, GmbH, Mannheim, Germany) or digoxigenin-11-dUTP (Roche Diagnostics, GmbH, Mannheim, Germany) by nick translation according to the standard protocol.
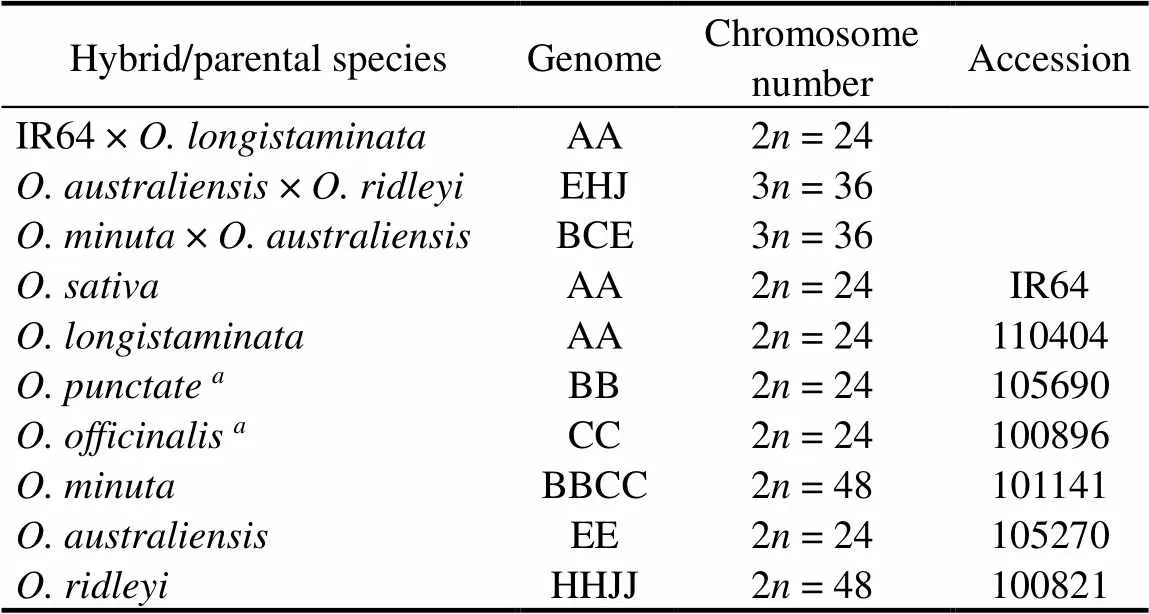
Table 2. Chromosome number and genomic composition of interspecific hybrids and their parental species.
Genomic DNA was labeled as probes in genomichybridization.
Chromosomes were denatured with 70% formamide in saline sodium citrate buffer (2× SSC: 300 mmol/L sodium citrate and 30 mmol/L NaCl, pH 7.0) at 80oC for 90 s, dehydrated in ice-cooled 70%, 90% and 100% ethanol for 5 min each, then air-dried. The hybridization mixture, which contained 50% deionized formamide, 2× SSC, sheared salmon sperm DNA (1 μg/μL), 10% dextran sulfate and probe DNA (10–100 ng), was denatured at 80oC for 10 min and immediately quenched on ice for 5 min. An aliquot of 20 μL hybridization mixture was dropped onto each slide, covered with a coverslip, and incubated in a moisture box at 37oC overnight. The post-hybridization washing, signal detection and image processing were performed as described (Chung, 2015). The meiotic stages are characterized by the chromosome morphology described for normal meiosis of rice (Chen et al, 2005).
Acknowledgements
This work was funded by the Institute of Plant and Microbial Biology, Academia Sinica, and Ministry of Science and Technology, Taiwan, China (Grant No. MOST103-2313-B-001- 001). We sincerely thank Dr. Darshan Brar at International Rice Research Institute (IRRI) for providing plant materials and encouragements.
Abbasi F M, Brar D S, Carpena A L, Fukui K, Khush G S. 1999. Detection of autogenetic and allosyndetic pairing among A and E genomes ofthrough genomichybridization., 16:24–25.
Abbasi F M, Ahmad H, Sajid M, Inamullah, Brar D S. 2009. Detecting nature of chromosome pairing in A and E genomes of.,74: 229–233.
Abbasi F M, Shah A H, Perveen F, Afzal M, Sajid M, Masood R, Nawaz F. 2010. Genomic affinity betweenandas revealed byhybridization and chromosome pairing., 9: 3068–3072.
Amante-Bordeos A, Sitch L A, Nelson R, Damacio R D, Oliva N P, Aswidinnoor H, Leung H. 1992.Transfer of bacterial blight and blast resistance from the tetraploid wild riceto cultivated rice,., 84: 345–354.
Ammiraju J S S, Luo M Z, Goicoechea J L, Wang W M, Kudrna D, Mueller C, Talag J, Kim H, Sisneros N B, Blackmon B, Fang E, Tomkins J B, Brar D, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith D W, Arumuganathan K, Rao K, Walling J G, Gill N, Yu Y, San Miguel P, Soderlund C, Jackson S, Wing R A. 2006. Thebacterial artificial chromosome library resource: Construction and analysis of 12 deep coverage large-insert BAC libraries that represent the 10 genome types of the genus., 16:140–147.
Anamthawat-Jonsson K, Schwarzacher T, Leitch A R, Bennett M D, Heslop-Harrison J S. 1990. Discrimination between closely relatedspecies using genomic DNA as a probe., 79(6): 721–728.
Anamthawat-Jonsson K, Reader S M. 1995. Preannealing of total genomic DNA probes for simultaneous genomic in situ hybridization.,38(8): 814–816.
Angeles-Shim R B, Vinarao R B, Marathi B, Jena K K. 2014. Molecular analysis ofDesv. (CCDD genome)-derived introgression lines and identification of value-added traits for rice (L.) improvement., 105(5): 676–689.
Baghyalakshmi K, Jeyaprakash P, Ramchander S, Radhamani T, Raveendran M. 2018. Comparative study on backcross inbred lines of IR64 rice (L.) introgressed with drought QTLs under varied moisture regimes over different seasons.,7(1): 2716–2725.
Brar D S, Elloran R, Khush G S. 1991. Interspecific hybrids produced through embryo rescue between cultivated and eight wild species of rice., 8:91–93.
Brar D S, Khush G S. 1995. Wide hybridization for enhancing resistance to biotic and abiotic stresses in rainfed lowland rice.: Fragile Lives in Fragile Ecosystems. Manila, the Philippines: International Rice Research Institute: 901–910.
Brar D S, Khush G S. 1997. Alien introgression in rice.,35: 35–47.
Chang K D, Fang S A, Chang F C, Chung M C. 2010. Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distantspecies., 96:181–190.
Chen C B, Xu Y Y, Ma H, Chong K. 2005. Cell biological characterization of male meiosis and pollen development in rice., 47: 734?744.
Chung M C, Lee Y I, Cheng Y Y, Chou Y J, Lu C F. 2008. Chromosomal polymorphism of ribosomal genes in the genus., 116:745–753.
Chung M C. 2015. Chromosome techniques and FISH.: Yeung ECT,Stasolla C, Summer M J, Huang B Q. Plant microtechniques and protocols. Switzerland: Springer International Publishing: 287–309.
Dawe R K. 1998. Meiotic chromosome organization and segregation in plants., 49:371–395.
Dong F, Miller J T, Jackson S A, Wang G L, Ronald P C, Jiang J. 1998. Rice () centromeric regions consist of complex DNA., 95: 8135–8140.
Dupl'áková N, Dobrev P I, Reňák D, Honys D. 2016. Rapid separation ofmale gametophyte developmental stages using a Percoll gradient., 11: 1817–1832.
Hue N T N, Ram T, Barrion A A, Brar D S. 2003. Characterization of monosomic alien addition lines ofthrough genomichybridization using meiotic chromosomes., 20:110–111.
Ikehashi H, Araki H. 2008. Genetics of F1sterility in remote crosses of rice.: Banta S. Rice Genetics I. Manila, the Philippines: IRRI: 119–130.
Iwata A, Gao D Y, Ohmido N, Jackson S A. 2014. Molecular cytogenetics of rice and its wild relatives.: Zhang Q F, Wing R A. Genetics and Genomics of Rice: Plant Genetics and Genomics: Crops and Models. New York, USA: Springer: 71–79.
Jauhar P P, Joppa L R. 1996. Chromosome pairing as a tool in genome analysis: Merits and limitations.: Jauhar PP. Methods of Genome Analysis in Plants. Boca Raton, FL: CRC Press: 9–37.
Jena K K, Khush G S. 1984. Embryo rescue of interspecific hybrids and its scope in rice improvement., 1:133–134.
Jena K K, Khush G S. 1989. Monosomic alien addition lines of rice: Production, morphology, cytology, and breeding behavior., 32(3): 449–455.
Jena K K, Multani D S, Khush G S. 1991. Monosomic alien addition lines ofand alien gene transfer.: Rice Genetics. II. Manila, the Philippines: International Rice Research Institute: 728.
Jena K K, Khush G S, Kochert G. 1992. RFLP analysis of rice (L.) introgression lines., 84: 608–616.
Jena K K. 2010. The species of the genusand transfer of useful genes from wild species into cultivated rice,.,60(5): 518–523.
Kalinka A, Achrem M, Rogalska S M. 2010. Cytomixis-like chromosomes/chromatin elimination from pollen mother cells (PMCs) in wheat-rye allopolyploids., 53:69–83.
Kalloo G. 1992. Utilization of wild species.: Kalloo G, Chowdhury J B. Distant Hybridization of Crop Plants. Berlin, Heidelberg, Germany: Springer: 149–167.
Khush G S, Brar D S. 1988. Wide hybridization in plant breeding.: Zakri AH. Plant Breeding and Genetic Engineering. SABRAO, Malaysia: 141–188.
Khush G S, Bacalangco E, Ogawa T. 1990. A new gene for resist to bacterial blight from.,7:121–122.
Kopecky D, Lukaszewski A J, Dole?el J. 2008. Cytogenetics of(hybrids)., 120:370–383.
Le H T, Armstrong K C, Miki B. 1989. Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe., 7: 150–158.
Lee Y I, Chang F C, Chung M C. 2011. Chromosome pairing affinities in interspecific hybrids reflect phylogenetic distances among lady’s slipper orchids ()., 108(1): 113–121.
Lu B R, Naredo M E B, Juliano A B, Jackson M T. 1998. Taxonomic status ofSteud. III. Assessment of genomic affinity among AA genome species from the New World, Asia, and Australia., 45: 215–223.
Mariam A L, Zakri A H, Mahani M C, Normah M N. 1996. Interspecific hybridization of cultivated rice,L. with the wild rice,Presl., 93:664–671.
Mason A S, Huteau V, Eber F, Coriton O, Yan G J, Nelson M N, Cowling W A, Chèvre A M. 2010. Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCABinterspecific hybrids., 18(6):655–666.
Multani D S, Jena K K, Brar D S, delos Reyes B G, Angeles E R, Khush G S. 1994. Development of monosomic alien addition lines and introgression of genes fromDomin. to cultivated riceL., 88:102–109.
Oka H I. 1974. Analysis of genes controlling F1sterility in rice by the use of isogenic lines., 77(3):521–534.
Ouyang Y D, Liu Y G, Zhang Q F. 2010. Hybrid sterility in plant: Stories from rice., 13(2):186–192.
Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Bra D S, Jackson S, Wing R A, Panaud O. 2006. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in, a wild relative of rice., 16(10):1262–1269.
Reddi T V V S, Rao D R M. 2000. Cytology of induced desynaptic mutants in rice., 65:35–41.
Sanchez P L, Wing R A, Brar D S. 2014. The wild relative of rice: Genomes and genomics.: Zhang Q F, Wing RA. Genetics and Genomics of Rice, Plant Genetics and Genomics: Crops and Models. New York, USA: Springer: 9–25.
Schwarzacher T, Leitch A R, Bennett M D, Heslop-Harrison J S. 1989.localization of parental genomes in a wide hybrid., 64: 315–324.
Sitch L A, Dalmacio R D, Romero G O. 1989. Crossability of wildspecies and their potential use for improvement of cultivated rice., 6: 58–60.
Sitch L A. 1990. Incompatibility barriers operating in crosses ofwith related species and genera.: Gustafson JP. Genetic Manipulation in Plant Improvement: II. New York, USA: Plenum Press: 77–93.
Stein J C, Yu Y, Copetti D, Zwickl D J, Zhang L, Zhang C J, Chougule K, Gao D Y, Iwata A, Goicoechea J L, Wei S R, Wang J, Liao Y, Wang M H, Jacquemin J, Becker C, Kudrna D, Zhang J W, Londono C E M, Song X, Lee S, Sanchez P, Zuccolo A, Ammiraju J S S, Talag J, Danowitz A, Rivera L F, Gschwend A R, Noutsos C, Wu C C, Kao S M, Zeng J W, Wei F J, Zhao Q, Feng Q, Baidouri M E, Carpentier MC, Lasserre E, Cooke R, da Rosa Farias D, da Maia L C, dos Santos R S, Nyberg K G, McNally K L, Mauleon R, Alexandrov N, Schmutz J, Flowers D, Fan C Z, Weigel D, Jena K K, Wicker T, Chen M S, Han B, Henry R, Hsing Y C, Kurata N, de Oliveira A C, Panaud O, Jackson S A, Machado C A, Sanderson M J, Long M Y, Ware D, Wing RA. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus., 50: 285–296.
Tikapunya T, Fox G P, Furtado A, Henry R J. 2016. Grain physical characteristic of the Australian wild rice., 15:1–12.
Tikapunya T, Zou W, Yu W W, Powell P O, Fox G P, Furtado A, Henry R J, Gilbert R G. 2017. Molecular structures and properties of starch of Australian wild rice., 172:213–222.
Uozu S, Ikehashi H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K. 1997. Repetitive sequences: Cause for variation in genome size and chromosome morphology in the genus., 35:791–799.
Vaughan D A. 1994. Wild Relatives of Rice: Genetic Resources Handbook. Los Banos, the Philippines: International Rice Research Institute.
Xie Q Z, Kang H N, Sparkes D L, Tao S T, Fan X B, Xu L H, Fan X Y, Sha L, Zhang H, Wang Y W, Zeng J, Zhou Y C. 2013. Mitotic and meiotic behavior of rye chromosomes in wheat:amphiploid ×progeny., 12(3):2537–2548.
Xiong Z Y, Tan G X, He G Y, He G C,Song Y C. 2006. Cytogenetic comparisons between A and G genomes inusing genomichybridization., 16:260–266.
Yan H H, Min S K, Zhu L H. 1999.Visualization ofchromosomes in intergenomic hybrid plants from×via fluorescenthybridization., 42(1): 48–51.
Zhao Z G, Zhu S S, Zhang Y H, Bian X F, Wang Y, Jiang L, Liu X, Chen L M, Liu S J, Zhang W W, Ikehashi H, Wan J M. 2011. Molecular analysis of an additional case of hybrid sterility in rice (L.)., 233:485–494.
Copyright ? 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2020.11.008
7 October 2019;
20 March 2020
Chung Mei-Chu (bomchung@gate.sinica.edu.tw)
(Managing Editor: Fang Hongmin)
- Rice Science的其它文章
- Different Hypotheses for Resistance Loss of Rice Varieties to Magnaporthe oryzae
- Development of Heat Tolerant Two-Line Hybrid Rice Restorer Line Carrying Dominant Locus of OsHTAS
- Identification of QTLs for Cadmium Tolerance During Seedling Stage and Validation of qCDSL1 in Rice
- Genetic and Geographic Patterns of Duplicate DPL Genes Causing Genetic Incompatibility Within Rice: Implications for Multiple Domestication Events in Rice
- Physiochemical Properties of Resistant Starch and Its Enhancement Approaches in Rice
- Anticancer Activities of Plant Secondary Metabolites: Rice Callus Suspension Culture as aNew Paradigm

