Brain Banks Spur New Frontiers in Neuropsychiatric Research and Strategies for Analysis and Validation
Le WangYan XiaYu ChenRujia DaiWenying QiuQingtuan MengLiz KuneyChao Chen*h
1Center for Medical Genetics&Hunan Key Laboratory of Medical Genetics,School of Life Sciences,Central South University,Changsha 410078,China
2Child Health Institute of New Jersey,Department of Neuroscience,Rutgers Robert Wood Johnson Medical School,New Brunswick,NJ 08901,USA
3Psychiatry Department,SUNY Upstate Medical University,Syracuse,NY 13210,USA
4Chinese Academy of Medical Sciences and Peking Union Medical College,Beijing 100101,China
5Affiliated Hospital of Guilin Medical University,Guilin 541000,China
6National Clinical Research Centre for Geriatric Disorders,Xiangya Hospital,Central South University,Changsha 410000,China
Abstract Neuropsychiatric disorders affect hundreds of millions of patients and families worldwide.To decode the molecular framework of these diseases,many studies use human postmortem brain samples.These studies reveal brain-specific genetic and epigenetic patterns via highthroughput sequencing technologies.Identifying best practices for the collection of postmortem brain samples,analyzing such large amounts of sequencing data,and interpreting these results are critical to advance neuropsychiatry.We provide an overview of human brain banks worldwide,including progress in China,highlighting some well-known projects using human postmortem brain samples to understand molecular regulation in both normal brains and those with neuropsychiatric disorders.Finally,we discuss future research strategies,as well as state-of-the-art statistical and experimental methods that are drawn upon brain bank resources to improve our understanding of the agents of neuropsychiatric disorders.
KEYWORDS Neuropsychiatric disorders;Brain bank;Postmortem brain;Expression quantitative trait loci;GWAS interpretation
Introduction
Neuropsychiatric and neurological disorders, such as schizophrenia(SCZ),bipolar disorder(BIP),major depression(MD),and Alzheimer’s disease(AD),are the leading cause of disability worldwide[1].However,for more than half a century,a stagnant understanding of their pathophysiology has blocked the development of effective and well-validated neuropsychiatric therapies.Yet,the characteristically high heritability of these disorders should inform us that an earnest understanding of the genetic mechanisms behind these diseases is essential[2,3].Genome-wide association studies(GWAS)are achieving huge successes in identifying disease-associated variants.For example,the Psychiatric Genomics Consortium(PGC;http://www.med.unc.edu/pgc)has identified hundreds of loci associated with SCZ[4],as well as dozens of loci associated with BIP[5]and MD[6,7].
Although many disease-associated variants have been identified,most have small effect sizes and are located in noncoding regions,which hinders interpretation of their functions and disease implications.Quantitative trait loci(QTL)analysis integrates population-based human variation with genomewide molecular information,such as gene expression[8],DNA methylation[9],histone modifications[10],or chromatin states[11].QTL is a possible solution for deciphering the function of non-coding variants[12].Interestingly,most QTL signals show strong tissue specificity[13].For example,the non-coding variant rs199347,associated with Parkinson’s disease exclusively,affects the expression of protein-coding gene GPNMB(Glycoprotein Nmb)in the human brain while sparing other tissues[14].Robust brain bank collections can facilitate the comprehensive molecular profiling needed to advance research in neuropsychiatric disorders.
Many prominent brain projects on neuropsychiatric disorders generated big data at multiple regulatory levels,including epigenetic markers and gene expression.Although these multidimensional data identified numerous functional genomic elements,challenges remain that impede our full understanding of the underlying molecular etiologies of neuropsychiatric disorders and limit our ability to translate this understanding into improving human health.Although brain tissue samples have become a critically valuable resource for neuropsychiatric studies,to our knowledge,there are only a few comprehensive reports on brain bank resources.Therefore,in this review,we present a summary of the most representative brain banks and brain projects,emphasizing how harnessing thesenew resources and technologies can refine our insight into the underlying mechanisms of neuropsychiatric disorders.For example,we will discuss brain expression quantitative trait loci(eQTL)analysis as a methodology to interpret the potential functions of GWAS signals identified in various brain disorders.We also discuss the insights and limitations of current brain studies.Finally,we propose best practices for analyzing postmortem brain samples to more accurately interpret the resulting multidimensional data,thereby augmenting future investigations.
Brain banks
A brain bank is a centralized resource that collects and stores postmortem brain tissues.Brain banks share samples and clinical information with qualified researchers worldwide to advance brain studies in both basic research and clinical trials.Currently,hundreds of human brain banks worldwide are dedicated to the collection of human post-autopsy brain tissues[15].These have been helpful in demystifying brain-related diseases,such as AD,SCZ,BIP,and MD.Although brain tissue collection is the cornerstone for brain studies,obtaining highquality brain tissues can be problematic.To counter this and enable better access,large networks such as the Australian Brain Bank Network,BrainNet Europe[16],NeuroBioBank[17],and the UK Brain Banks Network,share technologies and brain sample information.These brain banks have collectively standardized disease diagnosis and tissue collection procedures[18].Here,we introduce procedures for obtaining high-quality postmortem brain tissue followed by a brief overview of brain banks worldwide and in China.
Working with high-quality postmortem brain tissues
Various factors critically impact the quality of postmortem brain samples[19].For example,an extended time interval between death and acquisition,the postmortem interval(PMI),can lead to RNA degradation[20].Effective and rapid brain tissue acquisition and long-term preservation requires precise and unified manipulation using anatomical,cryopreservation,and slicing technologies.Rapid autopsy programs based on round-the-clock autopsy greatly shorten the PMI.Many important parameters are used to determine brain tissue quality,including brain pH,as well as the integrity of DNA,RNA,and proteins[19].In a strict autopsy environment,which often prolongs the process of sample acquisition,brain pH can notably affect the integrity of RNA and DNA[19].While formalin-fixed samples tender brain DNA relatively ef ficiently,the yields of high-quality RNA is somewhat problematic.It is clear that acquiring and preserving high-quality postmortem brain tissues requires great skill and adherence to standard procedures.
Accurately segmenting brain regions is critical,since biological functions vary by brain regions.There are several brain regions highly related to neuropsychiatric cognitive and emotional dysfunction.For example,the dorsolateral prefrontal cortex(DLPFC)and the hippocampus manage cognitive processes including working memory,planning,and cognitiveflexibility.The striatum can receive glutamatergic and dopaminergic inputs from multiple sources functional,in the cognitive and reward systems.Accurate definitions for landmarks and label boundaries are important based on our assumption of the close correspondence of brain function to anatomy.The human cerebral cortex is difficult to label due to the great anatomical variations in the cortical folds and the difficulties in establishing consistent and accurate reference landmarks across the brain.Brain banks classify brain regions according to the Brodmann atlas,which defines 52 cerebral cortex regions[21].Although there are no clear‘gold standards’for measuring the accuracy of anatomical assignments,it is common to measure consistency across trained human observers and variability across co-registered landmarks.
Brain banks worldwide
Although the study of human brains is as old as medicine,brain banks benefitting neuropsychiatric research today arise from international collaboration,guided by modern principles of ethics,quality,and safety with valid scientific aims.One of the most famous brain banks is the Netherlands Brain Bank(NBB)in Amsterdam(https://www.brainbank.nl/)[16].The NBB was established in 1985 to collect human brain tissues from donors with various neurological and psychiatric disorders and also non-diseased donors.NBB had collected brain samples from more than 4000 donors.Launched in 2001,the BrainNet Europe consortium(https://www.neuropathologie.med.uni-muenchen.de/funktionen/bne/index.html) has 19 members from across the continent.The brain tissues and the corresponding anonymized summary of each donor’s medical records support extensive national and international research projects.North America with a wealth of brain banking resources has over 50 brain banks including the Allen Institute for Brain Science(https://alleninstitute.org/),Harvard Brain Tissue Resource Center(https://hbtrc.mclean.harvard.edu/),and the Stanley Medical Research Institute(http://www.stanleyresearch.org/).Representative brain banks also include the New South Wales Tissue Resource Centre(Australia,https://nswbrainbank.org.au/about/nswbtrc),Tokyo Metropolitan Institute of Gerontology(Japan,http://www.tmig.or.jp/),and the Brain Bank of the Brazilian Aging Brain Study(Brazil,http://www2.fm.usp.br/gerolab_en/index.php).
Brain banks in China
In China,the number of brain samples is quite limited.The creation of Chinese brain banks has recently become a priority for researchers.China’s Han population represents the world’s largest ethnicity and roughly 80%of East Asia’s population;yet brain data from this population is currently understudied and will prove a valuable resource within the global survey.However,brain banking in China is slowly developing,with the China Human Brain Banking Consortium established in 2014 at the International Workshop on Human Brain Banking in China[22].So far,there are nearly one thousand brain samples from dozens of consortium members,including the Xiangya School of Medicine Brain Bank,the Zhejiang University of China Brain Bank,the Chinese Academy of Medical Sciences&Peking Union Medical College Human Brain Bank,and others.The consortium organizes conferences and workshops annually to build up a unified process for brain tissue acquisition and storage,discussing policy for sample sharing,and exchanging experiences and new findings[23].
Evolutionary perspectives can help us better understand the relationship between brain development and disease.Therefore,nonhuman primate(NHP)brain resources play an important role in distinguishing human brain-specific regions.The Nonhuman Primate Reference Transcriptome Resource(http://nhprtr.org/index.html)began in 2010[24].Its goal is to establish an NHP reference transcriptome consisting of transcriptome sequencing data from multiple nonhuman species,including Papio anubis,Pan troglodytes,Macaca fasicularis,Gorilla gorilla,and 11 other non-human primates.Within their protocol,22 tissue types are collected from four brain regions(i.e.,cerebellum,frontal cortex,hippocampus,and temporal lobe).By comparing brain regions of humans to those of non-human primates,Doan et al.was able to identify human-specific social and behavioral traits associated with autistic spectrum disorder(ASD)that are regulated by the human accelerated genomic regions[25].
Brain projects
The collective increase in brain banks globally has spurred a multitude of brain research projects.For most projects,samples are obtained from well-constructed brain banks[26].Brain research projects focus on many different dimensions,including brain development,spatiotemporal gene expression,epigenetic modification,and pathologicalcharacterization of neuropsychiatric disorders.Some of these efforts include,BrainSpan(http://www.brainspan.org/)[27,28],UK Brain Expression Consortium(UKBEC,www.braineac.org/)[29],Genotype Tissue Expression Project(GTEx,https://gtexportal.org/)[30],CommonMind Consortium(CMC,commonmind.org/)[31],BrainSeq(http://eqtl.brainseq.org/)[32],the Religious Orders Study and Memory and Aging Project(ROSMAP, http://www.radc.rush.edu/) [33], PsychENCODE(http://psychencode.org/)[34],and BrainCloud(http://braincloud.jhmi.edu/)[35].They aim to gather genotypic data and data at other regulatory levels for the human brain,to reveal the genetic regulatory mechanisms of the human brain at different levels(Figure 1 and Tables 1-3).
Benefitting from the continual production of data and strengthened by in-depth structured analyses,brain projects are valuable references revealing basic functions as well as molecular and cellular pathologies related to neuropsychiatric disorders.As a source of data,each brain project offers unique design features and advantages for specific research aims.For instance,the GTEx project,which collects samples from nondisease tissue sites,including but not limited to the brain,focuses on tissue specificity of gene expression,cross-tissue gene expression regulation,and genetic variations that contribute to complex diseases and quantitative traits in humans[30].The UKBEC,which collects samples from across a wide-range of brain regions,up to 12 regions per donor,focuses on the regulation and alternative splicing of gene expression[29].BrainCloud[35]and BrainSpan[27,28]focus on spatiotemporal gene expression regulation during the development of the human brain from embryonic to adult stages.Although BrainCloud is superior in terms of sample size,BrainSpan includes more brain regions and types of sequencing data,such as miRNA expression.
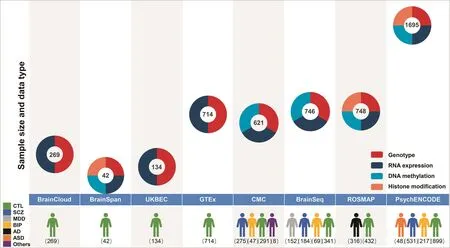
Figure 1 Overview of the representative brain projects

Table 1 Number of individuals across developmental stages per brain project
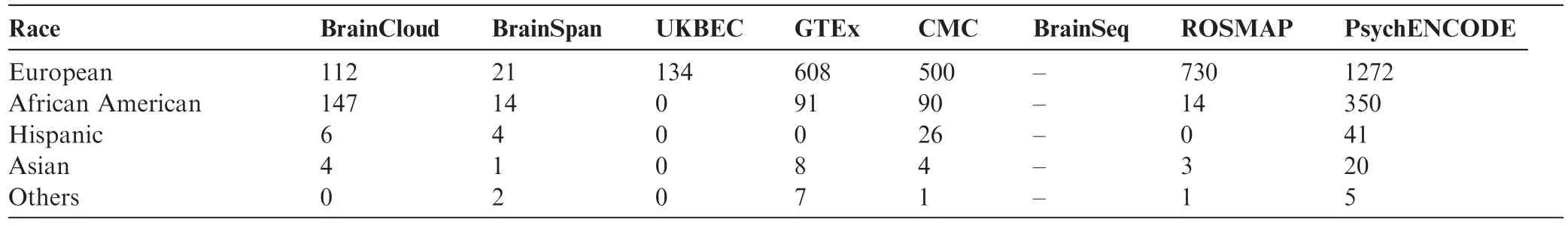
Table 2 Number of individuals by race per brain project
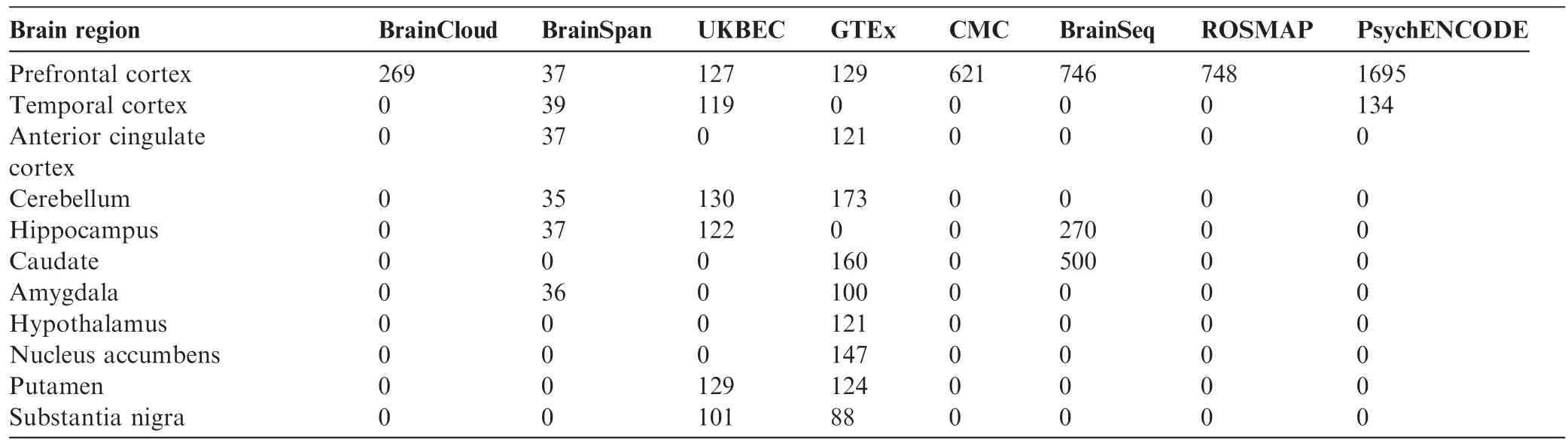
Table 3 Number of samples per brain region per brain project
Other brain projects include samples from donors with or without neuropsychiatric disorders,exploring the differences between brain features of patients and those of controls.The Religious Orders Study(ROS)[36]and the Memory and Aging Project(MAP)comprise the ROSMAP project[37],a longitudinal,clinical,and pathological cohort study of aging and dementia.The ROS component focuses on data from various conditions of dementia within a limited population,while the MAP project focuses on reduced cognitive and motor function and disease risk of those with AD within a more varied population.CMC and BrainSeq[31,32]focus on neuropsychiatric disorders,including SCZ,BIP,ASD,and MD,by comparing diseased samples with controls.The BrainSeq project seeks to identify therapeutic drug targets for neuropsychiatric disorders by understanding the genetic and epigenetic regulations across the human lifespan.The PsychENCODE project[34]makes an extensive,"multidimensional"genetic and epigenetic dataset available to the public,derived from the tissue samples of postmortem healthy and diseased human brains.The project characterizes disease-associated regulatory and genetic features within pathological models,focusing initially on ASD,BIP,and SCZ[38-40].Current data generated from the PsychENCODE project include:chromatin immunoprecipitation following next-generation sequencing(ChIP-seq),RNA-seq,whole-genome bisulfite sequencing(WGBS),miRNA sequencing(miRNA-seq),isoform sequencing(IsoSeq),assay for transposase accessible chromatin with high-throughput sequencing(ATAC-seq),enhanced reduced representation bisulfite sequencing(ERRBS),single nucleotide polymorphism(SNP)genotypes,array methylation,and reverse phase protein array(RPPA).
The major findings using postmortem samples from brain projects are summarized in Table S1.These data provide important insights into the contribution of genetic and epigenetic factors to mechanisms underlying neuropsychiatric disorders.Particularly,the BrainSeq Consortium performed RNA-seq on 495 postmortem brains with ages across the human lifespan,including 175 samples from SCZ patients and 320 controls[41].Through integrative analyses,this consortium demonstrates that 48.1%SCZ GWAS risk variants are associated with expression of nearby genes,and 237 differentially expressed genes implicated in synaptic processes are regulated in early brain development.The earlier study on the epigenetic landscape of frontal cortex in patients with SCZ[42]shows that SCZ-associated CpGs strongly correlate with fetal development stage rather than the adult stage of the brain.These results reveal potential SCZ pathogenesis in gene expression and DNA methylation during brain development and maturation.Moreover,recent studies by the PsychENCODE project have identified cell composition and maturation leading to spatiotemporal transcriptomic variation patterns in human and macaque brains[43].They also observe associations of neuropsychiatric diseases with epigenetic markers[38],QTLs[39],and isoform-level changes[44].For example,they have identified several interesting targets,including DGCR5 and POU3F2,which play essential roles in regulating SCZ-related genes at the network level[45,46].These postmortem studies provide important insights into the genetic architecture for robust and informative models of neuropsychiatric disorders,which will help in devising strategies for novel therapeutics interventions.
Strategies and execution
Unarguably,postmortem brain resources are valuable in revealing the biological underpinnings of neuropsychiatric disorders;however,unravelling the full potential of multidimensional brain data is still a great challenge.One promising strategy employs QTL analysis,which integrates populationbased human variations with genome-wide molecular information(e.g.,gene expression,DNA methylation,histone modi fication,and chromatin states).Widely used,QTL captures the associations between genetic variants and gene expression.For instance,QTL can beused to investigatevariantsat cis-regulatory elements,such as transcription factor-binding regions,which confer differential expression of target genes.Combined with GWAS,QTL studiesinterprethow disease-associated variants may contribute to molecular traits and disease susceptibility.In this section,we will discuss eQTL specifically,summarizing the key steps for pre-processing of brain gene expression data,highlighting important issues in eQTL analysis,explaining how to use eQTL to interpret GWAS signals,and finally,introducing cutting-edge experiments to validate regulatory signals(Figure 2 Overflow of the research strategies and methods).
Pre-processing brain gene expression data
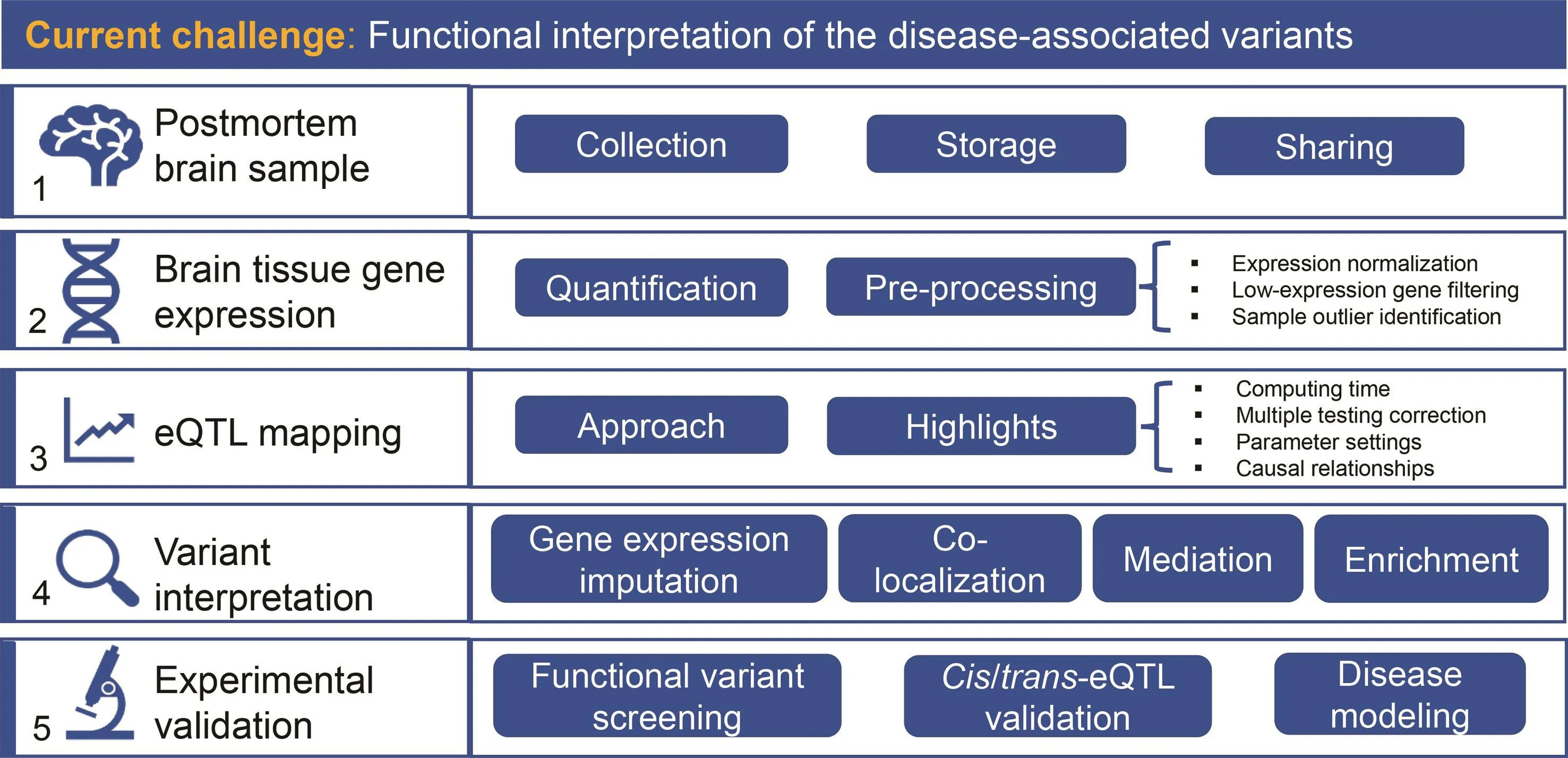
Figure 2 Overview of strategies and methods in neuropsychiatric studies
Although laborious,data pre-processing is essentially the first step to ensure proper and efficient data modelling.A clean,software-compatible format will ensure reproducible results and save hours,even days,of data analysis[47].Variable reporting of gene expression can arise from biological factors and technical variations.To distinguish biological variations from confounding factors,technical factors(e.g.,batch effects)must be removed or adjusted.Major pre-processing steps include gene expression normalization and filtering,sample outlier identification,and covariate correction.Because strategies in the human brain studies are the major focus of this article,we will only cover the key steps that may alter the quality of brain gene expression results.Comprehensive guidelines for gene expression data analysis are well discussed elsewhere[48,49]and are beyond the scope of this review.
The first key step is gene quantification and filtering.Tools for quantification are widely available,such as Cufflinks[50],eXpress[51],Flux Capacitor[52],kallisto[53],RSEM[54],Sailfish[55],and Salmon[56].Each tool can accurately assign reads to transcripts and quantify expression.These functions are vital for interpreting tissue-specific expression patterns in the brain[57].However,the criteria for poorly expressed genes vary across studies.For instance,PsychENCODE project filters genes with transcript per million(TPM)<0.1 in more than 25%of samples[58].
The second key step is sample outlier removal.Samples with a high degree of poorly expressed genes or gene expression patterns distinct from other samples are removed.This step can be carried out in dimension reduction analysis such as principal component analysis(PCA)and multidimensional scaling(MDS).Network concepts such as standardized connectivity(the overall strength of connections between a given sample and all of the other samples in a network)are also used to confirm sample outliers within a group[59].
The third key step is controlling covariates,including both known and unknown covariates.Known covariates can be either technical,such as batch effects,or biological,such as sex and age.Some biological covariates have been ignored by earlier research,leading to potentially confounding results.For instance,cell-type composition is one such common problem:since bulk-tissue RNA-seq only measures the average behavior,it is unable to capture cellular heterogeneity,which makes the observed changes in gene expression reflect only changes in cell-type composition,rather than fundamental changes in cell states[60].Therefore,cell numbers and ratios of multiple cell types are important biological covariates,that affect brain gene expression profiles,since different cell states rather than cell type composition reflect distinct biological activities and gene expression patterns.Another covariate that is critical but often neglected is drug treatment history.Gene expression can vary dramatically across therapeutic courses.The unknown factors,also called hidden determinants,can reduce the power to find eQTLs.Surrogate variable analysis(SVA)[34]or probabilistic estimation of expression residuals(PEER)[61]can calculate unknown sources of variation,followed by a linear regression model to remove them.One could choose ComBat[62](in R package sva)to remove the batch effects;finally,a linear regression model will remove the confounding factors.
Pitfalls and promises in eQTL analysis
The aim of eQTL analysis or eQTL mapping is to characterize associations between the expression of corresponding genes and SNPs,thereby isolating specific regulatory regions within the genome.A variety of approaches have been proposed,including using linear regression,ANOVA,and non-linear models.Some approaches also account for pedigree and other confounding factors[63],integrating known functional elements[64],or considering allelic imbalances[65].FastQTL,for instance,features expansive permutations that refine P values and reduce computational burden.
Several issues should be highlighted in eQTL analysis.Thefirst is computing time.Pairwise association compares up to one million genetic variants to tens of thousands of genes,making analysis computationally intensive,especially when employing a non-linear model on a larger dataset.Secondly,multiple testing corrections become necessary for many of the tests performed.One common solution is to calculate the false discovery rate for each SNP-gene pair.Furthermore,separating the cis-eQTLs and trans-eQTLs is crucial,since local variants may regulate gene expression much more than distal variants.However,this correction alone is too strict because thosetestsarenotbiologically independent.Therefore,permutation-based methods,which create the null distribution of associations by tens of thousands of permutations,were developed to set up an effective threshold for identifying statistically significant eQTLs.Third,parameter settings can be a critical factor when comparing eQTLs across multiple studies.For example,the distance between SNPs and gene locations is used to differentiate cis-eQTL and trans-eQTL signals,which could be defined as 1 Mb,5 Mb or 10 Mb in different studies.Varied distance settings may lead to different statistical burdens for SNPs located in regions ranging from 1 to 10 Mb and result in variable outcomes.The customized cut-off threshold for minor allele frequency(MAF)may also cause the loss of some true signals.Fourth,some eQTLs have such strong correlations with gene expression that they may not prompt gene expression changes.In other words,those genetic variants may be correlated with the causal variants due to linkage disequilibrium or other factors.Both statistical and experimental approaches have been proposed to solve this problem[66,67];either ways,it is critical to identify true causal variants when integrating eQTL and GWAS results[68].
Interpreting GWAS signals
GWAS variants can increase or decrease gene expression,a culprit behind the etiology of many diseases;QTL helps us interpret how non-coding GWAS variants work.Several kinds of methods,each with unique principles,have been developed to integrate GWASs and eQTL results(Table 4).One type of method is based on gene expression imputation,such as PrediXcan[70]and transcriptome-wide association study(TWAS/FUSION)[71].These methods estimate the genetically regulated component of expression using reference transcriptome datasets such as GTEx[30],GEUVADIS[8],and DGN[85]among others to build a database of prediction models.For each new genotype data,these methods impute gene expression and then correlate that gene expression to a trait of interest to identify trait-associated genes.The second group investigates the co-localization of GWAS causal variants and eQTL causal variants.For example,COLOC[72],MOLOC[73],ENLOC[86],HyPrColoc[74],and Sherlock[75]use a Bayesian statistical framework to integrate GWAS summary data and eQTLs to estimate the causal variants,and eCAVIAR[78]considers multiple causal variants within one locus.Other groups include enrichment methods,such as S-LDSC[82]and eQTLEnrich[81],and mediation methods.Summary data-based Mendelian Randomization(SMR)[66]and generalized SMR(GSMR)[84]test whether the effect of a GWAS SNP on a specific trait has been mediated by the expression of a gene.
While using eQTL to interpret GWAS results is a good way to understand gene regulatory mechanisms,it is not without limitations.First,for some diseases if the most relevant tissue/-cell types or developmental stages are not available in eQTL analysis,we can find neither the true genetic regulation nor the related genes.Second,gene expression is only one dimension of genetic regulation.If the biological mechanism is independent of gene expression levels but affects other regulatory cascades,such as splicing,chromosome accessibility,or ribosome profiling,eQTL alone will not be enough to explain the underlying processes.Third,QTL and GWAS focus on common variants,therefore they cannot capture rare variants with higher effect sizes in gene expression[87].
Experimental approaches to characterize functional variants
After identifying disease risk variants or regulatory elements using the aforementioned bioinformatics analysis methods,the next step is to characterize the function of the variants.
To validate risk variants as the eQTL signal,using highthroughput and sensitive methods to measure their effect on gene expression is a widely adopted approach.As a favored method,reporter gene assay screening validates whether functional elements with eQTL signals regulate target gene expression,by cloning the regulatory elements into an expression reporter vector[74].Whereas reporter assays validate regulatory functions of variance in vitro,CRISPR can be used to validate regulatory functions of the variance within native chromosome regions in vivo.For instance,Diao et al.used a CRISPR tiling-deletion-base genetic approach to identify some cis-regulatory elements in mammalian cells[88].Furthermore,high throughput CRISPR screening systems,such as the CRISPR-Cas9,have been used to investigate the effect of the regulatory variance on the downstream target genes[75,78,81,82,84].Recently,studies have refined the resolution of this technique,including the dCas9 fusion APOBEC1(Apolipoprotein B mRNA Editing Enzyme Catalytic Subunit 1)/TadA(tRNA-specific adenosine deaminase)-mediated ef ficient single base mutation system[69,87].While CRISPR technology has these advanced capabilities,it is not without limitations.For instance,inconsistencies such as off-target genome editing(i.e.,inducing unwanted allelic variances)have been problematic to date[89].Nonetheless,CRISPR has tremendous potential for single base screening and clinical applications.We are confident that CRISPR will mature into a dependable tool for correcting genetic variation in the future.
To understand the influence of risk variants on gene expression,several productive tools have been developed.For the chromatin states,ChIP-seq is an efficient genome-wide method to identify the transcription factor binding sites in open chromatin regions,including promoter,enhancer and other transcription active elements.Based on the principle of ChIP-seq,a series of targeted chromatin DNA sequencing technologies have been developed(e.g.,DNase-seq,MNase-seq,FAIRE-seq and ATAC-seq).For example,Forrest et al.revealed the function of non-coding GWAS risk variants using ATAC-seq data from neurons derived from SCZ patient induced pluripotent stem cells(iPSCs)[90].Chip-related technology can help us toannotate and interpret the functionality ofdisease-associated non-coding variants.Data on DNA-protein binding generated by sequencing technologies requires validation using in vitro methods,including the electrophoretic mobility shift assays(EMSAs).However,the throughput of the EMSA-based experiments is limited.To improve the throughput of this in vitro validation,mass spectroscopy proteome-wide analysis of SNPs(PWAS)can be applied for screening genetic variants for differential transcription factor binding[91].
Risk variants located in the untranslated region(UTR)and intronic regions may also contribute to disease through posttranscriptional regulation,such as splicing,RNA stability,or non-coding regulation. High-throughput analysis of RNA isolated by cross-linking immunoprecipitation sequencing (CLIP-Seq) could be used to map protein-RNA binding site or RNA modification site in vivo [92-94]. This technique can reveal risk variants that affect gene expression at the posttranscriptional level. For example, Eric T. Wang used RNAseq and CLIP-seq to reveal the transcriptome-wide regulation of pre-mRNA splicing and mRNA localization in myotonic dystrophy [95].
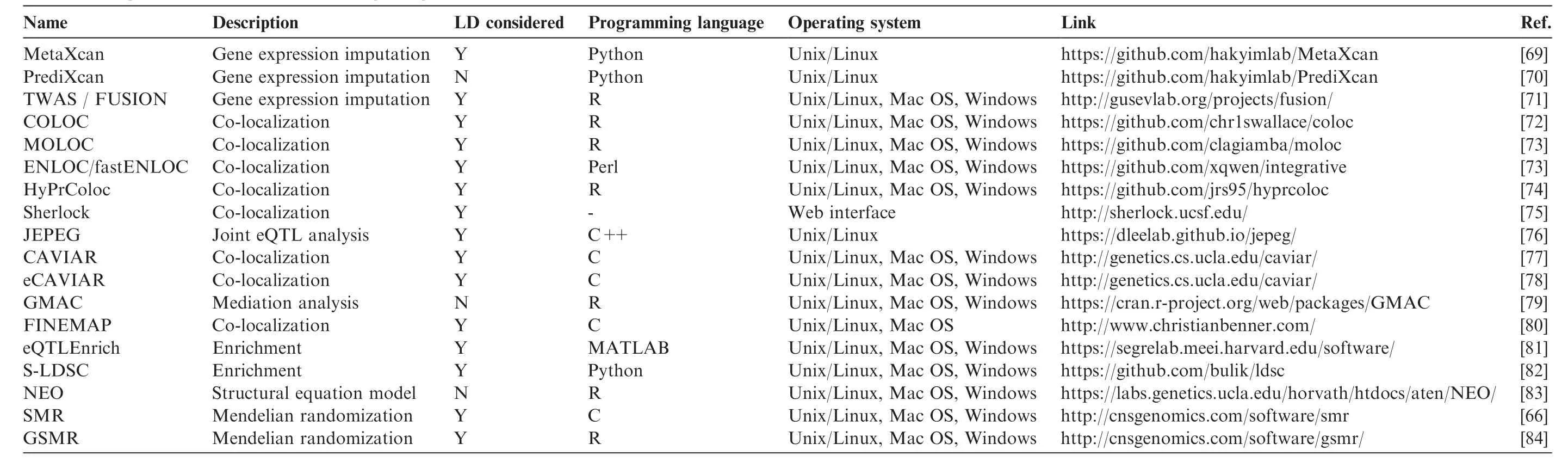
Table 4 Algorithms and software for integrating GWAS and eQTL data
It is important to note that risk variants may not necessarily affect expression of the nearest gene.Disease risk variants may also affect expression of distal genes through long-range chromatin interactions[96-98].The interaction of chromatinspecific regions can be explored by classic chromatin conformation capture(3C)techniques.This 3C-based technology involves cross-linking chromatin interaction sites,using genome DNA cleavage with a restriction enzyme and a ligation reaction to join cross-linked DNA fragments.Chromatin interactions at specific candidate loci could be further validated by polymerase chain reaction(PCR)[99].For example,Panos Roussos et al.demonstrated physical interactions between the CACNA1C eQTL risk locus and distal regulatory elements using 3C techniques in prefrontal cortex[100].
The next step is to explore disease-associated phenotypes of genetic risk variants by establishing cellular models or animal models.For example,human iPSCs(hiPSCs)research detects molecular and cellular phenotypes(e.g.,migration,proliferation,and electrophysiology)together with the genetic background of specific patients.Moreover,the 3D culturing of pluripotent stem cells produces organoids,demonstrating their remarkable capacity for self-organization and differentiation.This approach can be used to study human brain specific features and the mechanism of neurodevelopment and neuropsychiatric disorders.For example,Marina Bershteyn et al.used human-derived cerebral organoids to model the cellular features of Miller-Dieker syndrome caused by 17p13.3 deletion[101].While animal models differ from humans in terms of genetic background,they resemble the spectrum of human disease phenotypes,ranging from tissue and organ to behavior.Those two models,when combined with postmortem brain data,may unlock the mysteries of risk variant function and increase the probability of decoding the pathology of neuropsychiatric diseases.
Future directions
In this review, we summarized the most representative brain banks and brain projects worldwide, supporting a multidimensional understanding of neuropsychiatric disorders from pathology, genetic, and gene expression perspectives. Brain banks and projects are establishing research resources and building coalitions to reduce the incidence and impact of neuropsychiatric disorders. Multidimensional data collected using brain bank resources facilitate the study of complex neuropsychiatric disorders, as brain banks are increasingly linked to important sources of clinical information. Different brain projects use brain bank samples to generate a wide spectrum of data types and serve as an important resource to promoting brain research. Developing advanced research methods and experimental validation of findings increases our capability of finding true causal signals of neuropsychiatric illnesses.
Postmortem brain samples have lent profound insight into genomic,transcriptomic and epigenomic studies,however brain disorder research faces many challenges.Various cell types from different brain regions form specific neural circuits that govern complex behaviors.Most brain studies include samples from different brain regions and use the bulk brain tissue as a whole,which obviously contains many cell types,such asneurons,astrocytes,microglia,and oligodendrocytes.Single-cell studies are increasingly needed to achieve higher resolution in detailed genomic insights.Some recent studies have been used single-cell methods to isolate specific cell types from healthy human brain tissue to characterize human brain development[102,103].Heterogeneity in medical treatment is one confounding factor that can affect gene expression profiles and some epigenetic marks.Almost every psychotic patient has a long history of drug therapy,but individuals without neuropsychiatric disorders may not,which may result in possible false-positive findings.Furthermore,integrating the drug history relies on obtaining hospital medical records or selfreporting,both of which can be unreliable.For example,patients may refuse to take prescribed medications,while others may not be able to accurately recall their medication history.Directed toxicology testing for each sample is the best solution but may not be practical due to the many types of antipsychotic drugs available and the high expense involved.Moreover,smoking and drinking history,state of death(e.g.,unexpected death,expired while asleep,unconsciousness,fever and hypoxia)are also confounding factors for postmortem gene expression and other studies[104,105].Consider this necessary information when collecting samples.
One vital but challenging aspect of brain collection is the use of fetal and infant brains.In most banks,donated brains come from aged individuals,appropriate for the research of neurodegenerative diseases.For neurodevelopmental diseases,such as autism,SCZ,and intellectual disability,however,fetal and infant brain samples are critical for investigating disease etiology.So far,only a few banks have prenatal samples,and their samples sizes are relatively small.Including fetuses with lethal defects and those with defects not affecting brain function,identified through prenatal genetic screening,could increase available resources.Another solution would be using iPSC-derived neurons or other brain cells to model the very early stages of brain development.Combining these strategies,we can characterize the temporal regulatory landscape of brain development and genomic aberrations related to psychiatric illnesses.
Recently,it has been suggested that all postmortem brain studies are underpowered to correct for genetic and phenotypic heterogeneity[106].This begs the question,how can these studies derive from the brain banks with limited sample sizes achieve enough statistical power?One solution is in more accurately defining disease-related phenotyping and levels of disease taxonomy.For example,in BIP,only about 30%of patients respond to lithium[107,108],and a portion of patients have DLPFC or hippocampal volume abnormalities[109-112].Classification of these disease subtypes improves the understanding of disease phenotype.Availability of shared data is another big issue often limiting the power needed for research into neuropsychiatric disorders.With more and more data generated and released,an open public and user-interactive data center is needed to collect and to manage all the repositories.Our group established the Brain EXPression Database(BrainEXP,http://www.brainexp.org/)focusing on brain gene expression patterns in various regions,by sex and age[113].This database currently includes 4567 brain samples of 2863 normal individuals and will integrate approximately the same number of patient samples in the near future.These combined efforts hold the promise of powering brain studies adequately.
In conclusion,given the expanding framework of brain bank and brain project networks,we can improve exploration into the molecular regulatory mechanisms of neuropsychiatric disorders and facilitate research toward new avenues of treatment.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China(Grant Nos.31571312,31970572,and 81401114),the National Key R&D Project of China(Grant No.2016YFC1306000),Innovation-driven Project of Central South University(Grant Nos.2015CXS034 and 2018CX033)to CC.We thank Dr.Chunyu Liu from SUNY Upstate Medical University,Dr.Xiaoxin Yan from Central South University,Dr.Chao Ma from the Chinese Academy of Medical Sciences and Peking Union Medical College,and Dr.Zhiping Pang from Child Health Institute of New Jersey for critical reading and comments.
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.02.002.
 Genomics,Proteomics & Bioinformatics2019年4期
Genomics,Proteomics & Bioinformatics2019年4期
- Genomics,Proteomics & Bioinformatics的其它文章
- Big Data and the Brain:Peeking at the Future
- GliomaDB:A Web Server for Integrating Glioma Omics Data and Interactive Analysis
- PsyMuKB:An Integrative De Novo Variant Knowledge Base for Developmental Disorders
- Identification of Cognitive Dysfunction in Patients with T2DM Using Whole Brain Functional Connectivity
- Machine Learning to Detect Alzheimer’s Disease from Circulating Non-coding RNAs
- Translational Informatics for Parkinson’s Disease:from Big Biomedical Data to Small Actionable Alterations
