Genetic and Geographic Patterns of Duplicate DPL Genes Causing Genetic Incompatibility Within Rice: Implications for Multiple Domestication Events in Rice
XuXun,GeSong, ZhangFumin
Research Paper
Genetic and Geographic Patterns of DuplicateGenes Causing Genetic Incompatibility Within Rice: Implications for Multiple Domestication Events in Rice
XuXun1, 2,GeSong1, 2, ZhangFumin1, 2
(State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China)
Strong genetic incompatibilities exist between two primary rice subspecies,and. However, the wild ancestors of rice,Sharma et Shastry andGriff., are genetically compatible. How this genetic incompatibility became established has not been clearly elucidated. To provide insights into the process, we analyzed a pair of hybrid sterility genes in rice,() and(). Either of the two loci can have one defective allele (and). Hybrid pollen carrying bothandalleles is sterile. To explore the origination ofand, we sequenced theandgenes of 811 individual plants, including(132),(296) and(383). We then obtained 20and 34sequences offrom online databases. Using these sequences, we analyzed the genetic and geographic distribution patterns ofgenes in modern rice and its wild ancestors. Compared with the ancestral populations,andshowed reduced diversity but increased frequency in modern rice. We speculated that the diversity reduction was due to a historic genetic bottleneck, and the frequency had likely increased because the defective alleles were preferred following this artificial selection. Such results indicated that standing variances in ancestral lines can lead to severe incompatibilities among descendants. Haplotype analysis indicated that thehaplotype of rice emerged from anpopulation in India, whereas thehaplotype emerged fromin South China. Hence, the evolutionary history of DPLs conforms to the presumed multiple domestication events of modern rice.
rice;gene; domestication; genetic incompatibility; phylogeography
It is well known that strong genetic incompatibilityexists between two rice subspecies (L.),and.In contrast, two wild rice ancestors,Sharma et ShastryandGriff., are genetically compatible (Cai et al, 2019). Modernrice and these two wild ancestors constitute an excellent system for studying the evolutionarydevelopment of incompatible genes. The crossbreeding barriers between the two modern subspecies represent a major problem for ricecultivation.Deeper knowledge of theirevolutionary history therefore has substantial practical significance for improving rice cultivation.
The genetic incompatibility betweenandis a complex biological event with many loci involved (Ouyang et al, 2010; Wang et al, 2014; Xie et al, 2019), such as(Chen et al, 2008; Yang J Y et al, 2012)leading to endoplasmic reticulum stress;(Long et al, 2008),() (Mizuta et al, 2010) and(Shen et al, 2017), resultingin hybrid pollen sterility. Additional loci have been identified by genetic maps (Li et al, 2017). Although many loci have been identified, the evolutionary history of the development of overall incompatibility remains poorly understood.
In the present study, we utilized thesystem asa suitable framework for exploring this challenge. This system contains only two genes:() and(), both of which cause pollen sterility via reciprocal gene loss (Mizuta et al, 2010). Both genes of thesystem have two alleles:like(, nonfunctional) andlike(,functional),like(,functional) andlike(, nonfunctional). Theoretically, a quarter of all pollen in an F1hybrid cross betweenandwould have- andalleles and therefore fail to germinate.
The genetic distribution pattern ofin modernremains unknown, let alone the pattern in its wild ancestors. Modern rice includes six groups (Wang et al, 2014).can be divided into,,and, whereasconsists ofand. Previous reports have indicated thatis fixed inbut relatively rare in, whileis common in(Mizuta et al, 2010; Craig et al, 2014). Among wild rice strains,has been reported buthas not. The absence ofin wild rice strains indicates that the DPLsystemis established during domestication and not inherits from wild ancestors. Nevertheless, previous studies have lacked population sampling of wild rice strains. In addition, previous studies focused only on the major groups of(and), andandwere excluded.
In this study, we used thesystem to elucidatethe establishment process of genetic incompatibility betweenandand attempted to answer the following questions: 1) Does thesystem exist in wild rice strains? and 2) How does thesystem develop and what role has domestication played in this process?
RESULTS
Allele distribution
A total of 695and 787sequences were obtained. In addition, we collected 20sequences and 34sequences from online databases(Mizuta et al, 2010; Craig et al, 2014) (Table S1). Alignments with gaps produced lengths ofandare 1407 bp and 439 bp, respectively.
The frequency ofin(10.2%)was between those of(37.0%) and(5.0%) (Table 1).was present only in one wild rice strain (), with a much lower frequency(4.9%) overall than that in(30.4%). In contrast to previous studies (Mizuta et al, 2010; Craig et al, 2014), we identified bothandin wild rice strains, indicating that thesystem was established prior to the domestication of rice.
andexhibited clear frequency differences among rice groups. Only,andcarried, with frequencies of 70.6%, 3.0% and 4.8%, respectively.was detected in all the six groups of, with the highest frequency in(87.8%) and the lowest frequency inrice (4.8%) (Table 1). We speculated that thesystem more likely causes reproductive barriers between the two groups,and, rather than the two subspecies,and.
Phylogeography
The distribution of nonfunctional alleles among different populations confirmed the strong geographic nature ofand(Table 1).was found in bothandbut clustered largely inof South Asia (India and Nepal) and Southeast Asia (Thailand, Cambodia and Laos).was only present in twopopulations in southern China (GDGZ and GXHZ). Considering the geographic isolation between these strains, we speculated that the incompatible alleles failed to trigger hybrid fertility under natural conditions.
Theandnetworks (Figs. 1 and 2)contained 20 and 5 haplotypes, respectively. Thenetwork (Fig. 1-A) established that theallele inonly clustered in H_1, and the haplotype in rice had only one original source, the wild ancestors, which also contained H_1. These results stand in contrast to the multiple emergence hypothesis forpreviously suggested (Mizuta et al, 2010). The two most frequent haplotypes ofin rice (H_5 and H_36) were closely related with each other but separated from H_1, suggesting independent originations ofandin rice. Thehaplotype in wild rice strains showed discontinuous geographic distribution (Fig. 1-B). H_1 was present mostly infrom India, and also infrom Oceania. Previous studies have shown that South Asian countries play an important role in the domestication of rice (Kovach et al, 2007; Sweeney and McCouch, 2007; Vaughan et al, 2008). Therefore, we presumed that theofemerged in India. The reason underlying the discontinuous distribution of H_1 in wild rice strains remainedunclear. H_5 and H_36 were present in wildrice strains from almost every region, indicating thatwas established beforeand the two wild species diverged.

Table 1. Nucleotide polymorphism and results of neutral test in O. rufipogon, O. nivara and O. sativa.
, Number of segregating sites;, Nucleotide diversity;, Number of haplotypes;, Haplotype diversity. Sample names are composed of information of the species and their origins. Initial letters of sample names: n,; r,; s,. Origins of samples: IND, India; THA, Thailand; KHM, Cambodia; MMR, Myanmar; NEP, Nepal; LAO, Laos; LKA, Sri Lanka; GDGZ, Gaozhou, Guangdong Province, China; GXBH, Beihai, Guangxi Province, China; GXHZ, Hezhou, Guangxi Province, China; GXTD, Tiandong, Guangxi Province, China; HNCL, Chaling, Hunan Province, China; HNJY, Jiangyong, Hunan Province, China; HNDZ, Danzhou, Hainan Province, China; HNWC, Wenchang, Hainan Province, China; JXDX, Dongxiang, Jiangxi Province, China; YNJH, Jinghong, Yunnan Province, China; IDN, Indonesia; PNG, Papua New Guinea; ARO,rice; AUS,rice; INDI,rice; RAY,rice; TEJ,rice; TRJ,rice.*,< 0.05; **,< 0.02.
Thenetwork (Fig.2-A) indicated that allsequences in rice were concentrated in H_13 and theappeared in H_1 and H_5. Similar to, theandalleles were separated. We speculated thatand+ also originated separately in rice.H_13 included haplotypes from two adjoiningpopulations: rGDGZ and rGXHZ (Fig. 2-B). H_1 and H_5 included samples from all habitats of wild ancestor strains, indicating thathas likely existed in wild rice strains for a long time. Therefore, we speculated thatoriginated from rGDGZ and rGXHZ was subsequently inherited throughout rice.
In summary,andofare both immediate successor strains from wild ancestor strains that originated in geographically isolated areas.originated fromin India (nIND) whereasoriginated fromin South China (rGDGZ and rGXHZ). The definitive origination areas forand+ are still unclear because of their extensive geographic distributions.
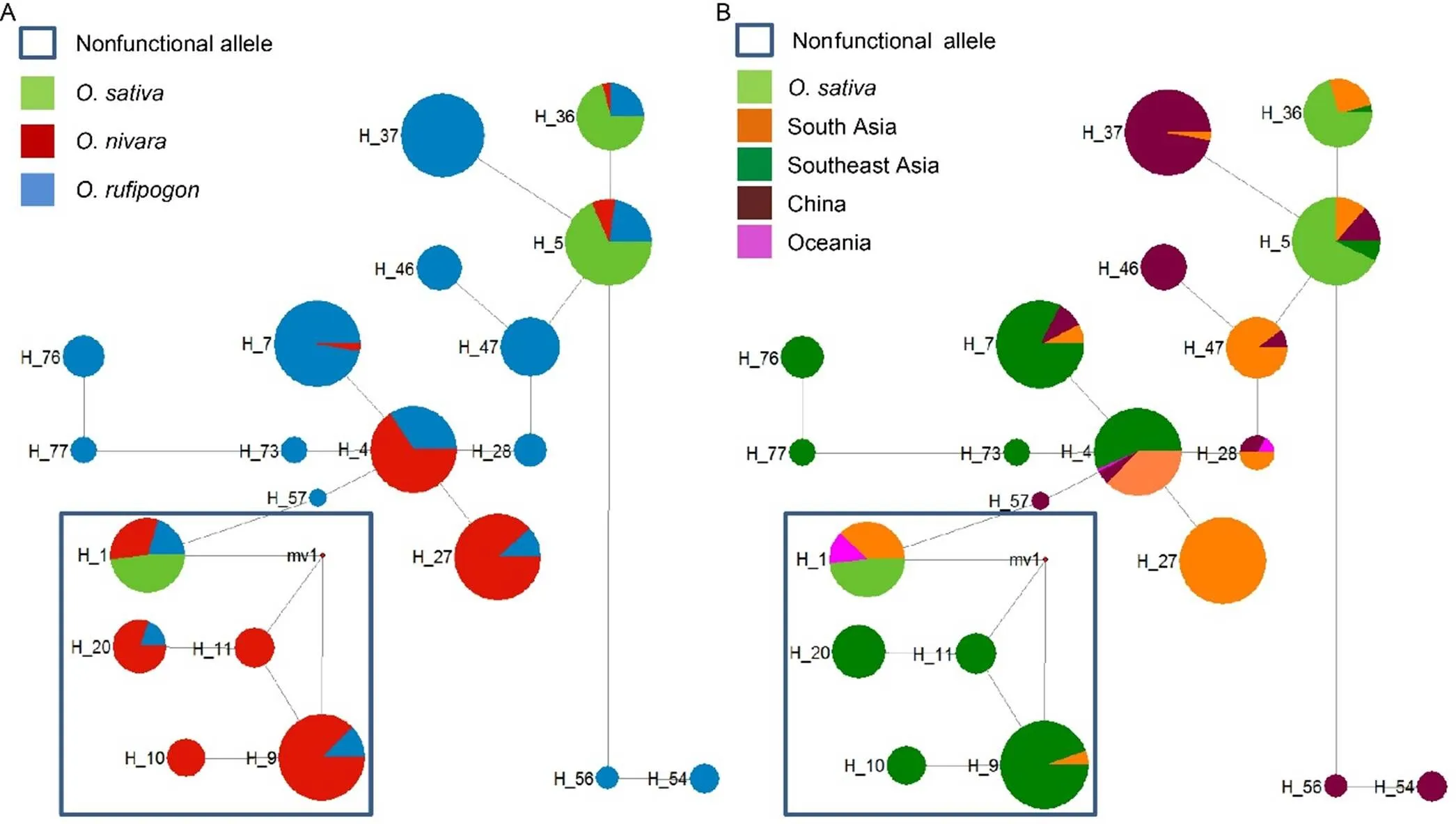
Fig. 1. Median-joining haplotype networks of.
Sections of each circle represent the proportion species (A) or locations (B) present in each haplotype. The size of each circle corresponds to the number of sequences of each haplotype. The lengths of the gray lines correspond to the number of mutations. Nonfunctional haplotypes are indicated by hollow squares.
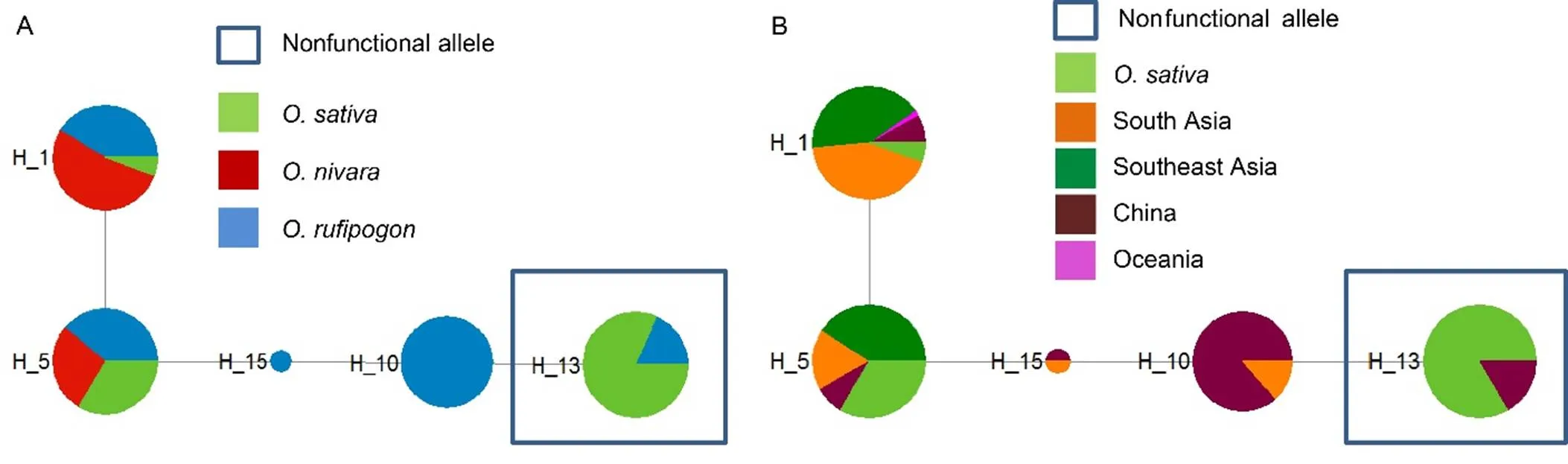
Fig. 2. Median-joining haplotype networks of.
Sections of each circle represent the proportion species (A) or locations (B) present in each haplotype. The size of each circle corresponds to the number of sequences of each haplotype. The lengths of the gray lines correspond to the number of mutations. Nonfunctional haplotypes are indicated by hollow squares.
Tests for selection
In addition to modern rice,was identified only in the two adjoiningpopulations (rGDGZ and rGXHZ), we therefore merged them into one ancestral population, rGDGX. We compared the frequencies ofandwith their ancestral populations (Table 1). The frequency ofinwas 70.6%, higher than that of nIND (45.0%). The frequency ofinwas 87.8%, which was also higher than that of rGDGX (41.2%). These results indicated that the frequencies ofandincreased after domestication, suggesting they might be preferred in the process of artificial selection.

Fig. 3. Ratios of nucleotide diversity () and haplotype diversity () between groups ofand ancestral populations.
AUS and TEJ indicateandrice, respectively. nIND and rGDGX indicate the ancestral populations:from India and the twopopulations carryingfrom South China. The ratio ofnucleotide diversity betweenand the ancestral populations is represented by a blank shape, sinceofhas no diversity.
We used the Tajima’sstatistic to assess whetherand/orunderwent selection, and if so, whether the selection differed among species, populations or groups (Table 1). In, significant and negative Tajima’svalues were detected in bothand. In,only the Tajima’sofwas significant negative. No significant resultwas tested in. At the population level, we found significantTajima’sononly in the populationrLAO1, and ononly in nLKA05. Although not significant, the values of Tajima’sfor most populations/groups were negative. One of the basic hypotheses of neutral tests is that the sample must be a population with internal gene flow (Tajima, 1989). However, the two wild species encompass many geographically isolated populations, and modern rice includes two subspecies that are severely incompatible.We speculated that the significance and lack of significant results of Tajima’stesting at the species level may be caused to this intraspecies structure. Methods other than neutral testing are likely required to understand the effects of artificial selection on rice.
An absence of polymorphic sites () was identified in several populations/groups (Table 1). This observationindicated possibly purifying selection at the population/ group level, as well as a divergence of selection within species.
We next estimated the nucleotide diversity () ofandin all the three species and each population/group (Table 1). In wild rice, we found that theofwas greater than that of(: 0.00336>0.00090;: 0.00421>0.00290). In modern rice, we found the opposite: theof(0.00118) was lower than that of(0.00363). After excluding recombined sequences, the number of recombined sequences and haplotype diversity () arealso listed in Table 1. Theofinwas 0.223, lower than that of(0.571). Inand, thevalues ofwere higher than those of. Therefore, we determined that artificial selection likely affected the genetic pattern ofin rice.
To further understand the effects of selection onand, we compared the nucleotide and haplotype diversities ofbetweenand the wild ancestral population nIND, then comparedbetween them. The same comparison was used forandbetweenand rGDGX. Theofcontained 12.37% nucleotide diversity and 72.43%haplotype diversity, but the number of haplotypes was equal to nIND (Fig.3). Theofshowed a more severe reduction in diversity, containing only one haplotype and no diversity (indicated via blanks in Fig. 3).andshowed marked reductions in diversity compared withand. In light of the high frequencies and low diversities, we speculated that the haplotypes ofandinwere randomly selected due to genetic bottleneck and the nonfunctional alleles were subsequently preferred by artificial selection, which led to their increase in frequencies in specific rice groups.
DISCUSSION
Domestication events have shaped DPL system derived from wild ancestor strains
The origin of the genetic incompatibility betweenandhas been unclear since the ancestor strainsandare compatible(Cai et al, 2019). In the present study, we wondered whether the incompatibility appeared during the process of domestication or whether the deficiency already existed in the ancestral strains.
To date, several major incompatible systems have been identified within rice:(Shen et al, 2017),(Chen et al, 2008; Yang J Y et al, 2012),(Mizuta et al, 2010) and(Long et al, 2008).Thesystem includes two alleles,typeandtype. High-expressedin sporophytic cells of hybrids leads to abortion of pollen carrying. Bothandare inherited respectively from diverged wild rice populations(Shen et al, 2017). However, the origin areas ofandare still unclear.Thesystem can be roughly classified into three types:like,like and wide compatibility.like andlike are incompatible, and the wide compatibility shows compatibility with both of the others(Du et al, 2011). All the three types are present in wild rice, which provides evidence of potential reproductive isolation (Du et al, 2011; Craig et al, 2014).causes hybrid pollen sterility and includes two adjacent genes,and.andcarryand, respectively.,andare necessary for pollen sterility and are present in wild rice (Long et al, 2008; Craig et al, 2014). These studies have established that several potential reproductive barriers betweenandare present in their ancestors. However, it is still unclear how the incompatible alleles have been maintained under natural conditions andthe influence of domestication. Ouyang and Zhang (2018) proposed three models to explain the establishment of incompatibilities within rice. The models focused on how the incompatible genes accumulated in each divergent lineage and may not be accurate for several loci due to the lack of clear evolutionary background information. For example, Ouyang and Zhang (2018) indicated thatdid not exist in wild rice strains and suggested that the incompatibility occurred during the divergence of the two subspecies.
Our study identified that bothandare present in wild rice. Therefore, the three major incompatibility systems are all inherited rather thanmutations that arose during domestication.Larger-scale studies involving more groups of rice are warranted. In a study including a typicaland two typical, 43 loci and 223 interactions involved in the fertility of embryo-sac, pollen and spikelet were identified (Li et al, 2017). The studyfailed to identifyandbutis consistent with our findings that thesystem was more likely found betweenandrice. We therefore hypothesized that the incompatibility betweenandmay contain a more complex genetic background than previously believed.
We found that domestication had affected the genetic background ofandin rice. After determining that theofwas severely reduced, we surprisingly found that the frequency ofwas much higher in(87.8%) than the ancestral population rGDGX (41.2%). Similarly, the frequency ofwas 45.0% in nIND, which then increased to 70.6% in, although theofwas reduced in(Fig. 3). These results indicated thatandwere preferred by artificial selection.
Significant and negative Tajima’svalues are a sign of extremely low polymorphisms and are considered a consequence of purifying selection, but population expansions or unknown structures can also lead to significant negative changes (Tajima, 1989). Neutral tests are generally designed for one population with fluid gene flow and are generally not suitable for widespread species.In the wild ancestors, we found significant and negative results of neutral testing. Considering the complex structures ofand(Londo et al, 2006; Liu et al, 2015), the significance was more likely caused by the intraspecies structure rather than selection. A similar phenomenon has been reported for: the neutral test results are also significantly negative in one wild ancestor () but not in(Du et al, 2011). We speculated this may also be due to the interspecies structure of the wild species, and detailed studies among the groups ofare warranted to clearly elucidate the role of artificial selection, as well as how this selection has formed thesystem.
We compared the frequencies and diversities ofandbetween groups of modern rice and populations of wild rice strains. At both loci, we found a decrease in diversity but an increase in frequency (Table 1 and Fig. 3). Therefore, we believed the effects of domestication can be divided into two parts: a sampling effect caused by genetic bottleneck and subsequent artificial selection. A few haplotypes ofandwere randomly preserved, then the frequencies of nonfunctional alleles arose during long- term artificial selection.
Ancestral structures contributed to establishment of DPL system
Mizuta et al (2010) suggested thatare a barrier between the two rice subspecies,and. However, their study did not considerseparately from, and their results actually indicated thatclustered in, not in. Craig et al (2014) and our results further established thathas a high frequency inbut a low frequency in.Therefore, we presumed thatinheritsdirectly from a wild ancestor and then spreadsit into the other groups. Our study indicated thathad the highest frequency in(87.8%). The groups belonging to(,,and) had higher frequencies thanthegroups (and) (Table 1). We therefore speculated that thegroups (most likely) inheritfirst and then spread it into other groups.
network analysis showed that the- haplotype of modern rice is related to nIND andin Oceania (rPNG). India is thought to be the primary potential domestication center of(Khush, 1997; Vaughan et al, 2008; Civan et al, 2015) and is closely connected with the domestication of(Morishima et al, 1992; Kovach et al, 2007; Sweeney and McCouch, 2007; Vaughan et al, 2008).network analysis indicated that the haplotype H_13 of rice2- was fromin South China (rGDGZ and rGXHZ), which is generally consideredthe origin center of(Khush, 1997; Londo et al, 2006; Vaughan et al, 2008; Civan et al, 2015). We speculated that the ancestral population ofwas nIND and those ofwere rGDGZ and rGXHZ. It appears that at least two domestication events are required forto obtain bothand. Our studyis therefore consistent with the ‘multiple domestications’ hypothesis, in whichis thoughtto have arisen as a result of more than one domestication event (Sang and Ge, 2007; Yang C C et al, 2012).
These results raise an interesting question regarding the discontinuous distribution ofH_1, which is present in wild rice strains not only from India but also from New Guinea. Similar to, the study of another incompatible system inand,, has also indicated the presence of anlike haplotype (H9) in New Guinea and South Asia but nowhere else (Du et al, 2011). Repeated extinction and colonization throughout the history ofhas previously been reported (Liu et al, 2015), therefore, we suspected the discontinuous distribution ofis a consequence of historical population dynamics.
Thehaplotype () in thesystem has been proved to originate from anpopulation in southern China, but there is no evidence for the origination area ofthehaplotypes (Long et al, 2008). Du et al (2011) provided the evolutionary history and geographic distribution of. Nevertheless, they did not definitively locate the origin areas of the incompatible alleles. Our study confirmed the origination centers of both parts and established that the genetic incompatibility betweenandis formed from an ancestral geographic pattern.
Not solely limited to modern rice, the effects of ancestral geographic patterns on incompatible genes have been widely reported.The same mechanism occurs in otherspecies./causes hybrid pollen sterility betweenand. This system is influenced by an ancestral geographic pattern and rapid sampling effects during the divergence of the two species (Yamagata et al, 2010).
DPLs are maintained in wild ancestors because of the absence of selection pressure and geographic isolation
Reproductive isolation genes lead to hybrid income- patibility, which means a certain number of hybrid descendants fail to survive or breed. Therefore, income- patible genes are generally deleterious for populations and are expected to result in loss, especially in gene flow conditions (Gavrilets, 1997; Bank et al, 2012). In contrast, incompatible genes are commonly found to be polymorphic, especially in plants(Rieseberg and Blackman, 2010; Bank et al, 2012; Cutter, 2012; Corbett-Detig et al, 2013; Lindtke and Buerkle, 2015). An interesting question is how incompatible alleles are maintained in nature.
Our results indicated that geographic isolation is a possible explanation.mainly clustered in Southeast and South Asia, whereaswas only found in a small area of South China (Table 1). The geographic distance prevents incompatible pairing ofand. Furthermore, neithernoris deleterious individually. Inand its wild ancestor,lossesare due to the lack of the longest exon (Mizuta et al, 2010). In twopopulations from Laos in our study,had high frequencies (100.0% and 90.5%).also showed 34.8% and 47.6% frequencies in twopopulations in South China. However, none of the species/populations mentioned above showed any limitations from the lack of one gene copy. Thus, it is possible thatoralone exists in wild rice.
Incompatibility caused by ancestral geographic structure can be found in many species. Genes leading to male sterility are variable within grasshopper populations, establishing that selection of incompatiblegenes can be weak or even absent(Shuker et al, 2005). Furthermore, the incompatible genes ofspecies are stable under balanced selection (Sicard et al, 2015). In the ancestor, balanced selection maintains the polymorphisms of two potentially incompatible genes,and, facilitating the establishment of genetic incompatibility between two selfing descendants,and. A pair of closely linked genes causes a globally distributed incompatibility inthat is maintained by balanced selection (Seidel et al, 2008). It is generally thought that purifying selection may be helpful for polymorphisms.andcause hybrid lethality betweenand, and both genes are polymorphic in the two species (Zuellig and Sweigart, 2018).andhave been maintained for decades, presumably because continual gene flow continues purging lethal combinations and reestablishing polymorphisms at both loci.
Our results further suggested that the establishment of thesystem relies on the increase in frequency of nonfunctional alleles. In this hypothesis, an ancestral polymorphism is a crucial precondition. In addition to the nonfunctional alleles reported by Mizuta et al (2010), we have identified other alleles that may possibly result in a loss of function. For example, a 7 bp insertion in the longest exon occurs only in wild rice strains from Sri Lanka. It is plausible that the hybrid sterility caused bymay be established in a more complex way under natural conditions. We speculated thatthe frequency of any other nonfunctional alleles increased after domestication, and genetic incompatibilitywould still have been established betweenand. We therefore hypothesized that incompatible systems of rice, or even other species, may arise in similar ways as. To clarify the evolutionary process of incompatible systems, large-scale analyses at the population level are warranted.
METHODS
Rice materials
We sampled 296and 383individual plants, represented as ‘n’ and ‘r’, respectively.The entire natural distribution range of the wild ancestors was covered (Fig. 4). Detailed material notes are listed in Table S2. These individuals were collected from different populations.Twenty- onenatural populations were from Southeast Asia and South Asia, including Sri Lanka, Nepal, Myanmar, Cambodia and Laos. The number of sampled individuals in these populations ranged from 29 to 36, except for Sri Lanka (9?10). We further collected a total of 115 individual samples from the International Rice Research Institute (IRRI), including 6 populations from India, Thailand, Indonesia and New Guinea.of New Guinea is a distinct line and is generally regarded as aspecialtype (Oka, 1988).Ten Chinesepopulations were sampled, each of which included 10?11 individuals. Of the Chinese populations,one was from Jiangxi Province (JXDX), the northern-most frontier of the natural range of(Morishima et al, 1992); two from Hunan Province (HNCL and HNJY); one from Guangdong Province (GDGZ); three from Guangxi Province (GXHZ, GXBH and GXTD); one from Yunnan Province (YNJH); and two from Hainan Province (HNDZ and HNWC).China and South Asia countries are commonly regarded as areas that are closely related with the domestication of rice (Morishima et al; 1992;Garris et al, 2005; Sang and Ge, 2007; Huang et al, 2012). In addition to these wild samples, 132(represented by ‘s’) landraces were sampled from all 6 groups:,,,,and.
Sequencing and genotype analysis
We extracted genomic DNA from fresh or silica gel-desiccated leaves using a DNAsecure Plant Kit (TIANGEN, Beijing, China). The primers and thermal cycling procedure used was previously described by Craig et al (2014). The genomic DNA and PCR products were analyzed via gel electrophoresis. PCR products were Sanger sequenced (Majorbio, Shanghai, China). When any double peaks in a sequencing string occurred, indicating heterozygous individuals.The PCR products were cloned into pEASY-T1 vectors (TransGen Biotech, Beijing, China). We consulted previous studies to confirm singletons (Zheng and Ge, 2010). All sequences are available in GenBank (http://www.ncbi.nlm.nih.gov). We also downloaded 20and 34sequences forfrom the National Centre for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) (Table S1).
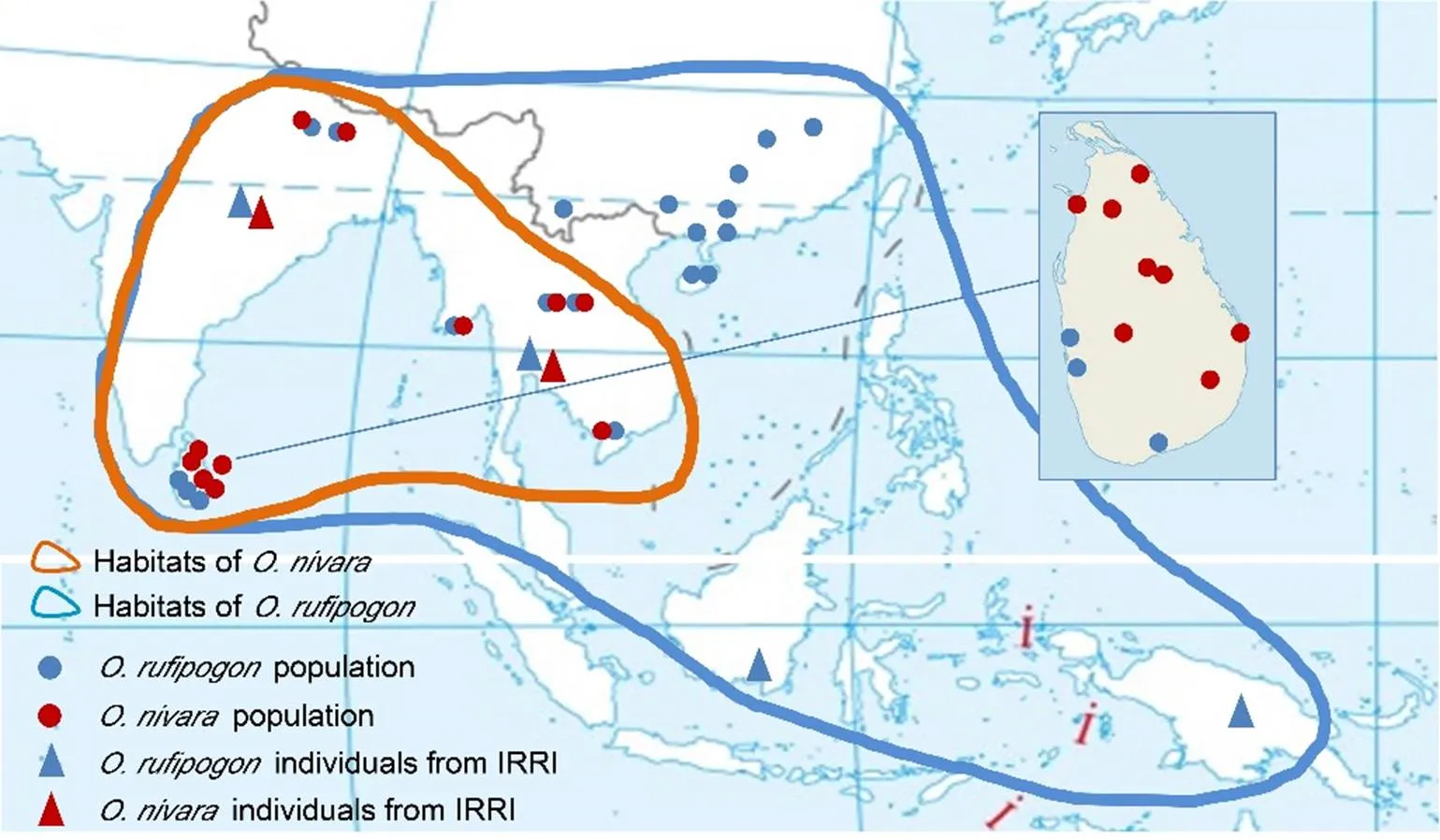
Fig. 4.Distribution of wild rice samplings, as well as functional and nonfunctional alleles of.
Populations ofandare indicated with spots. Triangles indicate individual samples from the International Rice Research Institute (IRRI). Red and blue lines indicateand, respectively. Circles indicate the frequencies of functional and nonfunctional alleles.
For simplicity,andwere defined according to Mizuta et al (2010).was defined by a 518 bp transposable element insertion in the first coding sequence that failed to translate.was defined by a single nucleotide transition from ‘A’ to ‘G’ in the intron, leading to a premature stop codon but producing a read-through protein (Fig. S1).
Genetic diversity and phylogeographic analysis
All obtained sequences were aligned using Clustal X 1.83(Thompson et al, 1997) and revised manually with BioEdit 7.0.9.0 (Hall, 1999). To detect potential traces of selection, we evaluated the number of polymorphic sites () and nucleotide diversity () and subsequently performed neutrality tests (Tajima’s) with DnaSP 5.1 (Librado and Rozas, 2009). The number of haplotypes () and haplotype diversity () were also calculated with DnaSP 5.1 after excluding recombined sequences via the online tool IMgc (Woerner et al, 2007). Insertions and deletions were excluded during analysis of diversities. Comparisons ofbetween each group ofand the wild ancestors were performed using the ratio ofof each group to thatof wild ancestor populations. The comparison ofwas performed in the same way.
We used networks of haplotypes to investigate the origination ofand2 in rice. The median-joining method (Bandelt et al, 1999) was used to construct networks of haplotypes via Network 5011 (FluxusTechnology Ltd., England). Recombined sequences were excluded by using IMgc (Woerner et al, 2007). Maximum Parsimony Calculation (Polzin and Daneshmand, 2003) was used to clarify the skeleton.
Data archiving
All sequence data used are available in GenBank (http://www. ncbi.nlm.nih.gov) under the accessions MN446022?MN446138, MN446139?MN446234, MN446235?MN446531, MN446532? MN446716, MN476103?MN476405, MN476406?MN476830, MN476831?MN476868 and MN476869?MN476888.
ACKNOWLEDGEMENTS
This work was supported by the Chinese Academy of Sciences (Grant Nos. XDA08020103 and XDB31000000) and the National Natural Science Foundation of China (Grant Nos. 31470332, 91731301 and 91231201). We thank Chen Chengbin of Guangxi Academy of Agricultural Sciences and Zheng Xiaoming of Institute of Crop Science of Chinese Academy of Agricultural Science for providing samples. We also thank the IRRI for providing samples and the members of Ge Song’s laboratory for field collections of wild rice.
Supplemental data
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/ 16726308; http://www.ricescience.org.
Fig.S1. Schematic representations ofand positions of study primers.
Table S1. List of downloadedsequences.
Table S2. Material list.
Bandelt H J, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies., 16(1): 37?48.
Bank C, Burger R, Hermisson J. 2012. The limits to parapatric speciation: Dobzhansky-Muller incompatibilities in a Continent- Island Model., 191(3): 845?345.
Cai Z, Zhou L, Ren N N, Xu X, Liu R, Huang L, Zheng X M, Meng Q L, Du Y S, Wang M X, Geng M F, Chen W L, Jing C Y, Zou X H, Guo J, Chen C B, Zeng H Z, Liang Y T, Wei X H, Guo Y L, Zhou H F, Zhang F M, Ge S. 2019. Parallel speciation of wild rice associated with habitat shifts., 36(5): 875?889.
Chen J J, Ding J H, Ouyang Y D, Du H Y, Yang J Y, Cheng K, Zhao J, Qiu S Q, Zhang X L, Yao J L, Liu K D, Wang L, Xu C G, Li X H, Xue Y B, Xia M, Ji Q, Lu J F, Xu M L, Zhang Q F. 2008. A triallelic system ofis a major regulator of the reproductive barrier and compatibility of-hybrids in rice., 105(32): 11436?11441.
Civan P, Craig H, Cox C J, Brown T A. 2015. Three geographically separate domestications of Asian rice., 1(11): 1?5.
Corbett-Detig R B, Zhou J, Clark A G, Hartl D L, Ayroles J F. 2013. Genetic incompatibilities are widespread within species., 504: 135?137.
Craig S M, Reagon M, Resnick L E, Caicedo A L. 2014. Allele distributions at hybrid incompatibility loci facilitate the potential for gene flow between cultivated and weedy rice in the US., 9(1): e86647.
Cutter A D. 2012. The polymorphic prelude to Bateson-Dobzhansky- Muller incompatibilities., 27(4): 209?218.
Du H, Ouyang Y, Zhang C, Zhang Q. 2011. Complex evolution of, a major reproductive barrier regulator, in the cultivated riceand its wild relatives., 191(1): 275?287.
Garris A J, Tai T H, Coburn J, Kresovich S, McCouch S. 2005. Genetic structure and diversity inL., 169(3): 1631?1638.
Gavrilets S. 1997. Hybrid zones with Dobzhansky-type epistatic selection., 51(4): 1027?1035.
Hall T A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT., 41: 95?98.
Huang X H, Kurata N, Wei X H, Wang Z X, Wang A, Zhao Q, Zhao Y, Liu K Y, Lu H Y, Li W J, Guo Y L, Lu Y Q, Zhou C C, Fan D L, Weng Q J, Zhu C R, Huang T, Zhang L, Wang Y C, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X P, Xu Q, Dong G J, Zhan Q L, Li C Y, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J Y, Han B. 2012. A map of rice genome variation reveals the origin of cultivated rice., 490: 497?501.
Khush G S. 1997. Origin, dispersal, cultivation and variation of rice., 35(1): 25?34.
Kovach M J, Sweeney M T, McCouch S R. 2007. New insights into the history of rice domestication., 23(11): 578?587.
Li G W, Li X T, Wang Y, Mi J M, Xing F, Zhang D H, Dong Q Y, Li X H, Xiao J H, Zhang Q F, Ouyang Y D. 2017. Three representative inter and intra-subspecific crosses reveal the genetic architecture of reproductive isolation in rice., 92(3): 349?362.
Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data., 25(11): 1451?1452.
Lindtke D, Buerkle C A. 2015. The genetic architecture of hybrid incompatibilities and their effect on barriers to introgression in secondary contact., 69(8): 1987?2004.
Liu R, Zheng X M, Zhou L, Zhou H F, Ge S. 2015. Population genetic structure ofand: Implications for the origin of., 24(20): 5211?5228.
Londo J P, Chiang Y C, Hung K H, Chiang T Y, Schaal B A. 2006. Phylogeography of Asian wild rice,, reveals multiple independent domestications of cultivated rice,., 103(25): 9578?9583.
Long Y M, Zhao L F, Niu B X, Su J, Wu H, Chen Y L, Zhang Q Y, Guo J X, Zhuang C X, Mei M T, Xia J X, Wang L, Wu H B, Liu Y G. 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes., 105(48): 18871?18876.
Mizuta Y, Harushima Y, Kurata N. 2010. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes., 107(47): 20417?20422.
Morishima H, Sano Y, Oka H. 1992. Evolutionary Studies in Cultivated Rice and Its Wild Relatives. Oxford: Oxford University Press.
Oka H I. 1988. Origin of Cultivated Rice. Amsterdam, Holland: Elsevier.
Ouyang Y D, Liu Y G, Zhang Q F. 2010. Hybrid sterility in plant: Stories from rice., 13(2): 186?192.
Ouyang Y D, Zhang Q F. 2013. Understanding reproductive isolation based on the rice model., 64(1): 111?135.
Ouyang Y D, Zhang Q F. 2018. The molecular and evolutionary basis of reproductive isolation in plants., 45(11): 613?620.
Polzin T, Daneshmand S V. 2003. On Steiner trees and minimum spanning trees in hypergraphs., 31(1): 12?20.
Rieseberg L H, Blackman B K. 2010. Speciation genes in plants., 106(3): 439?455.
Sang T, Ge S. 2007. The puzzle of rice domestication., 49(6): 760?768.
Seidel H S, Rockman M V, Kruglyak L. 2008. Widespread genetic incompatibility inmaintained by balancing selection., 319: 589?594.
Shen R X, Wang L, Liu X P, Wu J, Jin W W, Zhao X C, Xie X R, Zhu Q L, Tang H W, Li Q, Chen L T, Liu Y G. 2017. Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice., 8: 1310.
Shuker D M, Underwood K, King T M, Butlin R K. 2005. Patterns of male sterility in a grasshopper hybrid zone imply accumulation of hybrid incompatibilities without selection., 272: 2491?2497.
Sicard A, Kappel C, Josephs E B, Lee Y W, Marona C, Stinchcombe J R, Wright S I, Lenhard M. 2015. Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in., 6: 7960.
Sweeney M, McCouch S. 2007. The complex history of the domestication of rice., 100(5): 951?957.
Tajima F. 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism., 123(3): 585?595.
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools., 25(24): 4876?4882.
Vaughan D A, Lu B R, Tomooka N. 2008. The evolving story of rice evolution., 174(4): 394?408.
Wang C H, Zheng X M, Xu Q, Yuan X P, Huang L, Zhou H F, Wei X H, Ge S. 2014. Genetic diversity and classification ofwith emphasis on Chinese rice germplasm., 112: 489?496.
Woerner A E, Cox M P, Hammer M F. 2007. Recombination- filtered genomic datasets by information maximization., 23(14): 1851?1853.
Xie Y Y, Shen R X, Chen L T, Liu Y G. 2019. Molecular mechanisms of hybrid sterility in rice., 62(6): 737?743.
Yamagata Y, Yamamoto E, Aya K, Win K T, Doi K, Sobrizal, Ito T, Kanamori H, Wu J, Matsumoto T, Matsuoka M, Ashikari M, Yoshimura A. 2010. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice., 107(4): 1494?1499.
Yang C C, Kawahara Y, Mizuno H, Wu J Z, Matsumoto T, Itoh T. 2012. Independent domestication of Asian rice followed by gene flow fromto., 29(5): 1471?1479.
Yang J Y, Zhao X B, Cheng K, Du H Y, Ouyang Y D, Chen J J, Qiu S Q, Huang J Y, Jiang Y H, Jiang L W, Ding J H, Wang J, Xu C G, Li X H, Zhang Q F. 2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice., 337: 1336?1340.
Zheng X M, Ge S. 2010. Ecological divergence in the presence of gene flow in two closely relatedspecies (and)., 19(12): 2439?2454.
Zuellig M P, Sweigart A L. 2018. A two-locus hybrid incompatibility is widespread, polymorphic, and active in natural populations of., 72(11): 2394?2405.
Copyright ? 2021, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2020.11.007
7 April 2020;
20 June 2020
Zhang Fumin(zhangfm@ibcas.ac.cn)
(Managing Editor: Wu Yawen)
- Rice Science的其它文章
- Different Hypotheses for Resistance Loss of Rice Varieties to Magnaporthe oryzae
- Development of Heat Tolerant Two-Line Hybrid Rice Restorer Line Carrying Dominant Locus of OsHTAS
- Identification of QTLs for Cadmium Tolerance During Seedling Stage and Validation of qCDSL1 in Rice
- Physiochemical Properties of Resistant Starch and Its Enhancement Approaches in Rice
- Anticancer Activities of Plant Secondary Metabolites: Rice Callus Suspension Culture as aNew Paradigm
- Visualizing Meiotic Chromosome Pairing and Segregation in Interspecific Hybrids of Rice by Genomic in situHybridization

