Overview of noncoding RNAs involved in the osteogenic differentiation of periodontal ligament stem cells
Wei Qiu, Bu-Ling Wu, Fu-Chun Fang
Wei Qiu, Bu-Ling Wu, Fu-Chun Fang, Department of Stomatology, Nanfang Hospital, Southern Medical University, Guangzhou 510515, Guangdong Province, China
Abstract
Key words: Noncoding RNAs; Periodontal regeneration; Periodontal ligament stem cells;Osteogenic differentiation
INTRODUCTION
Periodontal diseases are infectious diseases characterized by progressive destruction of the periodontium (tooth-supporting tissue), which includes the periodontal ligament (PDL), cementum, alveolar bone, and gingiva[1]. Tooth loss mainly results from periodontal diseases in adults, which adds a substantial burden to public health worldwide[2,3]. Periodontal treatment is not as easy as only controlling inflammation and preventing disease development; the reconstruction of a healthy periodontium destroyed by diseases deserves equal attention[4,5]. Current therapies for periodontal diseases in the clinic, including conservative approaches, radicular conditioning,bioactive bone grafting/substitution, and guided tissue regeneration (GTR),encounter difficulty in regenerating the periodontium completely[6]. Therefore, the stem cell-based tissue regeneration approach involving transplantation of stem cells to enhance periodontal tissue regeneration has gradually taken the place of guided bone/tissue regeneration[7-9].
PDL is a specialized soft connective tissue that connects the cementum and alveolar bone; it shows the function of maintaining and supporting teethin situ, preserving tissue homoeostasis and repairing damaged periodontal tissue[1]. In the 1980s, Bordinet al[10]reported that PDL tissue possessed periodontal regenerative properties due to its resident cells, which were considered to be seed cells and a reliable source for periodontium regeneration. In 2004, Seoet al[11]first identified and characterized multipotent stem cells in human PDL and termed them periodontal ligament stem cells (PDLSCs). PDLSCs show similar features to other postnatal mesenchymal stem cells (MSCs): Multilineage differentiation potential and potent self-renewal ability.PDLSCs can further differentiate into cementoblasts/osteoblasts, chondrocytes and adipocytesin vitroand regenerate cementum/PDL-like tissuesin vivo[12]. As a consequence, PDLSC-mediated periodontium tissue regeneration is likely to be a practical cellular-based treatment for periodontal diseases[13]. However, what determines and regulates the multilineage differentiation potential of PDLSCs warrants further research.
Instead of the potential to encode proteins or peptides, noncoding RNAs (ncRNAs)are a category of unique RNAs that are widely present in eukaryotic cells[14-16].Following the development of this field, scientists have determined that ncRNAs play a significant role in the regulation of gene expression by controlling the expression levels of protein-coding RNAs and are involved in diverse cellular processes,including cell proliferation, cell differentiation, and ontogenesis, and are thus closely related to embryonic development and disease pathogenesis[17-19]. However, there are currently no uniform criteria for ncRNA classification. ncRNAs can be divided into cytoplasmic and nuclear ncRNAs based on their subcellular localization. In addition,ncRNAs are generally categorized into structural and regulatory ncRNAs, as well as regarding their function in cellular processes[20]. Structural ncRNAs include ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), whereas regulatory RNAs can be further divided into categories based on their length, such as long noncoding RNAs(lncRNAs) (size from > 200 nt to 100 kb) and several types of small RNAs, which include small interfering siRNAs (18-30 nt), piwiRNAs (24-30 nt) and microRNAs(miRNAs, 20-24 nt)[21]. Circular RNAs (circRNAs) are covalently linked to the end of RNA molecules, in which the 3′ and 5′ ends are connected in a non-collinear way through the back-splicing process[22]. CircRNAs, which are a type of competing endogenous RNA (ceRNA), can act as miRNA sponges. Recently, growing research has indicated that circRNAs are involved in embryonic development, cellular activities, and many other human diseases[23-25]. In summary, the cell differentiation of PDLSCs is collectively or individually regulated by ncRNAs. This review focuses on the three most important ncRNAs, namely, miRNAs, lncRNAs and circRNAs,currently identified to play a role in osteogenic differentiation (Tables 1 and 2).

Table 1 Expression profile of ncRNAs involved in the osteogenic differentiation of periodontal ligament stem cells
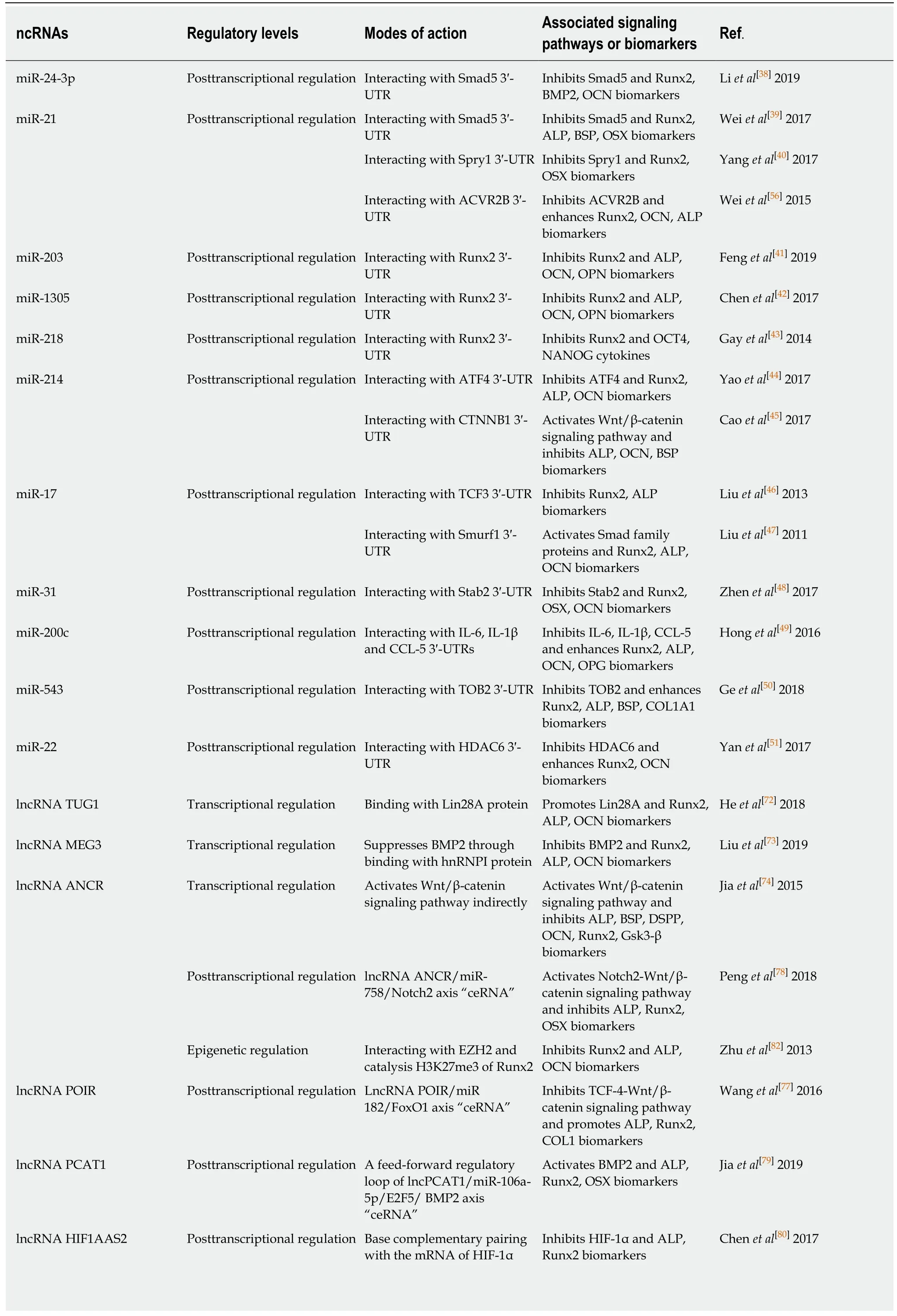
Table 2 Overview of ncRNAs involved in the osteogenic differentiation of periodontal ligament stem cells

miRNAs: MicroRNAs; lncRNAs: Long non-coding RNAs; circRNAs: Circular RNAs; hPDLSCs: Human periodontal ligament stem cells; PDLSCs:Periodontal ligament stem cells; Smad5/: SMAD family member 5; Runx2: Runt-relatedtranscriptionfactor 2; BMP2: Bone morphogenetic protein-2; OCN:Osteocalcin; ALP: Alkaline phosphatase; BSP: Bone Sialoprotein; OSX: Osterix; Spry1: Palmitate phosphoprotein Sprouty1; ACVR2B: Activin A receptor type 2B; OPN: Osteopontin; OCT4: Octamer-binding transcription factor-4; NANOG: Homeobox transcription factor nanog; ATF4: Activated transcription factor 4; CTNNB1: Catenin beta 1; TCF3: Transcriptional factor 3; Smurf1: Smad ubiquitin regulatory factor 1; Satb2: Special AT-rich sequence-binding protein 2; IL: Interleukin; CCL-5: Chemokines-5; OPG: Osteoprotegerin; TOB2: Transducer of ERBB2; COL1A1: Collagen type I alpha 1 chain; HDAC6:Histone deacetylase 6; Lin28A: Lin-28 homolog A; hnRNP I: Heterogeneous nuclear ribonucleoprotein I; DSPP: Dentin sialophosphoprotein; GSK3β:Glycogen synthase kinase 3β; ANCR: Anti-differentiation noncoding RNA; Notch2: Neurogenic locus notch homolog protein 2; ceRNA: Competing endogenous RNAs; EZH2: Enhancer of zeste homolog 2; H3K27me3: Histone H3 trimethylated at lysine 27; POIR: Osteogenesis impairment-related lncRNA of PDLSCs; FoxO1: Forkhead box O1; TCF4: Transcription factor 4; COL1: Collagen type I; PCAT1: Prostate cancer-associated ncRNA transcript-1;HIF1AAS1/2: HIF1A antisense RNA 1/2; HIF-1α: Hypoxia-inducible factor-1α; CDR1as: Antisense to the cerebellar degeneration-related protein 1 transcript; GDF5: Growth differentiation factor 5; MAPK: Mitogen-activated protein kinase; ACVR2B: Activin A receptor type 2B; SP7: Transcription Factor Sp7.
HISTORY OF PDLSCS
The progenitor cells residing within the PDL (periodontal ligament progenitor,PDLPs) were first described in seminal studies by Melcher in 1994[26]. Seoet al[11]described the identification and characterization of multipotent stem cells in human PDL in 2004, although these cells had been suspected to be present in the PDL for a long time. Nevertheless, there is no uniform standard for defining the features of PDLSCs. Often, reports suggest that the isolation of particular subsets of cells from bulk explant cultures is far less rigorous and was too liberal for the use of the term PDLSC[27]. Prateeptongkumet al[28]reported that the isolation methods of PDLPs and PDLSCs from PDL tissues are different and demonstrated that PDLPs could be isolated using outgrowth methods, while PDLSCs need single-cell isolation methods for isolation. PDLSCs can be further characterized by their cell surface expression of CD29, CD44, STRO-1, STRO-4, CD146, CD73, CD90, CD105 and CD166 and the lack of expression of endothelial (CD31), haematopoietic (CD14, CD34, CD45, and CD79a),and helper immune antigens (HLA-DR, CD40, CD54, CD80, and CD86)[10,29].Functionally, PDLSCs have been determined to fulfil all of the criteria of identifiable MSC-like properties, including self-renewal capacity, multipotencyin vitro, tissue regenerative capacityin vivo, and immunomodulation[30,31]. These processes are illustrated in Figure 1.
MICRORNAS IN PDLSCS: ORCHESTRATING CELLULAR OSTEOGENIC DIFFERENTIATION
miRNAs are endogenous, single-stranded noncoding RNAs derived from genomic sequences[32]. The lengths of mature miRNAs are typically 20~24 nucleotides, 8 of which are identified as the “seed sequences” (with nt positions 2 to 7 that were 99%conserved)[33,34]. miRNAs have been extensively investigated in the past two decades,and the underlying mechanism is relatively clear. Mature miRNAs are targeted to a sequence in the 3′ UTR (untranslatedregion) of mRNAs matching the seed sequence and further influence the stability of mRNAs or inhibit their translation to eventually downregulate protein expression[35]. miRNAs are a leading representative of small ncRNAs, and they are closely associated with diverse biological and pathological processes.
miRNA microarrays are a widely accepted high-throughput method and are very effective in analysing miRNA expression levels during osteogenic differentiation of PDLSCs[36]. Our team used a miRNA microarray to detect the different expression profiles of miRNAs in PDLSCs during the osteogenic differentiation processin vitro[37].The results showed a significant change in the expression level of 116 miRNAs, 30 of which were increased, while 86 miRNAs were downregulated in PDLSCs after 14 d of osteogenic induction. The results probably suggested an important regulatory role that miRNAs might play in the osteogenic differentiation of PDLSCs.
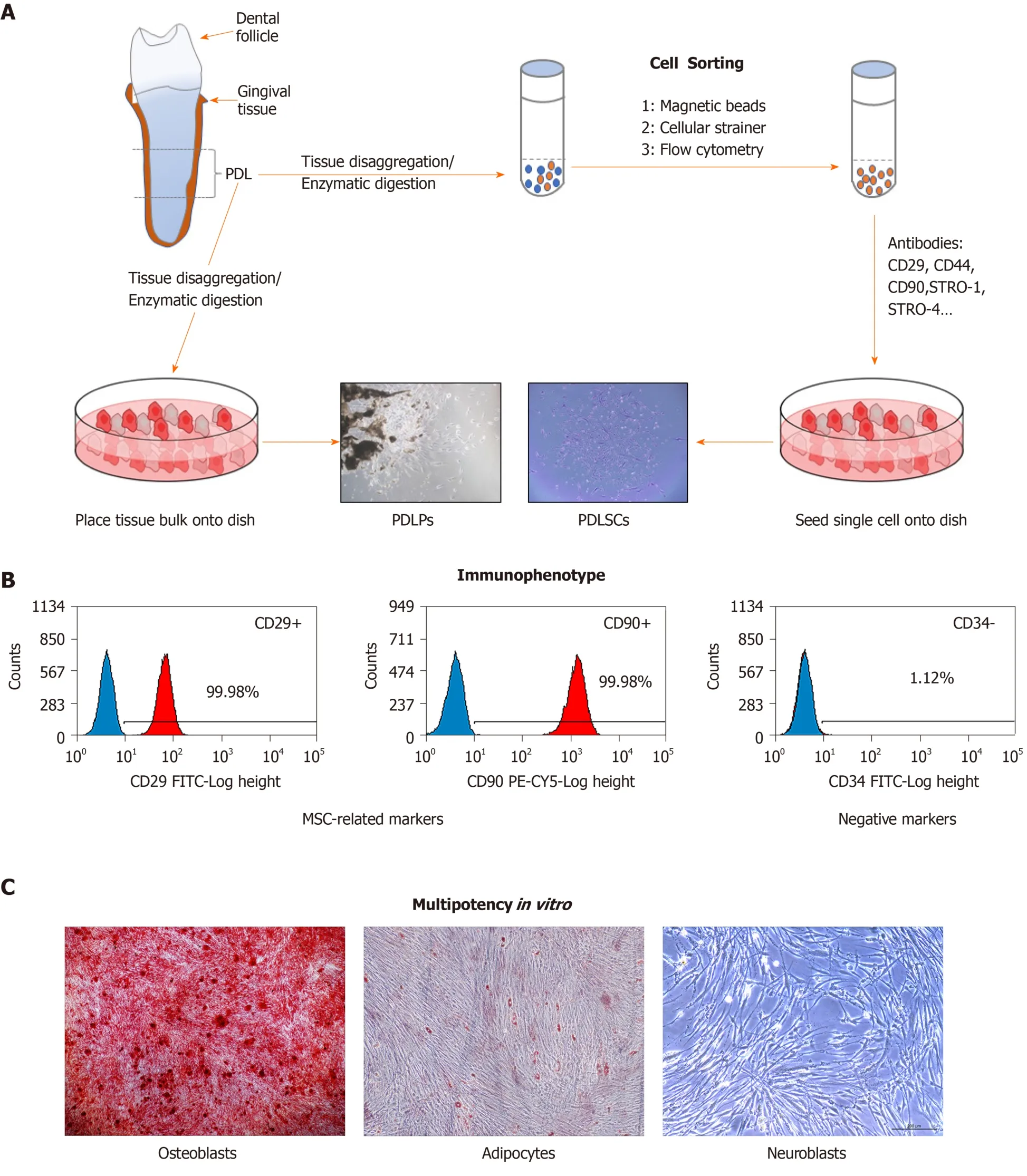
Figure 1 lsolation and characterization of periodontal ligament stem cells.
Similarly, a microarray was used in the study of Liet al[38]to detect the expression level of miRNAs in differentiated and undifferentiated PDLSCs and demonstrated that the expression level of miR-24-3p was significantly downregulated in osteogenically differentiated PDLSCs. Furthermore, double luciferase reporter assays and genetic engineering experiments demonstrated that miR-24-3p directly bound to the 3’-UTR of transduction protein 5 (SMAD family member 5, Smad5) and inhibited the transcription of the target gene.
The inhibition of the osteogenic differentiation of PDLSCs was the result of miR-21 downregulating the expression of Smad5 in the research of Weiet al[39]. Yanget al[40]found that the inhibition was attributed to the regulation of the miR-21/palmitate phosphoprotein Sprouty1 axis by tissue tumour necrosis factor-α. In addition, miR-203, miR-1305, and miR-218 have all been confirmed to target runt-related transcriptionfactor 2 (Runx2) and play important inhibitory roles in the osteogenic differentiation of PDLSCs[41-43]. miR-214 not only targeted activated transcription factor 4[44]but also bound with catenin beta 1 to modulate the Wnt/β-catenin signalling pathway, which is involved in osteogenic differentiation of PDLSCs[45]. Liuet al[46]found that miR-17 regulated the osteogenic differentiation of PDLSCs by reducing the expression of transcriptional factor 3 and inhibiting the Wnt signalling pathway. In contrast, Liuet al[47]demonstrated that miR-17 promoted differentiation by binding to the Smad ubiquitin regulatory factor one 3’-UTR in PDLSCs isolated from PDL tissue from periodontitis patients. miR-31 plays a regulatory role by targeting special ATrich sequence-binding protein 2 in osteogenic differentiation mediated by a high dose of glucose in PDLSCs[48]. All of the abovementioned miRNAs exerted inhibitory effects on osteogenic differentiation by targeting osteogenesis-related transcription factors through the classic miRNA regulatory mechanism.
Although several miRNAs suppress the osteogenic differentiation of PDLSCs,recent research has revealed that miRNAs promote the osteogenic differentiation of PDLSCs, including miR-200c, miR-543 and miR-22. Honget al[49]demonstrated that miR-200c decreased the levels of interleukin-6, interleukin-8 and chemokines-5 and increased the osteogenic differentiation of PDLSCs and BMSCs. miR-200c is a potentially effective means of preventing periodontitis-associated bone loss by arresting inflammation and osteoclast/osteogenesis and regenerating bone tissue.Previous research by our team found that miR-543 directly interacted with the 3’-UTR of transducer of ERBB2 and promoted osteogenesis in PDLSCs[50]. Yanet al[51]claimed that miR-22 promoted the osteogenic differentiation of PDLSCs by inhibiting the expression of histone deacetylase 6.
According to previous work, one of the factors affecting osteogenic differentiation of PDLSCs is mechanical stretch[52-54]. To investigate miRNA expression specifically in stretched PDLSCs, a microarray assay was utilized by Weiet al[55]to describe the differential expression of miRNAs in normal and stretched PDLSCs by using a tension system to achieve external mechanical stimulation. The results showed that 53 miRNAs were differentially expressed in stretched PDLSCs, and 26 of the miRNAs were upregulated, while 27 were downregulated. Noticeably, miR-21 directly targeted the 3’-UTR of activin A receptor type 2B (ACVR2B), thereby reducing the expression of ACVR2B and repressing the osteogenic differentiation of stretched PDLSCs[56].
The main regulatory mechanism of microRNAs is the posttranscriptional repression of target genes. However, several studies have reported that miRNAs function in other unconventional ways, including pri-miRNAs coding for short peptides and miRNAs interacting with non-AGO proteins, activating toll-like receptors, upregulating protein expression, targeting mitochondrial transcripts,directly activating transcription, and targeting nuclear ncRNAs[57-59]. To date, research has mainly focused on the classic regulatory mechanism of miRNAs in PDLSCs, and other regulatory mechanisms require further in-depth exploration.
LNCRNAS INVOLVED IN OSTEOGENIC DIFFERENTIATION OF PDLSCs
LncRNAs are a family of RNA molecules with transcript lengths of 200 nt to 20000 nt.These RNAs are unable to encode proteins or are only translated into small peptides at a very low level. Initially, lncRNAs were identified as a by-product of RNA polymerase II transcription and thought to be the “noise” of genomic transcription(referred to as “the dark matter” of the genome) with no biological function[36,60,61].However, research on lncRNAs has rapidly developed in recent years. According to their position relative to protein-coding genes in the genome, lncRNAs can be divided into five types: Sense, antisense, bidirectional, intronic, and intergenic lncRNAs[62].Recent studies show that lncRNAs act as novel and important regulators of numerous biological, developmental, and numerous cellular processes, including chromatin modification, X-chromosome silencing, genomic imprinting, transcriptional activation or interference, and intranuclear transport[63], and act through such mechanisms as transcriptional regulation, posttranscriptional regulation, and epigenetic regulation[64-66].
Guet al[67]compared the lncRNA profiles of PDLSCs on the 7th day with or without osteogenic differentiation medium using RNA sequencing. The results showed that 17 lncRNAs were upregulated and 31 were downregulated during osteogenic differentiation in PDLSCs. Zhanget al[68]also used RNA sequencing to detect the different expression profiles of lncRNAs in PDLSCs at different time points during osteogenic differentiation. These results indicated that 48 lncRNAs had significant changes on days 3, 7 and 14, of which 17 lncRNAs were upregulated and 31 were downregulated in PDLSCs. Our team used a lncRNA microarray to determine the expression levels of lncRNAs in osteoblast-induced and noninduced PDLSCs, and the results showed that 994 lncRNAs were upregulated and 1177 lncRNAs were downregulated at 14 d of osteogenic differentiation in PDLSCs. Further GO analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that a total of 83 signalling pathways were involved in the osteogenic differentiation of PDLSCs, including the mitogen-activated protein kinase (MAPK) and transforming growthfactor-β signalling pathways. In addition, coding-noncodinggenecoexpression analysis indicated a potential regulatory relationship between the differentially expressed lncRNAs and the osteogenesis-related mRNAs, of which 131 pairs of lncRNAs and mRNAs had negative correlations and 262 pairs had positive correlations[69]. Huanget al[70]used RNA sequencing to describe the lncRNA landscape during osteogenic differentiation in PDLSCs subjected to compressive force. The results indicated that 90 lncRNAs and 519 mRNAs were differentially expressed in PDLSCs under compressive stress. Of the lncRNAs, 72 were upregulated, and 18 were downregulated. To summarize, investigations of the regulatory mechanisms of lncRNAs in the osteogenic differentiation of PDLSCs are mainly focused on transcriptional regulation and posttranscriptional regulation.
Transcriptional regulation by lncRNAs in PDLSCs
Studies of the underlying mechanisms indicated that lncRNAs can specifically bind with a specific family of proteins called RNA binding proteins (RBPs) to regulate the biological functions of these proteins and then affect the transcriptional activation of downstream genes[71]. As a novel protein type, RBPs have been demonstrated to interact with lncRNAs and have attracted increasing attention from researchers. Heet al[72]identified the novel lncRNA- taurine upregulated gene 1 (TUG1), which was significantly upregulated in osteogenically induced PDLSCs. Gain/loss-of-function experiments showed that lncRNA-TUG1 was positively correlated with the osteogenic differentiation of PDLSCs following induction. Meanwhile, bioinformatic analysis demonstrated that lin-28 homologue A (Lin28A, a member of the RBP family)containing multiple binding sites of lncRNA-TUG1 was a potential target in the process of osteogenic differentiation of PDLSCs. Further investigation demonstrated that suppression of Lin28A inhibited the expression of several osteogenic-related gene markers, such as alkaline phosphatase, osteocalcin, and Runx2, in PDLSCs. These results suggested that lncRNA-TUG1 might interact with Lin28A to form a positive regulatory network of osteogenic-related genes to promote osteogenic differentiation of PDLSCs. However, there is insufficient evidence about the direct binding between lncRNA-TUG1 and Lin28A in this study. In a study by Liuet al[73], lncRNA maternally expressed gene 3 (MEG3) was markedly downregulated in osteogenically differentiated human PDLSCs compared with undifferentiated cells, as determined by microarray analysis. Overexpression of lncRNA MEG3 inhibited the activation of bone morphogenetic protein-2 (BMP2) and reversed osteogenic differentiation induced by mineralizing solution in PDLSCs. Furthermore, RNA-binding protein immunoprecipitation (RIP) assays verified that lncRNA MEG3 suppressed BMP2 through direct interaction with heterogeneous nuclear ribonucleoprotein I (hnRNPI)during osteogenic differentiation in hPDLCs. These results indicated that lncRNA MEG3 could modulate the osteogenic differentiation of hPDLCs by interacting with hnRNPI and thus inhibiting the transcriptional activity of BMP2. Moreover, lncRNAs can also interfere with the transcriptional activation of mRNAs or other ncRNAs. Jiaet al[74]found an inhibitory effect of ANCR (anti-differentiation noncoding RNA) on the gene expression of glycogen synthase kinase 3β and Runx2 and the classic Wnt/βcatenin signalling pathway, thereby suppressing osteogenic differentiation of PDLSCs.
Posttranscriptional regulation of lncRNAs in PDLSCs
Recent studies revealed that lncRNAs might act as miRNA sponges to compete with target genes for miRNA binding sites, thereby affecting the activity of the targeted genes in various biological processes[75]. These regulatory mechanisms involving lncRNAs, miRNAs and mRNAs are one type of ceRNA[76]mechanism. Wanget al[77]successfully identified the novel lncRNA-POIR (osteogenesis impairment-related lncRNA of PDLSCs), which is an osteogenesis impairment-related lncRNA of PDLSCs, and the gradual reduction in the expression of this lncRNA in PDLSCs was recorded among periodontitis patients, as demonstrated by lncRNA microarray analysis. Overexpression or knockdown of lncRNA-POIR was performed, and lncRNA-POIR was shown to positively regulate osteogenic differentiation of PDLSCs bothin vitroandin vivo. Further luciferase reporter assays and quantitative real-time PCRs demonstrated that lncRNA-POIR was likely to act as a ceRNA for miR-182,thereby leading to derepression of the target gene Forkhead box O1 (FoxO1).Activated FoxO1 could compete with transcription factor 4 for β-catenin binding and inhibit the classic Wnt signalling pathway, thereby promoting osteogenic differentiation of inflammatory PDLSCs. On the other hand, Penget al[78]suggested that lncRNA-ANCR targeted miR-758 directly as a molecular spongeviaRNA immunoprecipitation, and a dual luciferase reporter assay was also performed to demonstrate that miR-758 modulates the transcript expression of neurogenic locus notch homolog protein 2 (Notch2) by targeting the 3’-UTR of Notch2. These findings suggest that the lncRNA-ANCR/miR-758/Notch2 axis plays an essential role in regulating the osteogenic differentiation of PDLSCs. In addition, prostate cancerassociated ncRNA transcript-1 (lncPCAT1) was significantly increased in osteogenically induced PDLSCs and could positively regulate osteogenic differentiation bothin vitroandin vivoaccording to the study of Jiaet al[79]. Thereafter,these researchers inferred a predicted interaction and then confirmed the direct binding sites of miR-106a-5p on lncPCAT1. In conclusion, lncRNA-PCAT1 promoted osteogenic differentiation of PDLSCs by sponging miR-106a-5p to upregulate the miR-106a-5p-targeted gene BMP2. Interestingly, the authors also found that another target of miR-106a-5p, E2F5, could bind to the promoter of lncPCAT1 and then form a feed-forward regulatory network targeting BMP2. Chenet al[80]studied two lncRNAs,HIF1A antisense RNA 1 and HIF1A antisense RNA 2, that regulated the mRNA expression of HIF1α. The results showed that HIF1A-AS2 exerted a remarkable negative regulatory function on hypoxia-inducible factor-1α (HIF-1α) through complementary base pairing with HIF-1α mRNA in PDLCs under hypoxia.
Epigenetic regulation of lncRNAs in PDLSCs
Epigenetics plays a central role in regulating many critical cellular processes. From the perspective of epigenetics, several lncRNAs can interact with protein complexes and modulate the levels of DNA methylation as coordinators of chromatin modification,thereby regulating the expression of related genes at the epigenetic level[81]. Studies have claimed that lncRNAs affect the osteogenic differentiation of PDLSCs by epigenetic regulation. Zhuet al[82]found that lncRNA ANCR posed a physical interaction with enhancer of zeste homologue 2, and Runx2 expression was suppressed due to the catalysis of H3K27me3 in the Runx2 gene promoter, leading to the inhibiting effect on osteoblast differentiation of hFOB1.19 cells. At present, further exploration is needed to explore the function of lncRNAs in regulating the osteogenic differentiation of PDLSCs at the epigenetic level.
The discovery of lncRNAs greatly broadened the understanding of molecular regulatory mechanisms. LncRNAs regulate the expression of related genes via three different regulatory mechanisms at the levels of transcription, posttranscription, and epigenetics and participate in many important cellular processes. Current studies on lncRNAs regulating the osteogenic differentiation of PDLSCs have mainly focused on the transcriptional and posttranscriptional levels. However, most studies have not determined the subcellular localization of lncRNAs. Generally, the localization of an lncRNA in the cytoplasm is important when considering its functions, especially as a ceRNA sponge[83]. Due to the numerous lncRNAs and their complexity, the mechanisms of lncRNAs regulating osteogenic differentiation in PDLSCs need to be explored further.
CIRCRNAS ASSOCIATED WITH OSTEOGENIC DIFFERENTIATION OF PDLSCS
circRNAs are a novel class of endogenous lncRNAs that are characterized by a structure of covalently closed continuous loops lacking 5’ or 3’ polarities and are widely present in eukaryotic cells[84]. The circRNAs are more stable than linear RNAs because of their covalently circular structure, making them more resistant to RNase R digestion. The majority of circRNAs are conserved across species and often exhibit cell type-specific, tissue-specific or developmental stage-specific expression[85]. Evidence is increasing that circRNAs might act as miRNA sponges and play critical roles in signal transduction in a posttranscriptional manner. The circRNA-miRNA axis is involved in several cellular processes, such as proliferation, differentiation and apoptosis[86].
Guet al[67]conducted high-throughput sequencing to detect the different expression profiles of circRNAs in PDLSCs after 7 d of osteogenic differentiation. These researchers found that 766 circRNAs were significantly upregulated and 690 circRNAs were downregulated in PDLSCs. Furthermore, the authors predicted the potential functions of circRNAs as ceRNAs based on miRanda analysis and further investigated them using GO and KEGG analysis. The results showed that a total of 1382 circRNAs (including circRNA PTPRG, EXOC4, PRKCA, and SETBP1) were predicted to be able to interact with 148 miRNAs and compete for miRNA binding sites with 744 mRNAs, which were predicted to be significantly associated with osteoblast differentiation and the MAPK and Wnt signalling pathways regulating pluripotency of mesenchymal stem cells. Among these circRNAs, one circRNA could bind with multiple miRNAs, and the same miRNA could also interact with multiple circRNAs. Liet al[87]revealed that circRNA antisense to the cerebellar degenerationrelated protein 1 transcript (CDR1as) could act as a miR-7 adsorption sponge and then induce the upregulation of growth differentiation factor 5 and activate the Smad1/5/8 and p38MAPK signalling pathways, thereby promoting osteogenesis in PDLSCs. Wanget al[88]found that circRNA expression patterns were responsive to mechanical force in PDLSCs. Bioinformatic analysis showed that one circRNA could modulate one or several miRNA/miRNAs and vice versa. Importantly, the authors found that circRNA3140 was widely and highly related to microRNA-21, which played a key role in mechanical force-induced osteogenic differentiation of PDLSCs.These findings revealed that mechanical force induced the differential expression of circRNAs in PDLSCs, which might regulate the orthodontic tooth movement process and alveolar bone remodelling.
Overall, as a novel class of endogenous lncRNAs, circRNAs may modulate many pathophysiological processes, serve as diagnostic or predictive biomarkers for several diseases, and represent a novel and useful therapeutic method[89]. Compared with research on miRNAs and lncRNAs, research on circRNAs is in its infancy, and the potential functions and regulatory mechanisms of circRNAs are diverse. Investigating the regulatory mechanisms and functions of circRNAs in the osteogenic differentiation of PDLSCs may provide exciting potential therapies in periodontal regeneration.
CONCLUSION
Numerous ncRNAs are associated with the osteogenic differentiation of PDLSCs(Figure 2). ncRNAs offer an additional and promising possibility of osteogenesisrelated gene regulation that has not been fully elucidated to date. With increasing numbers of miRNAs, lncRNAs and circRNAs discovered in this process, it has become possible to use these ncRNA-related therapeutic methods in the field of periodontium repair and regeneration.
To date, ncRNA-related research on the osteogenic differentiation of PDLSCs has mainly focused on miRNAs. The demonstrated regulatory mechanism of miRNAs(miR-24-3p, miR-21, miR-203, miR-1305, miR-218, miR-214, miR-17, miR-31, miR-200c,miR-543 and miR-22) is to inhibit the mRNA levels or protein expression of targets.Other unconventional mechanisms could impact osteogenic differentiation. Currently,miRNAs are considered to be potential therapeutic targets based on their defined regulatory mechanism and clear functioning mode. miRNA-based therapeutic methods could become valuable in promoting periodontium repair and regeneration.Similarly, there are several studies on the role of lncRNAs during this process. Among these lncRNAs, lncRNA TUG1, MEG3 and ANCR regulate the osteogenic differentiation of PDLSCs in a transcriptional manner. The lncRNAs POIR, PACT1,HIF1A-AS2 and ANCR act as miRNA sponges and play critical roles during osteogenic differentiation of PDLSCs in a posttranscriptional manner (Figure 3).However, lncRNA-ANCR has been demonstrated to suppress osteogenic differentiation of PDLSCs in an epigenetic regulatory manner. To date, few studies have investigated circRNAs during osteogenic differentiation in PDLSCs. The demonstrated mechanism of circRNAs is to act as miRNA sponges to inhibit the mRNA levels of target genes (circRNA CDR1as and circRNA3140). Other types of ncRNAs involved in the osteogenic differentiation of PDLSCs warrant further exploration.
With the development of sequencing and microarray technologies, numerous novel ncRNAs have been screened out and identified in the past few years, and their regulatory mechanisms have also been predicted and explored, benefiting from the advancement of related bioinformatics databases. Subsequently, standard molecular biology experiments and genetic engineering methods, such as quantitative real-time PCR, western blotting, dual luciferase reporter assays, RNAi and overexpression plasmid transfections, have been used to characterize ncRNA functions and explore their regulatory mechanisms. In addition, some new experimental methods have emerged, such as RIP, RNA pull-down, chromatin isolation by RNA purification,cross-linking immunoprecipitation, cross-linking, ligation, and sequencing of hybrids,and capture hybridization analysis of RNA targets, which provide an ideal research platform for elucidating the signalling transduction mechanisms of ncRNAs. In addition, the regulatory mechanisms of ncRNAs, especially lncRNAs and circRNAs,in cellular processes and diseases are highly complex. However, there are few studiesconcerning the nonconventional mechanisms of ncRNAs during the osteogenic differentiation of PDLSCs.

Figure 2 Regulatory mechanisms of noncoding RNAs associated with the osteogenic differentiation of periodontal ligament stem cells.
Due to their osteogenic differentiation capability, PDLSCs show effective potential in the clinical application of periodontium repair and regeneration (Figure 4).However, reports have appeared that are less rigorous in the isolation and identification of PDLSCs. In addition, most current studies of ncRNAs involved in osteogenic differentiation in PDLSCs have focused on the cell levelin vitro; therefore,in vivoexperiments in this field warrant further in-depth exploration. Therefore,whereas interest and investigation in the contribution of ncRNAs to the osteogenesis of PDLSCs have increased considerably, the field is still a long way from understanding the full extent of the contribution of ncRNAs and the mechanisms by which ncRNAs exert their potential effects in this field.

Figure 3 Overview of the role of long noncoding RNAs and circular RNAs during osteogenic differentiation of periodontal ligament stem cells.

Figure 4 Schematic diagram of noncoding RNAs genetic modification-based periodontal ligament stem cells transplantation therapy applications for periodontium regeneration of periodontal disease.
 World Journal of Stem Cells2020年4期
World Journal of Stem Cells2020年4期
- World Journal of Stem Cells的其它文章
- Human umbilical cord derived mesenchymal stem cells in peripheral nerve regeneration
- Ability of human umbilical cord mesenchymal stem cells to repair chemotherapy-induced premature ovarian failure
- lnsights of stem cell-based endogenous repair of intervertebral disc degeneration
- Bone marrow-derived products: A classification proposal - bone marrow aspirate, bone marrow aspirate concentrate or hybrid?
