Development of novel microsatellite markers for Holothurian scabra (Holothuriidae), Apostichopus japonicas(Stichopodidae) and cross-species testing in other sea cucumbers*
SHANGGUAN Jingbo (上官靜波) , LI Zhongbao (黎中寶) ,
1 Fujian Provincial Key Laboratory of Marine Fishery Resources and Eco-Environment, Xiamen 361021, China
2 Fisheries College, Jimei University, Xiamen 361021, China
1 INTRODUCTION
Sea cucumbers (Echinodermata; also known as bêche-de-mer, trepan, or gamat) are soft-bodied,flexible, elongated and worm-like marine organisms that have been harvested for food and medicinal purposes over many centuries in Asian countries including China and Malaysia (Taiyeb-Ali et al.,2003; Bordbar et al., 2011). Sea cucumbers tend to live on the sea floor in deep waters and usually feed on plankton and seabed algae (Conand, 1990).Increasing demand for trepang and a steady increase in price have led to a worldwide intensification of artificial breeding and wild harvesting of sea cucumbers (Conand, 2006).
Apostichopusjaponicasis the only temperate sea cucumber found in Chinese waters, yet the species has a wider distribution along the coasts of Japan,Korea, and Far Eastern Russia. Over the past decade,large-scale aquaculture of the species throughout China has developed (Li, 2009). Conversely,Holothuriascabrais an abundant species, is widely distributed in shallow soft-bottom habitats throughout the Indo-Pacific, and is the only tropical holothurian species currently mass-produced in hatcheries(Battaglene and Seymour, 1998; Xia et al., 2016). The body of research regarding population genetics and aquaculture of these two species is increasing in size in response to a commercial over-exploitation of most wild stocks and an increasing market demand (Conand and Bryne, 1993; Tian et al., 2008). Selection of brood stocks with optimal traits, such as rapid growth and disease resistance, is urgently needed to alleviate multiple conservation concerns and achieve sustainable sea cucumber aquaculture.
Microsatellite DNA markers, also called simple sequence repeats (SSR), consist of multiple copies of 1–6 base pairs (bp) that occur as highly repetitive elements in all eukaryotic genomes, as well as in some prokaryotes and eubacteria (Tautz, 1989; Liu and Cordes, 2004). These molecular markers are suitable for use in species with relatively limited existing genomic information and for which collection might be limited due to protected status (for example,endangered species). Although Single Nucleotide Polymorphism (SNP) markers are considered the most powerful for population studies involving genome mapping and identification of candidate genes for Quantitative Trait Locus analysis, they are expensive to develop. It is currently not clear that SNP markers will become popular in aquaculture genetics due to the great financial investment (Liu and Cordes, 2004). We believe that microsatellite markers are appropriate for genetics research, especially for primary genome mapping, and thus use them here.
2 MATERIAL AND METHOD
2.1 Sample materials and DNA extraction
Thirty adults each ofH.scabraandA.japonicaswere collected from Chinese waters in October 2014(H.scabrafrom Sanya in Hainan Province andA.japonicasfrom Qingdao in Shandong Province). A total of thirty genomic DNA samples of high quality were extracted from the body wall of freshly collected individuals using the TIANamp Marine Animals DNA Kit (Tiangen Biotech, Beijing, China). The resulting DNA was purified using the EZNATMCycle-Pure Kit (Omega Bio-Tek, Norcross, GA, USA).
2.2 SSR-enriched library construction and primer design
Twenty μL of high-quality genomic DNA were sampled from a 100-ng/μL gene pool (a mixture of five randomly selected samples from the 30 collected)and used for the construction of a microsatellite library. The library was constructed following a modified version of the fast isolation method using Amplified Fragment Length Polymorphisms of sequences containing repeats (Zane et al., 2002). Two sets of SSR primers forH.scabraandA.japonicaswere developed. The microsatellite fragments were then captured by streptavidin-coated magnetic beads(Promega Corporation, Madison, WI, USA) and purified by eluent. SSR-enriched libraries were constructed after amplification by polymerase chain reaction (PCR) and then ligated into a PMD19-T vector (4:1 library volume:vector volume) (TaKaRa,Shiga, Japan). Next, the products were transformed intoEscherichiacoliDH5a strains (Tiangen Biotech)for positive monoclone selection on LuriaBertani agar medium with ampicillin. Satisfactory fragments with sizes 400–1 000 bp were selected and sent for sequencing to Life Technologies (Carlsbad, CA,USA). The reads containing microsatellite motifs of at least five repetitions of 1-6 bp were screened by the software SSR hunter 1.3 (Li and Wan, 2005). Finally,the SSR primers were designed from applicable flanking sequences by the software Primer Premier 5.0 (Lalitha, 2000).
2.3 Specific and polymorphic primers screening and verification
The PCR annealing temperature (Ta) for each primer was optimized by a gradient temperature PCR.PCR amplification was conducted in a total volume of 10 μL and using a mixed genomic DNA pool. After preliminary screening of all primers, the amplificationspecific primers with matched target strips were tested for polymorphisms using a panel of genomic DNA from 30 individuals at the specificTa(Tables 1 and 2).The amplicons were checked using 6% polyacrylamide gels in a vertical Sequi-Gen Sequencing Cell (Bio-Rad, Hercules, CA, USA) at a constant temperature of 50–55°C. Additionally, a 10-bp DNA ladder(Invitrogen, Carlsbad, CA, USA) was used as a size standard. The gels were visualized by silver staining and analyzed by visually counting the alleles.
2.4 Cross-species amplification
A total of 90 microsatellite loci were selected for cross-species amplification (Table 3). Four individuals from each of the four species were used in the amplification test. The PCR conditions were performed as previously described and the results were visualized using 2% agarose gel with a 50-bp DNA ladder (Invitrogen, Carlsbad, CA, USA). The loci were considered successfully amplified when one band of the expected target size was present.

Table 1 Basic genetic information for 16 microsatellite primers in H. scabra (sample size=30 individuals)
2.5 Relevant data analysis
The basic population genetic information about each microsatellite locus inH.scabraandA.japonicaswas quantified using a combination of software.Specifically, MICRO-CHECKER (v.2.2.3, van Oosterhout et al., 2004) was used for null alleles and scoring error assessments; CERVUS (v.3.0,Kalinowski et al., 2007) was used for to calculate the number of alleles (Na) per locus and the polymorphism information content (PIC); and POPGENE 32 (v.1.32,Yeh et al., 2000) was used to estimate observed heterozygosity (Ho), expected heterozygosity (He),genotypic linkage disequilibrium (LD), and deviation from Hardy-Weinberg equilibrium (HWE) for each locus. Critical significance values were adjusted for multiple comparisons by a modified false discovery rate (B-Y FDR) correction when necessary (Narum,2006).
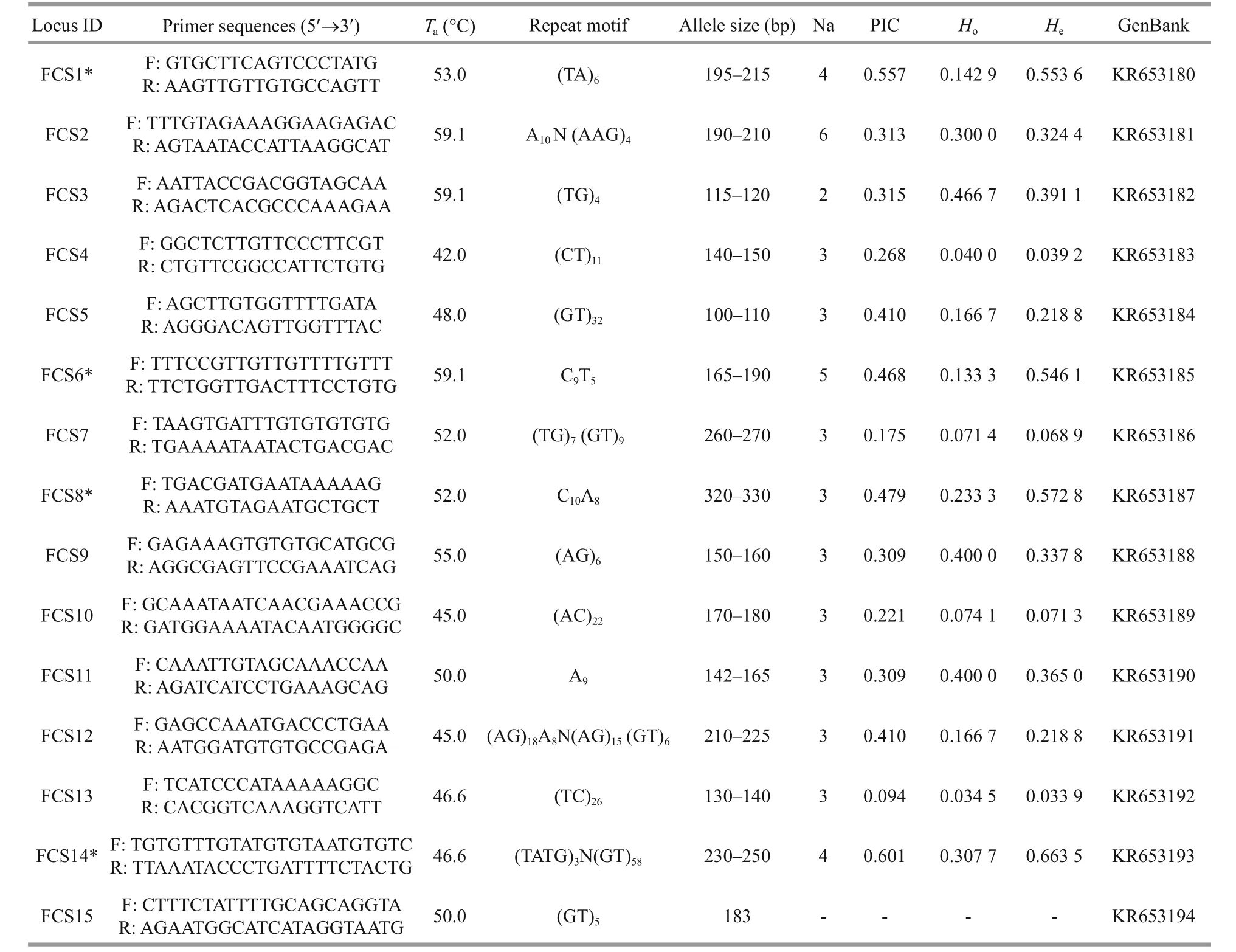
Table 2 Basic genetic information for 15 microsatellite primers in A. japonicus

Table 3 The microsatellite loci used in the cross-species amplification
3 RESULT AND DISCUSSION
3.1 Development and sceening of SSR markers in H. scabra and A. japonicas
H.scabra: Three-hundred positive clones were randomly select to sequence from the microsatellite enrichment library ofH.scabraand 240 successful sequences were achieved. The positive rate of cloning was therefore 80%. Twenty microsatellite loci,including 16 polymorphic loci and 4 monomorphic loci, were isolated from the designed 70 pairs of specific primers. Among the 16 polymorphic loci, the Na ranged from three to eight.Howas 0.033 3–0.517 2 andHewas 0.032 8–0.690 1. Fitch et al. (2013) also developed 18 microsatellite markers forH.scabra,and found Na ranging from two to 28. The reason for the difference in the number of alleles may be the geographic origins of the samples or the numbers of individuals used to estimate Na. Across all the loci,PIC ranged from 0.032 to 0.712. Six polymorphic loci were in the middle of the range (0.25<PIC<0.50) and five loci were highly polymorphic (PIC>0.50),according to the judgment standard (Botstein et al.,1980). Finally, 14 of the polymorphic loci were shown to not deviate from HWE after B-Y FDR correction,whereas two of the loci did deviate from HWE(CHS2* and CHS11*,P<0.014 790).
A.japonicas: One-hundred and forty-two positive clones (400–1 000 bp) were randomly selected from the microsatellite enrichment library ofA.japonicusand generated 80 successful sequences, for a rate of positive cloning of 56.3%. Fifteen microsatellite loci(14 polymorphic, one monomorphic) were isolated from 36 designed pairs of specific primers. Among the 14 polymorphic loci, the Na range was 2–6 and the mean was 3.43, which is lower than in previous studies, such as Chen et al. (2013) (5.53), Peng et al.(2012) (7.00), Zhan et al. (2007) (5.27) and Kanno et al. (2005) (6.65). The number of repetitions or the length of the SSR repeat unit may have led to this phenomenon. Furthermore, the polymorphisms of SSR loci in noncoding regions tend to be higher because reduced evolutionary constraints (Serapion et al., 2004), and thus genome location may have played a role.Howas 0.034 5–0.466 7 andHewas 0.033 9–0.663 5. PIC ranged from 0.094 to 0.679, and 11 of the polymorphic loci had either medium or high rates of polymorphisms. Finally, 10 of the polymorphic loci did not deviate from HWE after B-Y FDR correction, but four of the loci did deviate from HWE(FCS1*, FCS6*, FCS8*, FCS14*;P<0.015 377).
Summing across both species, 30 polymorphic microsatellite loci were tested and 24 were found to be in accordance with HWE (Tables 1 and 2). An additional five monomorphic microsatellite loci were screened (Tables 1 and 2). The results of these analyses indicate the potential utility of these markers for sea cucumber population genetics and parentage research.
3.2 Cross-species Transferability of SSR markers
Microsatellite markers are co-dominant and highly polymorphic, and the methods used with microsatellites are reproducible and transferable to related species, making them a powerful tool for analyzing population structure and genetic diversity(Varshney et al., 2005). The transferability ratio for of the 90 sea cucumber SSR markers is presented in Table 4. The success rate of cross-species amplification of SSR loci differed among the four species, but was generally low (less than 50%). Only four loci(KR653157, KR653159, KJ875909 and KJ875911)developed forS.horrenswere successfully applicable in all three of the other species.
Overall, the universalities of the primers were poor due to the low transferability ratios (3.45%–46.67%,mean=24.88%). However, similarly low rates of transferability were found in other taxonomic groups,for example among species of theIndiranagenus(mean=21.2%) (Nair et al., 2012). Some taxonomic groups show high cross-species amplification success rates, e.g.Begonia(50%–100%) (Chan et al., 2015),Allium(about 50%) (Lee et al., 2011),Cedrela(62.8%), (Soldati et al., 2014), and Mullidae (67%–94%) (Vogiatzi et al., 2012). different levels of transferability may results from variable species diversity among taxonomic groups. In addition, the genetic distance between the source and target species has a negative effect on the transferability of SSRs(Luo et al., 2015). A low rate of success in crossspecies SSR amplification can also be due to the conservation of the flanking regions surrounding the SSR (Balloux et al., 1998), a large genome size(Barbará et al., 2007), or evolutionary divergence between the target and source species (Primmer et al.,2005; Nair et al., 2012). Conversely, the length of the microsatellite repeat in the source species may have a positive effect on the cross-species transferability(Primmer et al., 2005; Shikano et al., 2010).
In general, the SSR markers presented here could be used to identify individuals among these four species, resolve taxonomic uncertainties, and assess genetic diversity of threatened and endemic sea cucumbers.
4 CONCLUSION
Many taxonomic groups, including sea cucumbers,lack easy methods for identifying individuals to visually similar species. The microsatellite loci developed here have the potential to play an important role in aiding species identification of sea cucumbers,at least among the four species tested. These SSR loci provide a PCR-based, non-destructive, accurate, and simple test to identify individuals to genetically different but morphologically similar species of sea cucumbers. In addition, the microsatellite markers

Table 4 Cross-species amplifications of the microsatellite loci
To be continued
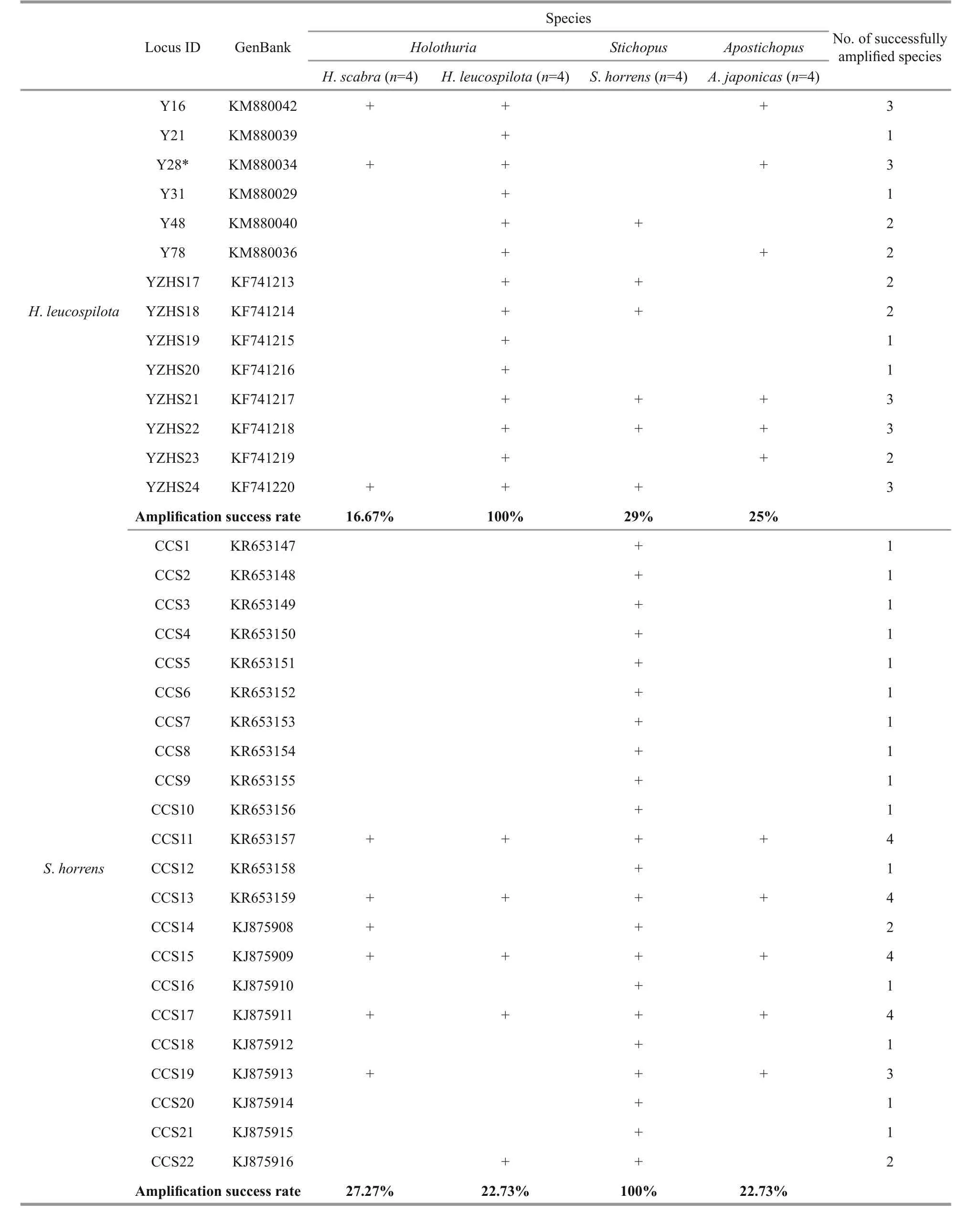
Table 4 Continued
To be continuedidentified here, including the polymorphic and monomorphic loci, will increase the limited population genetic information available forH.scabraandA.japonicus. These results will aid sea cucumber resource investigations and genetic resource conservation.
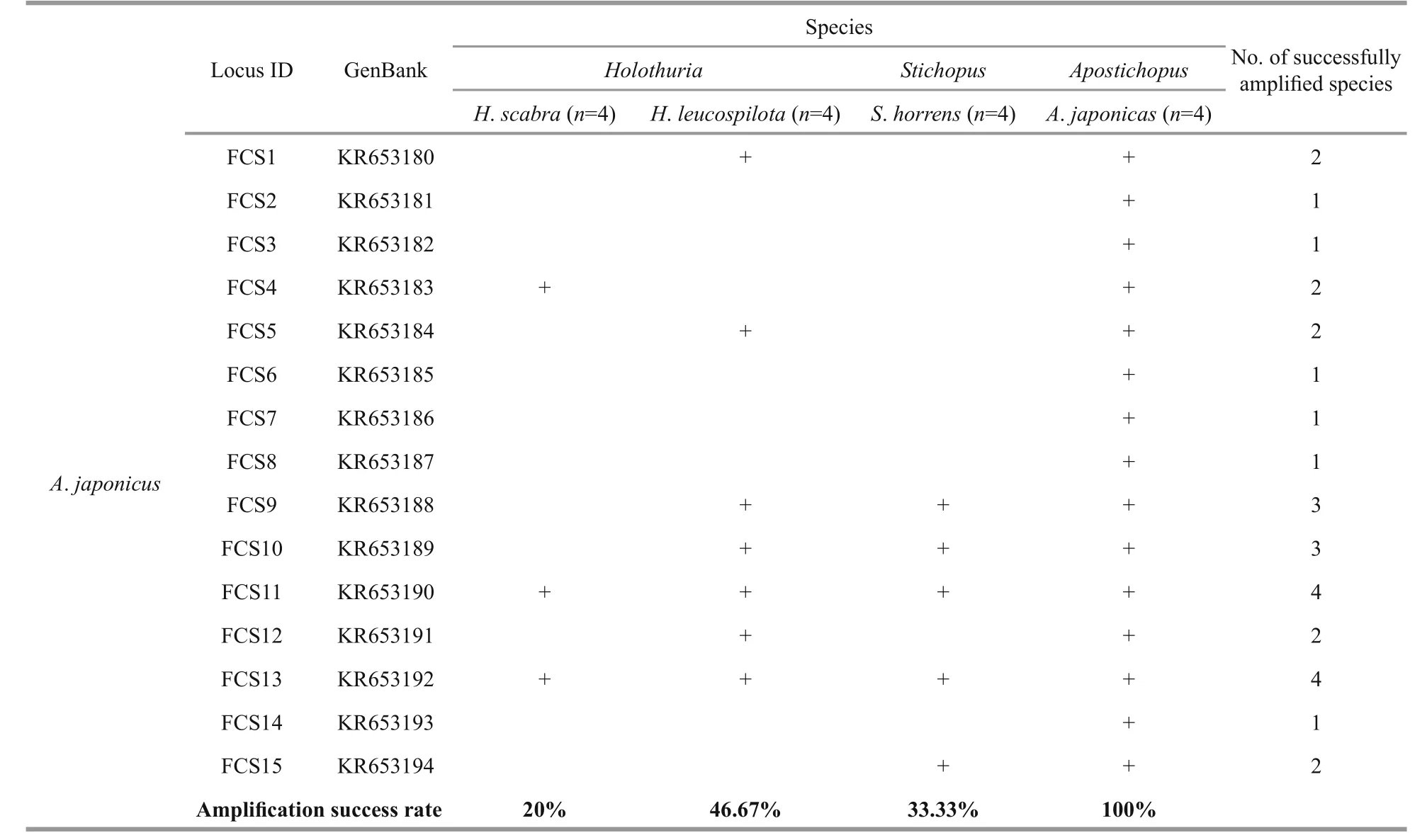
Table 4 Continued
5 DATA ACCESSIBILITY
Genbank accessions of DNA sequences:H.scabrafor KR653160–KR653179,A.japonicusfor KR653180–KR653194.
Balloux F, Ecoff ey E, Fumagalli L, Goudet J, Wyttenbach A,Hausser J. 1998. Microsatellite conservation,polymorphism, and GC content in shrews of the genusSorex(Insectivora, Mammalia).Mol.Biol.Evol.,15(4):473-475.
Barbará T, Palma-Silva C, Paggi G M, Bered F, Fay M F, Lexer C. 2007. Cross-species transfer of nuclear microsatellite markers: potential and limitations.Mol.Ecol.,16(18):3 759-3 767.
Battaglene S C, Seymour J E. 1998. Detachment and grading of the tropical sea cucumber sandfish,Holothuriascabra,juveniles from settlement substrates.Aquaculture,159(3-4): 263-274.
Bordbar S, Anwar F, Saari N. 2011. High-value components and bioactives from sea cucumbers for functional foods—A review.Mar.Drugs,9(10): 1 761-1 805.
Botstein D, White R L, Skolnick M, Davis R W. 1980.Construction of a genetic linkage map in man using restriction fragment length polymorphisms.Am.J.Hum.Genet,32(3): 314-331.
Chan Y M, Twyford A D, Tnah L H, Lee C T. 2015.Characterisation of EST-SSR markers forBegonia maxwelliana(Begoniaceae) and cross-amplification in 23 species from 7 Asian sections.Sci.Hortic.,190: 70-74.
Chen M, Gao L L, Zhang W J, You H Z, Sun Q, Chang Y Q.2013. Identification of forty-five gene-derived polymorphic microsatellite loci for the sea cucumber,Apostichopusjaponicus.J.Genet.92(2): e31-e35.
Conand C, Bryne M. 1993. A review of recent developments in the world sea cucumber fisheries.Mar.Fish.Rev.,55(4):1-13.
Conand C. 1990. The fishery Resources of Pacific Island Countries. Part 2: Holothurians. FAO Fisheries Technical Paper, No. 272.2. Food and Agriculture Organization of the United Nations, Rome, Italy. p.143.
Conand C. 2006. Sea Cucumber Biology, Taxonomy,Distribution: Conversation Status.In: Proceedings of the Convention on International Trade in Endangered Species of Wild Fauna and Flora Tech Workshop on the Conversation of Sea Cucumbers in the Families Holothuridae and Stichopodidae. Kuala Lumpur,Malaysia. p.1-3.
Dai G, Li Z B, Shangguan J B, Ning Y F, Deng H W, Yuan Y,Huang Y S, Yang H, Lu J. 2015. Development and characterization of polymorphic microsatellite loci in the sea cucumberHolothurialeucospilota.Genet.Mol.Res.,14(1): 538-541.
Fitch A J, Leeworthy G, Li X X, Bowman W, Turner L,Gardner M G. 2013. Isolation and characterisation of eighteen microsatellite markers from the sea cucumberHolothuriascabra(Echinodermata: Holothuriidae).Aust.J.Zool.,60(6): 368-371.
Kalinowski S T, Taper M L, Marshall T C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment.Mol.Ecol.,16(5): 1 099-1 106.
Kanno M, Li Q, Kijima A. 2005. Isolation and characterization of twenty microsatellite loci in Japanese sea cucumber(Stichopusjaponicus).Mar.Biotechnol.,7(3): 179-183.
Lalitha S. 2000. Primer Premier 5.BiotechSoftw.Int.Rep.,1(6): 270-272.
Lee G A, Kwon S J, Park Y J, Lee M C, Kim H H, Lee J S, Lee S Y, Gwag J G, Kim C K, Ma K H. 2011. Crossamplification of SSR markers developed fromAllium sativumto otherAlliumspecies.Sci.Hortic.,128(4): 401-407.
Li Q and Wan JM. 2005. SSRHunter: Development of a local searching software for SSR sites. Hereditas,27: 808-810.
Li Q, Chen L, Kong L. 2009. A genetic linkage map of the sea cucumber,Apostichopusjaponicus(Selenka), based on AFLP and microsatellite markers.Anim.Genet.,40(5):678-685.
Li Z B, Dai G, Shangguan J B, Ning Y F, Li Y Y, Chen R B,Huang Y S, Yuan Y. 2015a. Isolation and characterization of polymorphic microsatellite loci in the sea cucumberHolothuriascabra.Genet.Mol.Res.,14(2): 6 529- 6 532.
Li Z B, Dai G, Shangguan J B, Ning Y F, Li Y Y, Chen R B,Yuan Y, Huang Y S. 2015b. Isolation and characterization of microsatellite markers of sea cucumberStichopus horrens.Genet.Mol.Res.,14(3): 8 496-8 499.
Liu Z J, Cordes J F. 2004. DNA marker technologies and their applications in aquaculture genetics.Aquaculture,238:1-37.
Luo W, Qu H Y, Li J Y, Wang X, Lin Q. 2015. A novel method for the identification of seahorses (genusHippocampus)using cross-species amplifiable microsatellites.Fish.Res.,172: 318-324.
Nair A, Gopalan S V, George S, Kumar K S, Teacher A G F,Meril? J. 2012. High cryptic diversity of endemic Indirana frogs in the Western Ghats biodiversity hotspot.Anim.Conserv.,15(5): 489-498.
Narum S R. 2006. Beyond bonferroni: less conservative analyses for conservation genetics.Conserv.Genet.,7(5): 783-787.
Peng W, Bao Z M, Du H X, Yan J J, Zhang L L, Hu J J. 2012.Development and characterization of 70 novel microsatellite markers for the sea cucumber (Apostichopus japonicus).Genet.Mol.Res.,11(1): 434-439.
Primmer C R, Painter J N, Koskinen M T, Palo J U, Meril? J.2005. Factors affecting avian cross-species microsatellite amplification.J.AvianBiol.,36(4): 348-360.
Serapion J, Kucuktas H, Feng J N, Liu Z J. 2004. Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictaluruspunctatus).Mar.Biotechnol.,6(4): 364-377.
Shangguan J B, Li Z B, Ning Y F, Huang Y S, Yuan Y, Lu J, Li B B, Mao X Q. 2014a. Screening and characterization of novel polymorphic microsatellite markers from sea cucumberHolothurialeucospilota.Genet.Mol.Res.,14(2): 6 555-6 560.
Shangguan J B, Li Z B, Yuan Y, Huang Y S. 2014b.Identification and characterization of microsatellite markers from the tropical sea cucumber,Stichopus horrens(Selenka).Genet.Mol.Res.,14(4): 13 582-13 587.
Shikano T, Ramadevi J, Shimada Y, Meril? J. 2010. Utility of sequenced genomes for microsatellite marker development in non-model organisms: a case study of functionally important genes in nine-spined sticklebacks (Pungitius pungitius).BMCGenomics,11(1): 334.
Soldati M C, Inza M V, Fornes L, Zelener N. 2014. Cross transferability of SSR markers to endangeredCedrelaspecies that grow in Argentinean subtropical forests, as a valuable tool for population genetic studies.Biochem.Syst.Ecol.,53(8): 8-16.
Taiyeb-Ali T B, Zainuddin S L, Swaminathan D, Yaacob H.2003. Efficacy of ‘Gamadent’ toothpaste on the healing of gingival tissues: a preliminary report.J.OralSci.,45(3):153-159.
Tautz D. 1989. Hypervariabflity of simple sequences as a general source for polymorphic DNA markers.Nucleic AcidsRes.,17(16): 6 463-6 471.
Tian C Y, Li Q, Liang Y. 2008. Healthy Aquaculture Techniques of the Sea CucumberApostichopusjaponicus. Ocean University of China Press, Qingdao, China. (in Chinese)
van Oosterhout C, Hutchinson W F, Wills D P M, Shipley P.2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data.Mol.Ecol.Notes,4(3): 535-538.
Varshney R K, Graner A, Sorrells M E. 2005. Genic microsatellite markers in plants: features and applications.TrendsBiotechnol.,23(1): 48-55.
Vogiatzi E, Hanel R, Dailianis T, Lagnel J, Hassan M,Magoulas A, Tsigenopoulos C S. 2012. Description of microsatellite markers in four mullids based on the development and cross-species amplification of 18 new markers in red mullet (Mullusbarbatus).Biochem.Syst.Ecol.,44: 279-285.
Xia J J, Ren C H, Yu Z H, Wu X Y, Qian J, Hu C Q. 2016.Complete mitochondrial genome of the sandfishHolothuriascabra(Holothuroidea, Holothuriidae).Mitochondr.DNAPartA,27(6): 4 174-4 175.
Yeh F C, Yang R, Boyle T J, Ye Z, Xiyan J M. 2000. PopGene32,Microsoft Windows-Based freeware for Population.Genetic Analysis. Version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton.
Zane L, Bargelloni L and Patarnello T. 2002. Strategies for microsatellite isolation: a review.Mol.Ecol.,11: 1-16.
Zhan A B, Bao Z M, Lu W, Hu X L, Peng W, Wang M L, Hu J J. 2007. Development and characterization of 45 novel microsatellite markers for sea cucumber (Apostichopus japonicus).Mol.Ecol.Notes,7(6): 1 345-1 348.
 Journal of Oceanology and Limnology2018年2期
Journal of Oceanology and Limnology2018年2期
- Journal of Oceanology and Limnology的其它文章
- Editorial Statement
- Hydroacoustic estimates of fish biomass and spatial distributions in shallow lakes*
- A comparison between benthic gillnet and bottom trawl for assessing fish assemblages in a shallow eutrophic lake near the Changjiang River estuary*
- Morphological beak differences of loliginid squid, Uroteuthis chinensis and Uroteuthis edulis, in the northern South China Sea*
- Muelleria pseudogibbula, a new species from a newly recorded genus (Bacillariophyceae) in China*
- Planaxidae (Mollusca, Gastropoda) from the South China Sea*
