Novel predictive and prognostic strategies of hepatitis B virus related hepatocellular carcinoma
Department of Epidemiology, Second Military Medical University, Shanghai 200433, China. *Authors contributed equally.
Novel predictive and prognostic strategies of hepatitis B virus related hepatocellular carcinoma
Wen-Bin Liu*, Fan Yang*, Ding-Yi Shao*, Guang-Wen Cao
Department of Epidemiology, Second Military Medical University, Shanghai 200433, China. *Authors contributed equally.
How to cite this article:Liu WB, Yang F, Shao DY, Cao GW. Novel predictive and prognostic strategies of hepatitis B virus related hepatocellular carcinoma. Hepatoma Res 2016;2:331-40.
Article history:
Received: 14-09-2016
Accepted: 12-12-2016
Published: 23-12-2016
Hepatocellular carcinoma, hepatitis B virus, evolution, prediction, prognosis
Hepatocellular carcinoma (HCC) is a common malignancy and an important cause of cancer death worldwide. Chronic hepatitis B virus (HBV) infection is the major cause of HCC. Recent studies of HBV-induced carcinogenesis not only discovered many new biomarkers but also developed a novel theory: Cancer Evolution-Development (Cancer Evo-Dev). Cancer Evo-Dev provides an evolutionary insight of developing more reasonable predictive and prognostic strategies. Characterizing chronic infammatory microenvironment of cancer evolution, genetic polymorphisms of infammatory factors, and HCC-related HBV mutations that negatively selected by host immunity may help greatly in identifying HBV-infected individuals who are more likely to develop HCC or beneft from HCC prophylactic options. Gene expression signatures and somatic mutation profles refect the different patterns of signaling pathway networks underlying tumor heterogeneity and can be applied to improve the molecular classifcation and prognostic stratifcation of HCC patients. Mutant cells that survive the selection can retro-differentiate into tumor initial cells and aggressive sub-clones. Detection of mutants or their hallmarks in cell-free DNA in peripheral blood potentially improve the early diagnosis, prognosis prediction, and personalized treatment of HBV-caused HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most frequently diagnosed cancers and an important cause of cancer death worldwide. Annually, there are 782,500 HCC incident cases and 745,500 HCC-caused deaths worldwide.[1]Developing countries in East Asia and Sub-Saharan Africa contribute 80% of new HCC cases and related deaths.[2]Chronic infection of hepatitis B virus (HBV) is the major etiological reason for HCC in these areas, which contributes 80-90% of HCC patients.[3,4]According to a cohort study conducted in Taiwan, the cumulative lifetime (age 30 to 75 years) incidences of HCC for men and women that positive for hepatitis B surface antigen (HBsAg) were 27.38% and 7.99%, far more than those of men and women negativefor HBsAg and anti-hepatitis C virus (1.55% and 1.30%).[5]Besides, HBV infection is also responsible for the increasing trend of HCC in western countries because of the travel and immigration of HBV infected populations.[6]Most HCC patients are diagnosed at advanced stage and cannot accept resection operation or liver transplantation.[7]Approximately 70% patients that have curative hepatectomy will relapse within 5 years.[8]Both the narrow therapeutic window and the high recurrence rate highlight the importance of developing more rigorous surveillance and more active prevention for chronic HBV infected subjects with high HCC risk, and tailoring more suitable treatment options for HCC patients, which depend on continuously discovering promising biomarkers as well as developing carcinogenesis theory for the specifc prophylaxis.
Cancer Evo-Dev is a novel scientifc theory describing the mechanism of HBV-induced hepatocarcinogenesis.[9]The central aspects of its framework are as follows. Carcinogenesis is an evolutionary process under the microenvironment of chronic non-resolving infammation. This microenvironment is characterized by immune imbalance due to the interaction between the genetic predisposition of immune/proinfammatory molecules and HBV infection. Cytidine deaminases and their analogous are persistently activated by proinflammatory factors and subsequently induced mutations both in host and viral genomes. Mutant cells are mostly eliminated by selective pressures. Only a small proportion can survive in the inflammatory microenvironment because the somatic mutations alter signaling pathways. Those surviving clones usually share some characteristics of stem cells and gradually retro-differentiate into cancer initialing cells.
This theory was presented based on recent outcomes of HBV-related carcinogenesis researches, mainly including molecular epidemiological studies, cancer genomic mutation analyses, and signaling transduction researches.[10-20]Those breakthroughs not only improved the understanding of cancer evolution from different aspects but also discovered many novel biomarkers and therapeutic targets. Therefore, this theory can provide an evolutionary insight of predicting HCC risk and developing more reasonable predictive and prognostic biomarkers and therapeutic targets. Here, we summarize the important novel viral, infammatory, genetic, and protein biomarkers of HCC occurrence and prognosis and evaluate them through the lens of Evo-Dev theory.
EVALUATING THE MICROENVIRONMENT OF CANCER EVOLUTION
In the evolution process of HBV-induced hepatocarcinogenesis, inflammatory microenvironment plays an important role via facilitating the generation of viral and host genetic mutation and also providing selective pressure. Therefore, the characteristics of the microenvironment in different evolutionary phases and in different populations can be used to stratify HBV-infected individuals with different risk of developing HCC. Although inflammatory microenvironment is a complex system, it can be elucidated in two aspects: HBV itself and immune imbalance.
HBV
Despite the high incidence of HCC in HBV-infected population, only small percentages of chronic hepatitis B (CHB) patients develop HCC. HBV variables can serve as clues to identify distinctive outcomes of HBV-infected populations, and to guide the personalized preventive medication accordingly.
HBV replication
The level of HBV replication directly reflects the selective stress from the inflammatory environment, which can influence the evolution of HCC as well. Currently, HBV DNA load is regularly applied in clinic as an indicator of initiating antiviral treatment. It has been demonstrated by various studies that HBV DNA load increases the risk of HCC in CHB patients.[21-23]High level of HBV DNA load either in serum or liver tissue can also predict poor postoperative prognosis in HCC.[24]Hepatitis B e antigen (HBeAg), encoded by HBV precore region, is another marker for active replication of HBV. HBeAg positivity has been proved to be associated with an increased risk of HCC.[25]However, due to HBeAg seroconversion during the natural course of HBV infection, HBeAg expression is not usually high in HCC patients, explaining the reasons that HBeAg positivity is not significantly associated with an increased risk of HCC in some case-control studies.[14]Thus, HBV DNA load should be a more reliable indicator in the prediction of HCC.
HBV genotypes
According to a sequence divergence of no less than 8% in whole viral genome, HBV can be classifed into eight genotypes A to H, which can be further classifed into sub-genotypes if the sequence divergence is between 4% and 8%.[26]Variant genotypes are distributed unevenly around the world, and the predominant one in mainland China is genotype C (68.3%), followed by genotype B (25.5%).[27]Under selection pressure from inflammatory microenvironment, the fates of different genotypes/sub-genotypes are distinct in a given population. Genotype C HBV infection is an independent risk factor for HCC development.[16,21,28,29]Meanwhile, genotype B HBV infection was associatedwith the development of HCC in young patients (< 50 years old).[30]Our study further revealed that genotype B2 HBV infection was related to HCC recurrence, and that HBV genotype C2 HBV was predominant in HCC patients, which was related to its high prevalence.[31]As the HBV genotype is usually identified through a complex procedure that includes extracting HBV DNA, polymerase chain reaction, sequencing, and phylogenetic analysis, the wide application of HBV genotype/subgenotype for preliminary screening in community is limited.
HBV mutations
In the process of HBV-HCC evolution, one of the most prominent molecular events is the generation of HBV mutation, especially mutations in the preS region and basic core promoter (BCP) region of HBV genome. Due to lack of proof reading capacity, HBV genome has a higher mutation rate than other DNA viruses. Moreover, inflammatory factors induced by HBV infection can activate the expression of apolioprotein B mRNA editing enzyme catalytic polypeptides (APOBECs). HBV genome can be degraded and edited by APOBECs.[32]Most HBV mutants are cleared by host immune system, and only those that gained the ability to escape immune eradication survived. The mutant viruses, in return, keep on stimulating the immune system and maintain the inflammatory microenvironment. The HBV mutations refect, to some extent, the selection pressure of host immune system and serve as risk factors of HCC.
Our recent study of HBV mother-to-child transmission revealed that mutated viruses lost their advantages in infecting infants, whereas the wild-type HBV had advantage of infecting newborn’s hepatocytes, interestingly, the HCC-risk HBV mutations was being gradually selected since the establishment of chronic infection.[10]Mutations in HBV the preS region (including A2962G, A2964C, C3116T, C7A, T105C, and preS start codon mutation) and mutations in the BCP region (including C1653T, T1753V, and A1762T/G1764A) were independently associated with an increased risk of HCC.[11,15,21,33]Mutations in combination (combo mutations) can enhance the validity of predicting the occurrence of HCC.[21,33,34]HBV combo mutations of C1653T, T1753V, and A1762T/G1764A increase the validity of HCC prediction compared with single HBV mutation.[21]The HBV mutations can improve the sensitivity and specifcity of HCC prediction model based on age, gender, cirrhosis and HBV DNA loads.[21,25,35]
The carcinogenic effects of HBV can be blocked by antiviral treatments. In our prospective hospitalbased cohort study, antiviral treatment against HBV using interferon and nucleoside analogues (NAs) signifcantly reduced HCC occurrence (13.90/1,000 vs. 7.70/1,000 person-years, P = 0.005).[36]Furthermore, proved by a cohort study and randomized clinical trial, treatment with NAs can also signifcantly reduce the risk of early recurrence (hazard ratios, 0.41; P <0.001).[13]However, levels of those protective effects are distinct among HBV-infected subjects with different viral mutations. Antiviral treatment with NAs cannot reduce HCC risk in patients without A1762T/G1764A or C1653T and in those with T1753V.[36]The protective function of antiviral treatments for postoperative recurrence cannot be observed in the HCC patients expressing carboxylic acid-terminal truncated HBV X protein (Ct-HBx) in their liver remnants.[13]
Immune imbaIance
Immune imbalance is responsible for the maintenance of chronic non-resolving inflammation and subsequently provides a fertile microenvironment for cancer evolution. Immune imbalance can be refected by the proportion shift of immune cells, abnormal activation of inflammatory pathways, and genetic predisposition of inflammatory molecules, which can serve as biomarkers for HCC prediction and prognosis.
Immune cells
The liver is enriched with innate immune cells such as macrophages and natural killer (NK) cells, as well as adaptive immune cells such as CD8+cytotoxic T cells, CD4+T helper cells and B cells, playing an important role not only in host defenses against invading microorganisms and tumor transformation, but also in liver injury and repair. Their presence or enrichment can be seen as predictive or prognostic factors for HCC. CD8+T in liver tissues, for example, is the protective factor, while the enrichment of M2 macrophages and T helper 17 cells (Th17) as well as the imbalance between CD8+T cells and regulatory T (Treg) cells or between Th1 and Th2 are the risk factors of HCC.[37]Immune cells that infltrated into HCC tissues function distinctly on HCC prognosis. Intratumoral natural killer cells and CD8+T cells indicate good prognosis, while intratumoral Treg cells, neutrophils, and M2 macrophages indicate poor prognosis.[37]
Infammatory pathways
The abnormal alteration of inflammatory pathways can be reflected by hallmark cytokines. Biomarkers indicating the abnormal activation of inflammatory pathways can also predict the occurrence and recurrence of HCC.[38,39]For example, Wnt/β-catenin signaling pathway plays an important role in infammation-induced carcinogenesis via regulating the expression of cytokine-induced human inducible nitricoxide synthase.[40]Activation of Wnt/β-catenin pathway contributes to HCC development. The hallmarks of Wnt/β-catenin pathway, Wnt-1 and Wnt3a, have both predictive and prognostic value.[37,41,42]Likewise, signaling pathways such as phosphatidylinositol-3 kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway, and insulin-like growth factor pathway also play an important role in hepatocarcinogenesis.[43]
Genetic polymorphisms of immune/inflammatory molecules
Genetic polymorphisms of immune/inflammatory molecules can also serve as predictive biomarkers for HCC development. For example, genetic polymorphisms of signal transducer and activator of transcription 3 (STAT3), class II human leukocyte antigen DP (HLA-DP), HLA-DQ, miRNA-122-binding site, pre-miR-218, nuclear factor-kappaB (NF-κB), and its inhibitor IkappaBalpha are signifcantly associated with HCC risk.[12,17,18,44-47]
IDENTIFYING SIGNATURES OF SIGNALING PATHWAY ALTERATION
Gene signatures
The alteration of signaling pathways confers stemness characteristics and competitive advantages to cancer cells. These alterations usually affect complex signaling networks that cannot be represented by a signal gene. More than 300 published microarray studies of human HCC samples provide sufficient information regarding tumor gene expression profiles.[48]The accumulation of data regarding differentially expressing genes makes it possible to conduct metaanalysis and subsequently determine gene signatures. Recent gene signature studies are summarized in Table 1.[49-66]Gene signatures developed in those studies were used to separate patients into 2 or more subgroups with different clinical outcomes, phenotypes, and altered signaling pathways. The methods of developing gene signatures fall into two major groups. The first group of gene signatures was generated in case-control studies with the data of training cohort or published gene expression data. Most of the gene signature studies belong to this group.[50,52,53,55,57,59,61-65]The second group of gene signatures concerning defined phenotypes or signaling pathways was derived from the data of cell or animal model studies.[49,51,56,58,60]For examples, Lee et al.[49]developed a gene signature of stemness from the gene profling data of rat fetal liver tissue and Kaposi-Novak et al.[51]developed a gene signature of Met signaling pathway using the Met defcient mouse model. The predictive value of novel gene signatures was usually evaluated in cohort studies. High risk patients that were identified through cluster analysis or score model based on gene signatures were prone to have unfavourable clinical outcomes, such as poor overall survival and early recurrence.
Although the tumor gene signatures were identified by different studies with various comparison strategies, they shared some genes conferring cancer stemness. For instance, a group of genes related to proliferation and epithelial cell adhesion molecule (EpCAM)-positive phenotype were included in 8 gene signatures summarized in different studies and all associated with poor prognosis.[48]Gene signatures from adjacent non-tumor tissues were also reported to be significantly associated with HCC recurrence, indicating that the histological “normal” adjacent tissue may be at the early stage of cancer evolution. That highlights the need of biopsy-based gene signature detection for specific individuals, like HBV-infected patients. However, signatures from adjacent tissues obtained in different studies are lack of genes in common. Cross validations are needed to consolidate the criteria. Altered expression patterns of the genes in HCC are usually caused by epigenetic modifcations in their regulatory elements and somatic mutations of their repressors.
Somatic mutation profiIes
Somatic mutations are genetic basis of carcinogenesis. The values of somatic mutations depend on their impacts on related signaling pathways. By changing patterns of signaling transduction, somatic mutations on a small proportion of genes can promote cancer evolution, which are categorized as “driver mutations”.[19]As a matter of fact, some outstanding somatic mutations in HBV-HCC occur in the genes responsible for epigenetic modifications-chromatin remodeling including ARID1A and ARID2 and methylation such MLL4.[67,68]Due to survival competition and the positive selection of infammatory microenvironment, driver mutations accumulate sufficiently to promote malignant transformation of hepatocytes.
The distribution, combination, and dynamic patterns of driver mutations reflex the pressure of microenvironmental selection and growth advantage of hepatocyte subsets. The high frequent mutations can have clinical values as biomarkers for targeted therapy, classification, and prognostic prediction.[67-71]For instance, homozygous deletions were detected in 40% of HCC patients and were signifcantly associated with poor survival (P < 0.0001).[68]
Using next generation sequencing technology, somebasic patterns of HCC somatic mutations have been extensively investigated. The somatic mutations provide a novel genomic insight of molecular classification and prognostic prediction. Some genes including TP53, TERT, CTNNB1, ARID1A, and AXIN1 are proved to be hotspots of genetic alteration [Table 2]. However, specifc mutation in a single hot gene is not frequent, ranging from 5% to 20%. Such a low ratelimits the application of a single mutation. For example, RB1 somatic mutation can serve as an independent predictor for poor cancer-specific survival (HR 2.5, 95% CI: 1.05-5.93, P = 0.038) and early recurrence (OR 3.93, 95% CI: 1.29-11.90, P = 0.015). But the frequencies of RB1 somatic mutation were only 3.4% and 7% among different studies.[68,69]Similarly, somatic mutations of CDKN2A and FGF-CCND1 were proved to be significantly associated with overall survival (P = 3.0 × 10-4and P = 7.4 × 10-6respectively) and their frequencies were both less than 5%.[70]

Table 1: Representative gene signature studies of hepatocellular carcinoma
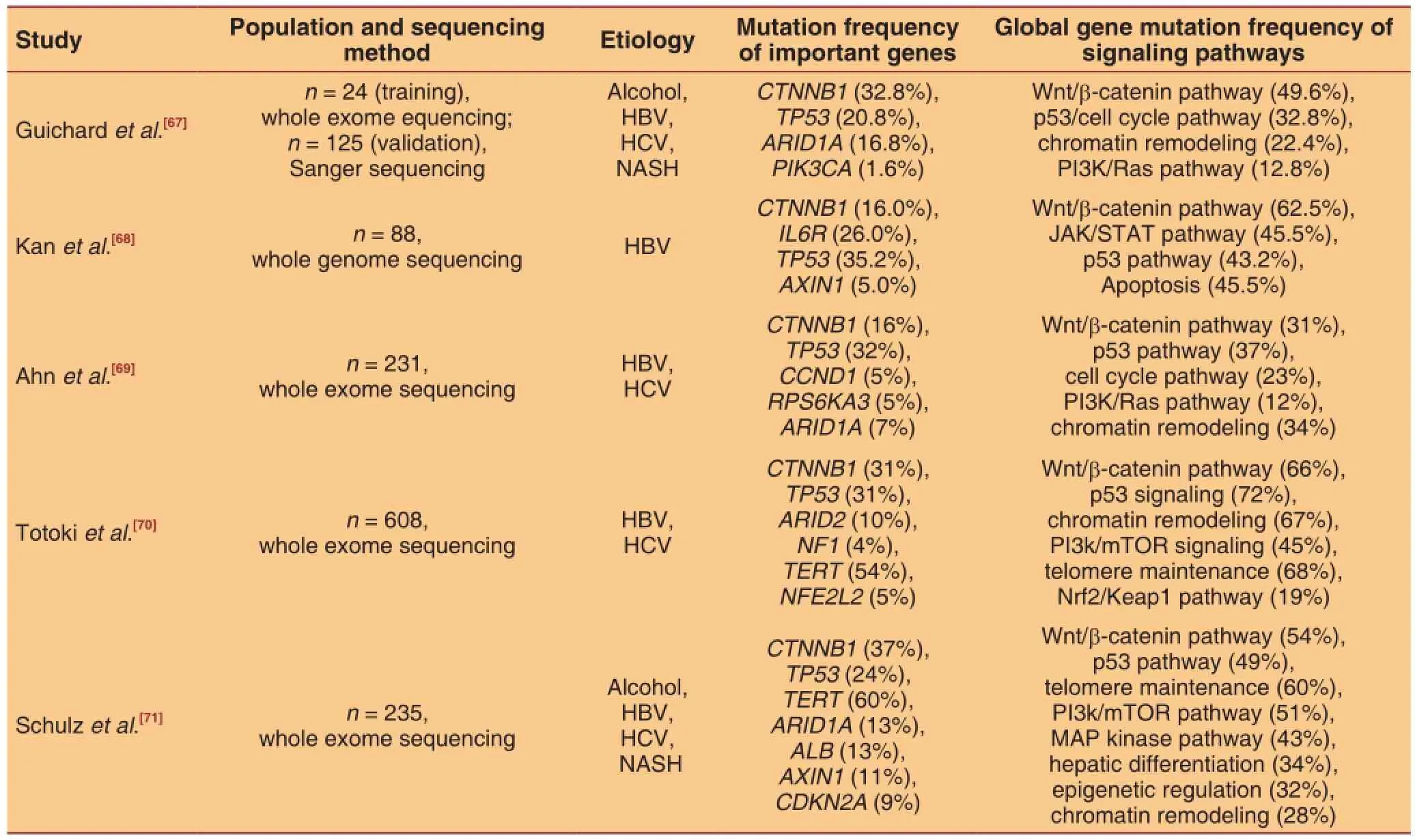
Table 2: Important somatic mutations and related signaling pathways of hepatocellular carcinoma
Although the spectrums and frequencies of altered genes vary greatly among individuals, they are clustered to pathways or function groups that are closely related with stemness and embryonic characteristics. In this regard, global mutation rates of functionally related genes are added together to defne the mutation rate of a given signaling pathway. Mutation rates of Wnt/β-catenin, p53/cell cycle control, JAK/STAT, PI3k/mTOR, and MAP kinas signaling pathways range from 12% to 72%. Similar outstanding outcomes are also observed in function gene groups of chromatin remodeling and telomere maintenance. Ahn et al.[69]developed a somatic mutation signature of cell cycle pathway which comprised 4 genes including RB1, MYC, CCND1, and RBL2. The total mutation rate of those 4 genes were 23% and the signature was significantly associated with poor cancer-specifc and recurrence-free survival (P = 0.002 and P = 0.007, respectively). Therefore, it is promising to use combo somatic mutations as predictive and prognostic biomarkers.
DETECTING CELLS WITH MALIGNANCY POTENTIAL AND THEIR HALLMARKS IN PERIPHERAL BLOOD
CircuIating tumor ceIIs
Release of cancer cells into the circulation is common in HCC patients. The appearance of circulating tumor cells (CTC) in the blood stream characterizes the intermediate stage of tumor metastasis process.[72]CTC test can be applied to monitor early metastasis, assess the effectiveness of therapeutic options, and predict the prognosis.[73]A study examining blood samples of 123 HCC patients one month before and after tumor resection indicated that EpCAM+CTCs were presented in 66.67% of patients and that CTCs count in 7.5 mL blood (CTC7.5) is an independent prognostic factorof tumor recurrence.[74]Therefore, EpCAM+CTCs may be used as a real-time parameter for monitoring treatment response. In addition, EpCAM+CTCs are positive in HCC patients with different BCLC stages and the positive rates of EpCAM+CTCs in patients of BCLA stage A, B, and C are 11.1%, 19.4%, and 57.9%, respectively.[75]Thus, EpCAM+CTC is prognostic and predictive in HCC.
CeII-free DNA
Biopsy of HCC may be restricted by the special position of tumors or the poor condition of patients, resulting in the limitation of HCC gene analysis for prognostic and predictive purposes.[76]The necrosis and apoptosis of tumor cells usually release cell-free DNA (cfDNA) into circulation. Based on sequencing technology, genetic and epigenetic information can be obtained from these cfDNA. Detecting cfDNA is a microinvasive method to find early HCC, termed as “l(fā)iquid biopsy”.[77]The abnormities including methylation changes and point mutations in cfDNA can be detected in peripheral blood even before the solid tumor nidus can be detected.
Hypermethylated RASSF1A within cfDNA sequence is present in the sera of 93% HCC patients. When combining RASSF1A methylation and AFP to diagnose HCC, the sensitivity and specifcity increase from 65% and 87% using AFP alone to 77% and 89%, respectively. Serum methylated RASSF1A is also prognostic and also reflects the tumor load in HCC patients.[78]A study with a cohort of 151 HCC patients indicated that 4 hypermethylation genes (RGS10, ST8SIA6, RUNX2, and VIM) in sera have weak correlation with each other but the combination of the 4 genes as a classifier successfully identified HCC patients from HBV-induced cirrhosis population, with the sensitivity of 85% and the specifcity of 96%.[79]
TP53 R249S mutation in cfDNA was proved to have a remarkable ecological correlation with HCC exposure in China and Africa.[80]In a retrospective study using short oligonucleotide mass analysis to exam R249S in the plasma ahead of cancer diagnosis, 9 (64%) of 14 patients who developed HCC during the followup were positive for R249S.[81]Genetic mutation in serum is related to the mutation in tumor tissue. Another study examining the mutations of CTNNB1, a gene encoding β-catenin, in HCC patients’ sera indicated that CTNNB1 mutation was not present both in serum and corresponding tumor tissues, although the average mutation rate of CTNNB1 was about 25% in previous researches.[82]This suggests that clinical application of cfDNA mutations should be mutation signatures rather than single gene mutation.
CONCLUSION
HBV-induced HCC is a common malignancy characterized by high mortality, high recurrence rate, and significant heterogeneity. Cancer Evo-Dev, a novel scientific theory of HBV-induced carcinogenesis, provides an evolutionary insight of HCC occurrence/recurrence prediction. From this point of view, recent development of HCC predictive and prognostic strategies can be categorized as three main directions: evaluating the inflammatory microenvironment of cancer evolution via investigating HBV variables and characteristics of immune imbalance, identifying alteration patterns of signaling transformation through signatures of gene expression and somatic mutation, and detecting cells with malignancy potential and their hallmarks in peripheral blood. To validate predictive or prognostic biomarkers, 4 steps should be taken: (1) exploratory research, to discover promising biomarkers; (2) case-control study, to evaluate statistical association between the occurrence/recurrence and biomarkers; (3) cohort study, to validate the sensitivity and specificity of biomarkers; (4) randomized clinical control trail, to determine if the screening and related prophylaxis/ treatment can reduce the occurrence/recurrence. Currently, most novel biomarkers were just validated in phase 2 or 3. Further validation and reasonable combination of novel biomarkers should be conducted under the direction of Cancer Evo-Dev theory.
FinanciaI support and sponsorship
This work was supported by grants from the National Key Basic Research Program (Grant No. 2015CB554006). The study sponsors had no role in the writing of the manuscript or in the decision to submit the manuscript for publication.
ConfIicts of interest
There are no conficts of interest.
Patient consent
There is no patient involved.
Ethics approvaI
This review is waived for ethical approval.
REFERENCES
1. Torre LA, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87-108.
2. Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat 2009;16:453-63.
3. Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol 2005;34 Suppl 1:S1-3.
4. E-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264-73.
5. Huang YT, Jen CL, Yang HI, Lee MH, Su J, Lu SN, Iloeje UH, Chen CJ. Lifetime risk and sex difference of hepatocellular carcinoma among patients with chronic hepatitis B and C. J Clin Oncol 2011;29:3643-50.
6. Sharma S, Carballo M, Feld JJ, Janssen HL. Immigration and viral hepatitis. J Hepatol 2015;63:515-22.
7. Du Y, Su T, Ding Y, Cao G. Effects of antiviral therapy on the recurrence of hepatocellular carcinoma after curative resection or liver transplantation. Hepat Mon 2012;12:e6031.
8. Hyder O, Marques H, Pulitano C, Marsh JW, Alexandrescu S, Bauer TW, Gamblin TC, Sotiropoulos GC, Paul A, Barroso E, Clary BM, Aldrighetti L, Ferrone CR, Zhu AX, Popescu I, Gigot JF, Mentha G, Feng S, Pawlik TM. A nomogram to predict long-term survival after resection for intrahepatic cholangiocarcinoma: an Eastern and Western experience. JAMA Surg 2014;149:432-8.
9. Liu WB, Wu JF, DuY, Cao GW. Cancer Evolution-Development: experience of hepatitis B virus-induced hepatocarcinogenesis. Curr Oncol 2016;23:e49-56.
10. Li ZX, Xie ZY, Ni HX, Zhang Q, Lu W, Yin JH, Liu WB, Ding YB, Zhao Y, Zhu YB, Pu R, Zhang HW, Dong HJ, Fu YF, Sun Q, Xu GZ, Cao GW. Mother-to-child transmission of hepatitis B virus: evolution of hepatocellular carcinoma-related viral mutations in the postimmunization era. J Clin Virol 2014;61:47-54.
11. Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst 2009;101:1066-82.
12. Xie J, Zhang Y, Zhang Q, Han Y, Yin J, Pu R, Shen Q, Lu W, Du Y, Zhao J, Han X, Zhang H, Cao G. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology 2013;57:2369-77.
13. Yin J, Li N, Han Y, Xue J, Deng Y, Shi J, Guo W, Zhang H, Wang H, Cheng S, Cao G. Effect of antiviral treatment with nucleotide/ nucleoside analogs on postoperative prognosis of hepatitis B virusrelated hepatocellular carcinoma: a two-stage longitudinal clinical study. J Clin Oncol 2013;31:3647-55.
14. Yin J, Xie J, Liu S, Zhang H, Han L, Lu W, Shen Q, Xu G, Dong H, Shen J, Zhang J, Han J, Wang L, Liu Y, Wang F, Zhao J, Zhang Q, Ni W, Wang H, Cao G. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol 2011;106:81-92.
15. Yin J, Xie J, Zhang H, Shen Q, Han L, Lu W, Han Y, Li C, Ni W, Wang H, Cao G. Signifcant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol 2010;45:1063-71.
16. Zhang HW, Yin JH, Li YT, Li CZ, Ren H, Gu CY, Wu HY, Liang XS, Zhang P, Zhao JF, Tan XJ, Lu W, Schaefer S, Cao GW. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut 2008;57:1713-20.
17. Zhang Q, Ji XW, Hou XM, Lu FM, DuY, Yin JH, Sun XY, Deng Y, Zhao J, Han X, Yang GS, Zhang HW, Chen XM, Shen HB, Wang HY, Cao GW. Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann Oncol 2014;25:2413-9.
18. Zhang Q, Yin J, Zhang Y, Deng Y, Ji X, Du Y, Pu R, Han Y, Zhao J, Han X, Zhang H, Cao G. HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol 2013;87:12176-86.
19. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi N C, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson J V, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, Laura van ‘t Veer, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome I, Consortium I B C, Consortium I M-S, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfster S M, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature 2013;500:415-21.
20. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24.
21. Chan HL, Tse C H, Mo F, Koh J, Wong VW, Wong GL, Chan SL, Yeo W, Sung JJ, Mok TS. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol 2008;26:177-82.
22. Yang HI, Yuen MF, Chan HL, Han KH, Chen PJ, Kim DY, Ahn SH, Chen CJ, Wong VW, Seto WK. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol 2011;12:568-74.
23. Lee MH, Yang HI, Liu J, Batrla-Utermann R, Jen CL, Iloeje UH, Lu SN, You S L, Wang LY, Chen CJ. Prediction models of longterm cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profles. Hepatology 2013;58:546-54.
24. Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai MW, Chen TC, Lin CY, Yeh T S, Lee WC. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology 2010;52:1922-33.
25. Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 2002;347:168-74.
26. Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 2004;47:289-309.
27. Yin J, Zhang H, He Y, Xie J, Liu S, Chang W, Tan X, Gu C, Lu W, Wang H, Bi S, Cui F, Liang X, Schaefer S, Cao G. Distribution and hepatocellular carcinoma-related viral properties of hepatitis B virus genotypes in Mainland China: a community-based study. Cancer Epidemiol Biomarkers Prev 2010;19:777-86.
28. Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut 2004;53:1494-98.
29. Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen D S, Chen CJ. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst 2005;97:265-72.
30. Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 2000;118:554-59.
31. Yin J, Zhang H, Li C, Gao C, He Y, Zhai Y, Zhang P, Xu L, Tan X, Chen J, Cheng S, Schaefer S, Cao G. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis 2008;29:1685-91.
32. Deng Y, Du Y, Zhang Q, Han X, Cao G. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation andhepatocellular carcinoma. Cancer Lett 2014;343:161-71.
33. Liu S, Xie J, Yin J, Zhang H, Zhang Q, Pu R, Li C, Ni W, Wang H, Cao G. A matched case-control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol 2011;83:45-53.
34. Xie JX, Zhao J, Yin JH, Zhang Q, Pu R, Lu WY, Zhang HW, Wang HY, Cao GW. Association of novel mutations and haplotypes in the preS region of hepatitis B virus with hepatocellular carcinoma. Front Med China 2010;4:419-29.
35. Yuen MF, Tanaka Y, Fong D Y, Fung J, Wong D K, Yuen J C, But D Y, Chan A O, Wong B C, Mizokami M, Lai C L. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol 2009;50:80-8.
36. Yin J, Wang J, Pu R, Xin H, Li Z, Han X, Ding Y, Du Y, Liu W, Deng Y, Ji X, Wu M, Yu M, Zhang H, Wang H, Thompson TC, Ni W, Cao G. Hepatitis B virus combo mutations improve the prediction and active prophylaxis of hepatocellular carcinoma: a clinic-based cohort study. Cancer Prev Res (Phila) 2015;8:978-88.
37. Han YF, Zhao J, Ma LY, Yin JH, Chang WJ, Zhang HW, Cao GW. Factors predicting occurrence and prognosis of hepatitis-B-virus-related hepatocellular carcinoma. World J Gastroenterol 2011;17:4258-70.
38. Chew V, Tow C, Teo M, Wong HL, Chan J, Gehring A, Loh M, Bolze A, Quek R, Lee VK, Lee KH, Abastado JP, Toh HC, Nardin A. Inflammatory tumour microenvironment is associated with superior survival in hepatocellular carcinoma patients. J Hepatol 2010;52:370-9.
39. Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, Xu Y, Li YW, Tang ZY. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007;25:2586-93.
40. Du Q, Zhang X, Cardinal J, Cao Z, Guo Z, Shao L, Geller DA. Wnt/ beta-catenin signaling regulates cytokine-induced human inducible nitric oxide synthase expression by inhibiting nuclear factor-kappaB activation in cancer cells. Cancer Res 2009;69:3764-71.
41. Pan L, Yao M, Zheng W, Gu J, Yang X, Qiu L, Cai Y, Wu W, Yao D. Abnormality of Wnt3a expression as novel specifc biomarker for diagnosis and differentiation of hepatocellular carcinoma. Tumour Biol 2016;37:5561-8.
42. Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2015;21:6317-28.
43. Fabregat I. Dysregulation of apoptosis in hepatocellular carcinoma cells. World J Gastroenterol 2009;15: 513-20.
44. He Y, Zhang H, Yin J, Xie J, Tan X, Liu S, Zhang Q, Li C, Zhao J, Wang H, Cao G. IkappaBalpha gene promoter polymorphisms are associated with hepatocarcinogenesis in patients infected with hepatitis B virus genotype C. Carcinogenesis 2009;30:1916-22.
45. Du Y, Han X, Pu R, Xie J, Zhang Y, Cao G. Association of miRNA-122-binding site polymorphism at the interleukin-1 alpha gene and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Front Med 2014;8:217-26.
46. Han Y, Pu R, Han X, Zhao J, Li W, Yin J, Zhang Y, Shen Q, Xie J, Zhang Q, Jiang S, Li J, Zhang H, Wang H, Cao G. Association of a potential functional pre-miR-218 polymorphism and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Liver Int 2014;34:728-36.
47. Ji X, Zhang Q, Li B, Du Y, Yin J, Liu W, Zhang H, Cao G. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol 2014;28:201-9.
48. Andrisani OM, Studach L, Merle P. Gene signatures in hepatocellular carcinoma (HCC). Semin Cancer Biol 2011;21:4-9.
49. Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, Nevens F, Roskams T, Thorgeirsson SS. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410-6.
50. Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 2006;10:99-111.
51. Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defnes a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest 2006;116:1582-95.
52. Wang SM, Ooi LL, Hui KM. Identifcation and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res 2007;13:6275-83.
53. Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, Herault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Transcriptome classifcation of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007;45:42-52.
54. Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, Kim W, Park SC, Chung YJ, Kim BG, Yoon JH, Lee HS, Kim CY, Yi NJ, Suh KS, Lee KU, Chu IS, Roskams T, Thorgeirsson SS, Kim YJ. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res 2008;14:2056-64.
55. Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, Reich M, Chan JA, Glickman JN, Ikeda K, Hashimoto M, Watanabe G, Daidone MG, Roayaie S, Schwartz M, Thung S, Salvesen HB, Gabriel S, Mazzaferro V, Bruix J, Friedman SL, Kumada H, Llovet JM, Golub TR. Gene expression in fxed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008;359:1995-2004.
56. Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 2008;47:2059-67.
57. Yoshioka S, Takemasa I, Nagano H, Kittaka N, Noda T, Wada H, Kobayashi S, Marubashi S, Takeda Y, Umeshita K, Dono K, Matsubara K, Monden M. Molecular prediction of early recurrence after resection of hepatocellular carcinoma. Eur J Cancer 2009;45:881-9.
58. Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES, Thorgeirsson S S, Kim Y J. Identifcation of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res 2010;70:3034-41.
59. Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in earlystage hepatocellular carcinoma patients. Cancer Res 2010;70:10202-12.
60. Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, Thung S N, Bruix J, Newell P, April C, Fan JB, Roayaie S, Mazzaferro V, Schwartz ME, Llovet JM. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011;140:1501-12.
61. Minguez B, Hoshida Y, Villanueva A, Toffanin S, Cabellos L, Thung S, Mandeli J, Sia D, April C, Fan J B, Lachenmayer A, Savic R, Roayaie S, Mazzaferro V, Bruix J, Schwartz M, Friedman S L, Llovet JM. Gene-expression signature of vascular invasion in hepatocellularcarcinoma. J Hepatol 2011;55:1325-31.
62. Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang D, Ling C. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer 2012;11:39.
63. Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, Lim JY, Heo J, Kim YJ, Kim DG, Kaseb A, Park YN, Wang XW, Thorgeirsson SS, Lee JS. Sixty-fve gene-based risk score classifer predicts overall survival in hepatocellular carcinoma. Hepatology 2012;55:1443-52.
64. Kim BY, Suh KS, Lee JG, Woo SR, Park IC, Park SH, Han CJ, Kim SB, Jeong S H, Yeom YI, Yang SJ, Kim CM, Cho SJ, Yoo YD, Cho MH, Jang JJ, Choi DW, Lee KH. Integrated analysis of prognostic gene expression profles from hepatitis B virus-positive hepatocellular carcinoma and adjacent liver tissue. Ann Surg Oncol 2012;19 Suppl 3:S328-38.
65. Lim HY, Sohn I, Deng S, Lee J, Jung SH, Mao M, Xu J, Wang K, Shi S, Joh JW, Choi YL, Park CK. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profling. Ann Surg Oncol 2013;20:3747-53.
66. Kim JH, Sohn BH, Lee HS, Kim SB, Yoo JE, Park YY, Jeong W, Lee SS, Park E S, Kaseb A, Kim BH, Kim WB, Yeon JE, Byun KS, Chu IS, Kim SS, Wang XW, Thorgeirsson SS, Luk JM, Kang KJ, Heo J, Park YN, Lee JS. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med 2014;11:e1001770.
67. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad I B, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8.
68. Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, Hauptschein R, Rejto PA, Fernandez J, Wang G, Zhang Q, Wang B, Chen R, Wang J, Lee NP, Zhou W, Lin Z, Peng Z, Yi K, Chen S, Li L, Fan X, Yang J, Ye R, Ju J, Wang K, Estrella H, Deng S, Wei P, Qiu M, Wulur IH, Liu J, Ehsani ME, Zhang C, Loboda A, Sung WK, Aggarwal A, Poon RT, Fan ST, Wang J, Hardwick J, Reinhard C, Dai H, Li Y, Luk JM, Mao M. Wholegenome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422-33.
69. Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, Baek D, Haq F, Ansari AA, Lee SY, Chun SM, Choi S, Choi HJ, Kim J, Kim S, Hwang S, Lee YJ, Lee J E, Jung WR, Jang HY, Yang E, Sung WK, Lee NP, Mao M, Lee C, Zucman-Rossi J, Yu E, Lee HC, Kong G. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratifcation. Hepatology 2014;60:1972-82.
70. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11.
71. Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H, Yamamoto S, Shinbrot E, Hama N, Lehmkuhl M, Hosoda F, Arai Y, Walker K, Dahdouli M, Gotoh K, Nagae G, Gingras MC, Muzny DM, Ojima H, Shimada K, Midorikawa Y, Goss JA, Cotton R, Hayashi A, Shibahara J, Ishikawa S, Guiteau J, Tanaka M, Urushidate T, Ohashi S, Okada N, Doddapaneni H, Wang M, Zhu Y, Dinh H, Okusaka T, Kokudo N, Kosuge T, Takayama T, Fukayama M, Gibbs RA, Wheeler DA, Aburatani H, Shibata T. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73.
72. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559-64.
73. Wu LJ, Pan YD, Pei XY, Chen H, Nguyen S, Kashyap A, Liu J, Wu J. Capturing circulating tumor cells of hepatocellular carcinoma. Cancer Lett 2012;326:17-22.
74. Sun Y F, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, Shi RY, Hu B, Zhou J, Fan J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013;57:1458-68.
75. Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, Riethdorf S, Wege H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer 2013;133:2165-71.
76. Tang JC, Feng YL, Guo T, Xie AY, Cai XJ. Circulating tumor DNA in hepatocellular carcinoma: trends and challenges. Cell Biosci 2016;6:32.
77. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37.
78. Chan KC, Lai PB, Mok TS, Chan HL, Ding C, Yeung SW, Lo YM. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem 2008;54:1528-36.
79. Wen L, Li J, Guo H, Liu X, Zheng S, Zhang D, Zhu W, Qu J, Guo L, Du D, Jin X, Zhang Y, Gao Y, Shen J, Ge H, Tang F, Huang Y, Peng J. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25:1376.
80. Villar S, Le Roux-Goglin E, Gouas D A, Plymoth A, Ferro G, Boniol M, Lereau M, Bah E, Hall AJ, Wild CP, Mendy M, Norder H, van der Sande M, Whittle H, Friesen MD, Groopman JD, Hainaut P. Seasonal variation in TP53 R249S-mutated serum DNA with afatoxin exposure and hepatitis B virus infection. Environ Health Perspect 2011;119:1635-40.
81. Szymanska K, Chen J G, Cui Y, Gong Y Y, Turner P C, Villar S, Wild C P, Parkin D M, Hainaut P. TP53 R249S mutations, exposure to afatoxin, and occurrence of hepatocellular carcinoma in a cohort of chronic hepatitis B virus carriers from Qidong, China. Cancer Epidemiol Biomarkers Prev 2009;18:1638-43.
82. Hosny G, Farahat N, Tayel H, Hainaut P. Ser-249 TP53 and CTNNB1 mutations in circulating free DNA of Egyptian patients with hepatocellular carcinoma versus chronic liver diseases. Cancer Lett 2008;264:201-8.
Prof. Guang-Wen Cao, Department of Epidemiology, Second Military Medical University, 800 Xiangyin Rd, Shanghai 200433, China. E-mail: gcao@smmu.edu.cn
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
For reprints contact: service@oaepublish.com

