A Carboxymethyl Cellulase from a Marine Yeast (Aureobasidium pullulans 98): Its Purification, Characterization, Gene Cloning and Carboxymethyl Cellulose Digestion
RONG Yanjun, ZHANG Liang, CHI Zhenming, and WANG Xianghong
UNESCO Chinese Center of Marine Biotechnology, Ocean University of China, Qingdao 266003, P. R. China
A Carboxymethyl Cellulase from a Marine Yeast (Aureobasidium pullulans 98): Its Purification, Characterization, Gene Cloning and Carboxymethyl Cellulose Digestion
RONG Yanjun, ZHANG Liang, CHI Zhenming*, and WANG Xianghong
UNESCO Chinese Center of Marine Biotechnology, Ocean University of China, Qingdao 266003, P. R. China
? Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2015
We have reported that A. pullulans 98 produces a high yield of cellulase. In this study, a carboxymethyl cellulase (CMCase) in the supernatant of the culture of A. pullulans 98 was purified to homogeneity, and the maximum production of CMCase was 4.51 U (mg protein)-1. The SDS-PAGE analysis showed that the molecular mass of the purified CMCase was 67.0 kDa. The optimal temperature of the purified enzyme with considerable thermosensitivity was 40℃, much lower than that of the CMCases from other fungi. The optimal pH of the enzyme was 5.6, and the activity profile was stable in a range of acidity (pH 5.0–6.0). The enzyme was activated by Na+, Mg2+, Ca2+, K+, Fe2+and Cu2+, however, it was inhibited by Fe3+, Ba2+, Zn2+, Mn2+and Ag+. Kmand Vmaxvalues of the purified enzyme were 4.7 mg mL-1and 0.57 μmol L-1min-1(mg protein)-1, respectively. Only oligosaccharides with different sizes were released from carboxymethylcellulose (CMC) after hydrolysis with the purified CMCase. The putative gene encoding CMCase was cloned from A. pullulans 98, which contained an open reading frame of 954 bp (EU978473). The protein deduced contained the conserved domain of cellulase superfamily (glucosyl hydrolase family 5). The N-terminal amino acid sequence of the purified CMCase was M-A-P-H-A-E-P-Q-S-Q-T-T-E-Q-T-S-S-G-Q-F, which was consistent with that deduced from the cloned gene. This suggested that the purified CMCase was indeed encoded by the cloned CMCase gene in this yeast.
marine yeast; Aureobasidium pullulans; gene cloning; carboxymethyl cellulase; purification; characterization
1 Introduction
Cellulose is the most abundant organic material on the earth. It has raised considerable economic interests of developing processes of the effective treatment and utilization of cellulosic wastes as an inexpensive carbon source (Lo et al., 2011). Cellulases are the enzymes that degrade crystalline cellulose to glucose. Three types of cellulases, endoglucanases (the carboxymethyl cellulase, CMCase) (EC 3.2.1.4, endo-1,4-β-D-glucanases), cellobiohydrolases (EC 3.2.1.91) and β-glucosidases (EC 3.2.1.21), are considered to be needed for synergistically degrading crystalline cellulose into glucose in vivo (Liu et al., 2013).
Cellulases have diverse applications in environmental, food and agricultural industries. In addition, cellulases are also potentially useful for converting cellulosic plant polymeric biomass into single-cell protein and biofuels (Deshpande et al., 1992; Chi et al., 2006). Cellulases produced by fungi such as those in genus Aspergillus, Rhizopus and Trichoderma have been extensively studiedearly (Akiba et al., 1995; Tang et al., 2012; Ma et al., 2013). Many fungal endoglucanases from members of the subdivision Deuteromycotina, such as Aspergillus sp., Fusarium sp., Humicola sp., Penicillium sp. and Trichoderma sp., have been purified and characterized (Singhania et al., 2010; Zhou et al., 2008). It has been reported that Saccharomycopsis fibuligera and Kluyveromyces marxianus produce also β-glucosidase (Liu et al., 2011b; Fonseca et al., 2008).
Aureobasidium pullulans is popularly known as a black yeast and widely distributed in the phyllosphere of many crops, on various tropical fruits, in fresh water, estuarine, marine sediments, hypersaline habitats, seawater and deep sea (Sugumaran et al., 2013). We found that some strains of A. pullulans isolated from marine environment contain over 30% of the crude protein in their cells that may be used as single cell protein (Chi et al., 2008). It has been observed that most cultures of A. pullulans usually fail to show any cellulolytic activity. For example, a total of 198 isolates studied in Italy did not show cellulolytic activity (Federici et al., 1982; Kudanga et al., 2005). However, Kudanga et al. (2005) reported that some isolates of A. pullulans of tropical origin produce CMCase (endoglucanase) and alpha-cellulase (exoglucanase).
In our previous studies (Zhang et al., 2007), the abilityto produce cellulase by different strains of A. pullulans isolated from different marine environments was determined. We found that A. pullulans 98 produces a high yield of cellulase. In the present study, the CMCase from the supernatant of A. pullulans 98 culture was purified and characterized. At the same time, CMC hydrolysis by the purified CMCase and cloning of the putative gene encoding the CMCase was also performed. In our knowledge, this is the first report of the CMCase purified and characterized from a marine black yeast.
2 Materials and Methods
2.1 Yeast Strain
The marine yeast 98 was isolated from the sea saltern of Yellow Sea, China, and was maintained on YPD medium containing 20.0 g L-1glucose, 20.0 g L-1peptone, 10.0 g L-1yeast extract, and 20.0 g L-1agar at 4℃ and subcultured each month at 28℃. It was a strain of A. pullulans (Zhang et al., 2007).
2.2 CMC and Other Chemicals
CMC was purchased from Amresco, and other analytical chemicals were bought from Sigma.
2.3 Cellulase Production
Two loops of the cells of the strain were transferred to 50.0 mL of YPD medium prepared with seawater in a 250-mL flask and aerobically cultivated for 24 h. Five milliliters of the cell culture with a final concentration of 105cells mL-1were transferred to 45.0 mL of the production medium (prepared with seawater) that contained 20.0 g L-1CMC, 20.0 g L-1yeast extract, and 5.0 g L-1peptone, pH 7.0, and the cells were grown by shaking at 160 r min-1and 28℃ for 24 h. The fermented broth was centrifuged at 2823 × g and 4℃ for 5 min, and the supernatant was collected for the crude enzyme preparation.
2.4 Determination of Cell Dry Weight
Ten milliliters of the cell cultures were collected and washed three times with sterile saline water by centrifugation at 4℃ for 10 min. The pellets obtained were dried at 80℃ until cell dry weight was constant. Determination of cell dry weight was performed by the methods described by Chi et al. (2007).
2.5 Determination of CMCase
The reaction mixture containing 0.8 mL of 5.0 g L-1CMC in 0.2 mol L-1acetate buffer (pH 5.6) and 0.2 mL of the supernatant was incubated at 40℃ for 60 min (Zhang et al., 2007). After appropriate dilution, the reducing sugar in the dilute was determined by the method of Nelson-Somogyi (Spiro, 1966). The similar mixture with the supernatant heated at 100℃ for 5 min was used as a control. One unit of CMCase activity was defined as the amount of enzyme causing release of reducing sugars equivalent to 1.0 μmol L-1of glucose from CMC per min under the assay conditions. The specific CMCase activity was defined as units per mg of protein in the supernatant. Protein concentration was measured by the method of Bradford, and bovine serum albumin served as standard (Bradford, 1976).
2.6 Cloning of the Putative Gene Encoding the CMCase from A. pullulans Strain 98
Genomic DNA of A. pullulans 98 was isolated according to the method described by Adams et al. (1998). To isolate the consensus sequence of the partial gene encoding CMCase from A. pullulans 98, two degenerated primers, forward CEL-1: 3’-CNGGHGGHCARAACCA RGRYTGGC-5’ and reverse CEL-2: 3’-TAGACTGGDC CVMAYTCRCCRTTCC-5’, were designed based on the sequence information of the CMCase genes of Neosartorya fischeri NRRL 181 (XP_001258152), Pichia stipitis CBS 6054 (XP_001387349), Phaeosphaeria nodorum SN15 (XP_001800923), Aspergillus terreus NIH2624 (XP_001216992), Debaryomyces hansenii CBS767 (XP_ 461827) and Pichia guilliermondii ATCC 6260 (XP_ 461827), and used to amplify the target gene. The reaction system in 25.0 μL was composed of 2.5 μL of 10 × buffer, 2.0 μL (2.5 mmol L-1) of dNTP (each), 1.0 μL (100 mmol L-1) of CEL-1 and CEL-2, 0.5 μL of Taq DNA polymerase, 1.0 μL (10.0 ng mL-1) of the template DNA, and 17.0 μL of H2O. The condition for the PCR amplification were as follows: initial denaturation at 94℃ for 10 min, denaturation at 94℃ for 1 min, annealing temperature at 53℃ for 1 min, extension at 72℃ for 2 min, final extension at 72℃ for 10 min. PCR was run for 32 cycles and the PCR cycler was GeneAmp PCR System 2400 (PerkinElmer, Waltham, MA). The PCR products were cloned into pMD19-T and sequenced. The amino acid sequence of the conserved DNA sequence was deduced by using the software on NCBI. Then, an inverse PCR was performed to clone the DNA fragment of the gene encoding the CMCase according to the methods described by Sambrook et al. (1989). The primers for the inverse PCR were designed according to the partial gene encoding the CMCase in the yeast strain. The genomic DNA of the yeast strain was digested with Bam HI. The DNA fragments were purified with TIANquick Mini Purification Kits (Tiangen Biotech Co., Ltd., Beijing). The DNA fragments were circulated by T4DNA ligase (MBI) at 22℃ for 4 h. The circulated DNA was purified again with TIANquick Mini Purification Kits. The inverse PCR reaction system was composed of 5.0 μL 10× La Taq buffer (TaKaRa, Japan), 8.0 μL 2.5 mmol L-1dNTP (each), 1.0 μL 20.0 μmol L-1of the primers (each), 1.0 μL the circulated DNA, 0.5 μL TaKaRa LA Taq DNA polymerase (5.0 U μL-1), and 33.5 μL double-distilled water. Touchdown PCR conditions were 94℃ 8 min; 94℃ 30 s, 66.5℃(0.5 each cycle) 30 s, 72℃ 2.5 min, 20 cycles; 94℃ 30 s, 56.5℃ 40 s, 72℃ 2.5 min, 20 cycles, final extension at 72℃ for 15 min. The PCR products were checked and separated by agarose gel electrophoresis and sequenced. After the sequence of the PCR products was analyzed with NCBI ORF finder program and aligned with theknown sequences of the genes encoding the CMCase from different fungi by NCBI BLASTn, the fragment of 954 bp of the CMCase gene (accession number: EU978473) from the yeast strain 98 was obtained. Multiple alignments of the amino acid sequences were produced using DNA- MAN 6.0.
2.7 Purification of CMCase
Enzyme purification was carried out at 0℃. One liter of the culture aerobically grown in the production medium for 24 h was used as the starting material for the CMCase purification. After removal of the cells by centrifugation at 14006 × g for 20 min, solid ammonium sulfate was added to the supernatant to 80% saturation at 0℃. This solution was stirred for 1 h at 0℃ and then centrifuged at 14006 × g (0℃) for 20 min. The precipitate formed was collected and dissolved in 5.0 mL of 50.0 mmol L-1sodium acetate buffer (pH 5.6). The enzyme solution was dialyzed against the same buffer at 4℃ for 48 h. The protein in the dialysate was concentrated by PEG (20000 MW, Tianjin Chemical Company, China). The concentrated enzyme solution was then applied to SephadexTMG-75 column (medium grade; Pharmacia 2.5 × 100 cm) and the column was eluted with 50.0 mmol L-1sodium acetate buffer (pH 5.6) by using ?KTATM prime with HitrapTM(Amersham, Biosciences, Sweden). At a flow rate of 0.5 mL min-1, 2.0 mL of the fractions was collected. The CMCase-positive fractions were combined and dialyzed in 20.0 mmol L-1Tris-HCl buffer (pH 8.0) overnight. The dialyzed CMCase-positive elute was applied to DEAE Sepharose Fast Flow anion-exchange column (2.5 cm × 30 cm) that had been equilibrated with 20.0 mmol L-1Tris-HCL buffer (pH 8.0) and the column was washed with the same buffer for 1 h at a flow rate of 1.0 mL min-1. The bound proteins were then eluted with a linear gradient of NaCl solution in the range of 0–1.0 mol L-1in the equilibrating buffer. The CMCase-positive fractions were concentrated by filtration through an AmiconYM3 (MW cut-off 10000) membrane.
2.8 Electrophoresis
The purity and molecular mass of the CMCase in the concentrated fractions were analyzed in non-continuous denaturing SDS-PAGE (Laemmli, 1970) with a Two Dimensional Electrophoresis System (Amersham, Biosciences, Sweden) following the instructions from the manufacturer, and the gel was stained by Coomassie Brilliant Blue R-250 (George et al., 1983). The molecular mass standards for SDS-PAGE comprised β-galactosidase (116.0 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45.0 kDa), lactate dehydrogenase (35.0 kDa), restriction endonuclease Bsp981 (25.0 kDa), β-lactoglobulin (18.4 kDa) and lysozyme (14.4 kDa).
2.9 Determination of N-terminal Amino Acid Sequence of the Purified CMCase
To determine the N-terminal amino acid sequence, the band of the purified CMCase on SDS-PAGE as described above was electroblotted to a poly vinylidene fluoride (PVDF) membrane (Bio-Rad) with a CAPS (3-(cyclohexylamino)-1-propanesulfonic acid) buffer system. Electroblotting was conducted at 200 mA for 1 h. The membrane was stained with Ponceau S to visualize the protein. The band was excised and submitted for N-terminal amino acid sequencing with an ABI PROCISETM492cLC protein sequencer (Liu et al., 2011).
2.10 Effects of Temperature and pH on CMCase Activity
The optimal temperature for activity of the enzyme was determined at temperatures of 30, 35, 40, 45, 50, 55 and 60℃ in the same buffer as described above. Temperature stability of the purified enzyme was tested by pre-incubating the enzyme at different temperatures ranging from 30 to 50℃ during 40 min, and the residual activity was measured as described above immediately (Song et al., 2010). Here, the relative CMCase activity of the pre-incubated sample at 4℃ was regarded as 100%.
The effect of pH on the enzyme activity was determined by incubating the purified enzyme between pH 4.0 and 7.0 using the standard assay condition. The buffers used were 0.1 mol L-1acetate buffer (pH 4.0–5.6) and phosphate buffer (pH 6.0–7.0). To test the enzyme stability under different pH values, the purified enzyme was mixed in the similar reaction buffer with pH values ranging from 4.6 to 6.6, and was kept at 4℃ for 12 h. Then the remaining activity of the CMCase was measured immediately after this treatment with the standard method as mentioned above (Song et al., 2010).
2.11 Effects of Different Metal Ions on CMCase Activity
To examine effects of different metal ions on the CMCase activity, enzyme assay was performed in the reaction mixture as described above with various metal ions at a final concentration of 1.0 mmol L-1. The activity assayed in the absence of metal ions was defined as a control. The metal ions tested included Na+, Mg2+, Ca2+, Fe3+, K+, Ba2+, Fe2+, Zn2+, Mn2+, Cu2+and Ag+(Song et al., 2010).
2.12 Determination of Kinetics Parameters
To obtain Kmand Vmaxfor CMC, 0.8 mL of 2.0 g L-1, 5.0 g L-1, 10.0 g L-1, 15.0 g L-1, 20.0 g L-1and 25.0 g L-1of CMC in 0.2 mol L-1acetate buffer (pH 5.6) was mixed with 0.2 mL of the purified CMCase, respectively, and the mixture was incubated at 40℃ for 60 min and the reaction was stopped immediately by heating at 100℃ for 5 min. Kmand Vmaxvalues were obtained from Lineweaver-Burk plot and expressed as the mean of the three different experiments (Song et al., 2010).
2.13 CMC Hydrolysis
The reaction mixture containing 0.2 mL of 3.0 U mL-1of the purified CMCase and 5.0 g L-1CMC in 0.8 mL of the acetate buffer (0.2 mol L-1, pH 5.6) was incubated at40℃ for 60 min, After that, the CMCase in the mixture was inactivated by heating at 100℃ for 10 min immediately (Gong et al., 2007). The end products of the hydrolysis were determined by thin layer chromatography (Li et al., 2012).
3 Results
3.1 Preparation of Crude CMCase
Under the optimal conditions used in this study, the maximum production of CMCase with a specific activity of 4.51 U mg-1protein was obtained when A. pullulans 98 growth reached the late logarithm phase (Table 1).
3.2 Purification of CMCase
CMCase isolated from the supernatant was purified by ammonium sulfate fractionation, gel filtration chromatography (SephadexTMG-75), and DEAE Sepharose Fast Flow anion-exchange chromatography. The elution profile of gel filtration chromatography indicated that peak 2 with the specific enzyme (with CMCase activity) activity from fraction numbers 30 to 45 showed one single peak (data not shown) while the elution profile of DEAE Sepharose Fast Flow anion-exchange chromatography showed that peak 2 with the specific enzyme (with CMCase activity) activity from fraction numbers 17 to 27 displayed one single sharp peak (data not shown). Therefore, the fractions were collected and concentrated by ultrafiltration. The results in Table 1 showed that the enzyme was purified to homogeneity with a 1.4-fold increase in the specific enzyme activity with a yield of about 15.3% as compared to that in the supernatant.
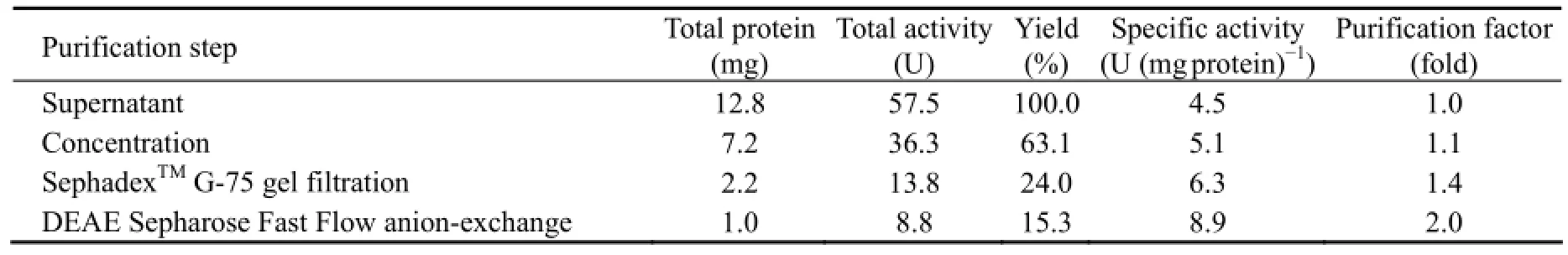
Table 1 Summary of the purification procedure of the enzyme with CMCase activity from A. pullulans 98
3.3 Gel Electrophoresis
The results (Fig.1) obtained from the SDS-PAGE indicated that there was one single protein band from the finally concentrated elute and the relative molecular mass of the purified enzyme was estimated to be 67.0 kDa by SDS-PAGE.
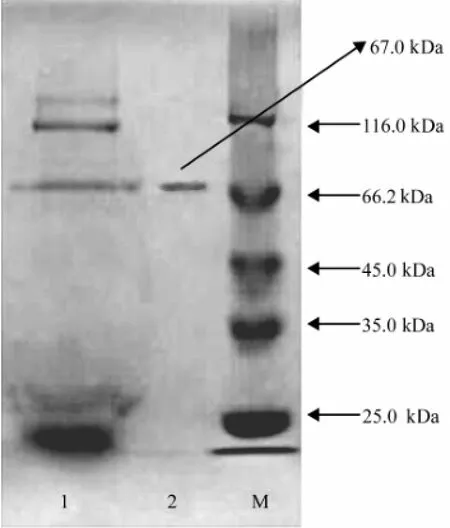
Fig.1 SDS-PAGE (120 g L-1) of the fractions showing the enzyme with CMCase activity obtained during the purification. The lanes are as follows: 1, the concentrated supernatant; 2, the elute from DEAE Sepharose Fast Flow anion exchange chromatography; M, marker proteins with relative molecular masses indicated on the right.
3.4 N-terminal Amino Acid Sequence of the Enzyme
After the determination of the N-terminal amino acid sequence of the purified enzyme with CMCase activity (Liu et al., 2011a), it was found that the sequence of the 20 amino acids at the N-terminus of the purified enzyme was M-A-P-H-A-E-P-Q-S-Q-T-T-E-Q-T-S-S-G-Q-F (data not shown). The N-terminal amino acid sequence was consistent with that of the deduced CMCase from the cloned CMCase gene as shown below. Therefore, the purified enzyme was confirmed to be a CMCase.
3.5 Optimum Temperature and Thermal Stability
The CMCase activity measured as a function of temperature from 30 to 60℃ showed that the activity was the highest at 40℃ (Fig.2). The thermostability was investigated by pre-incubating the enzyme in the same buffer as described in Materials and methods for 40 min and the remaining activity was determined. As shown in Fig.2, the residual CMCase activity still kept 95% of the control after treatment at 30℃ for 40 min, indicating that the en-zyme was stable up to 30℃. Fig.2 also revealed that the enzyme was inactivated rapidly at temperature higher than 30℃ and was inactivated totally at 50℃ within 40 min. From these results, the CMCase seemed to have considerable thermosensitivity.
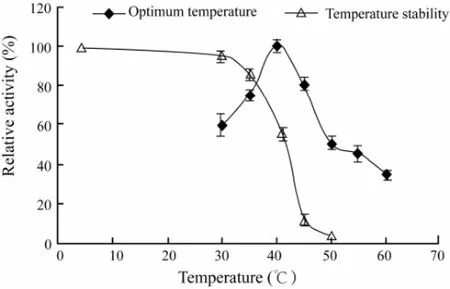
Fig.2 Effects of different temperatures on the purified CMCase activity and stability. Data are given as means ± SD, n=3.
3.6 Optimum pH and pH Stability
The CMCase activity was measured at various pH values in buffers with the same ionic concentration. Our results (Fig.3) showed that the maximum activity was observed at pH 5.6. The pH stability was tested by 12 h pre-incubation of the purified enzyme in appropriate buffers that had the same ionic concentrations at different pH values ranging from 4.6 to 6.6 at 4℃. The remaining activities of the CMCase were measured immediately after this treatment with the standard method as mentioned above. It can be seen from the results in Fig.3 that the activity profile of the enzyme was stable in the range of pH 5.0 to pH 6.0.
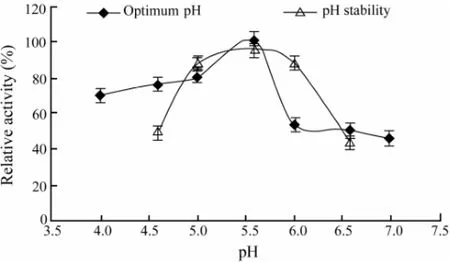
Fig.3 Effects of different pH values on the purified CMCase activity and stability. Data are given as means ± SD, n=3.
3.7 Effects of Different Cations on Activity of the Purified CMCase
It was found that Na+, Mg2+, Ca2+, K+, Fe2+and Cu2+(at the concentrations of 1.0 mmol L-1) activated the activity of the purified CMCase (Fig.4). However, Fe3+, Ba2+, Zn2+, Mn2+and Ag+(at the concentrations of 1.0 mmol L-1) acted as inhibitors in decreasing activity of the purified CMCase, with Ba2+(at the concentration of 1.0 mmol L-1) showing the lowest rank (65.0%) (Fig.4), suggesting that they were able to alter the enzyme conformation.
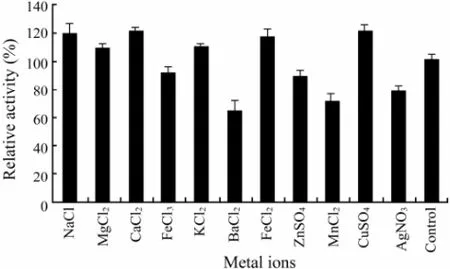
Fig.4 The effect of different metal ions on the purified CMCase activity. Data are given as means ± SD, n=3.
3.8 Kinetics Parameters
Lineweaver-Burk plots showed that apparent Kmand Vmaxvalues of the CMCase for CMC were 4.7 mg mL-1and 0.6 μmol L-1min-1(mg protein)-1, respectively (Fig.5).
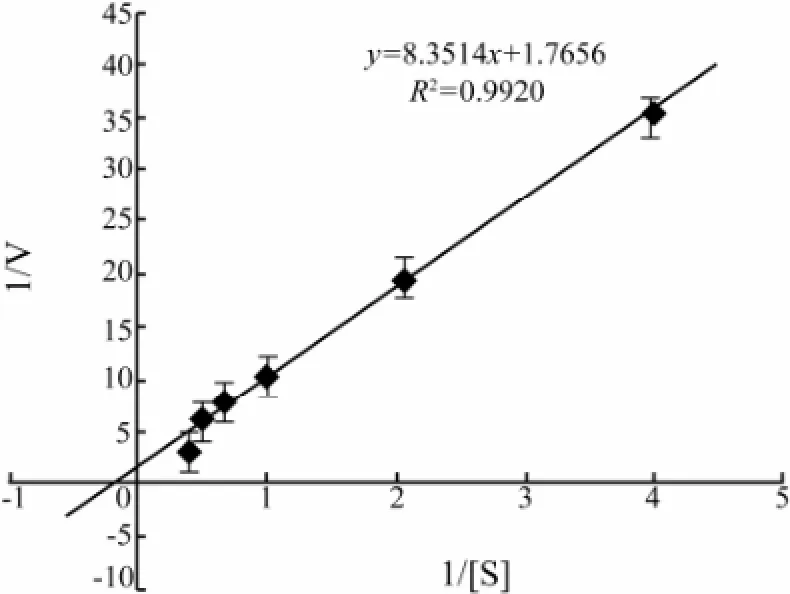
Fig.5 Lineweaver-Burk plot for Kmand Vmaxvalues of the CMCase in the presence of different concentrations of CMC. Data are given as means ± SD, n =3.
3.9 CMC Hydrolysis
The results (Fig.6) demonstrated that only oligosaccharides with different sizes were released from CMC after hydrolysis for 1 h with the purified CMCase.
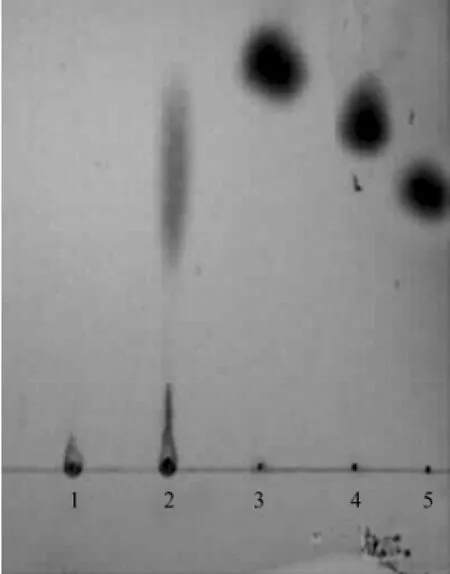
Fig.6 Thin-layer chromatogram of the hydrolysis products of CMC with the purified CMCase. Lane 1: Control (CMC + the inactivated CMCase); Lane 2: CMC + the purified CMCase; Lane 3: Glucose; Lane 4: Cellobiose; Lane 5: Kestose.
3.10 Cloning of the Putative Gene Encoding the CMCase from A. pullulans Strain 98

Fig.7 Multiple alignments of the deduced CMCase amino acid sequences from different fungi. 0: The amino acid sequence deduced from the cloned gene in the study. 1: Hypothetical protein SNOG_10661 [Phaeosphaeria nodorum SN15]. 2: Cellulase, putative [Neosartorya fischeri NRRL 181]. 3: Conserved hypothetical protein [Aspergillus terreus NIH2624]. 4: Hypothetical protein DEHA0G07095g [Debaryomyces hansenii CBS767]. 5: Hypothetical protein PGUG_00389 [Pichia guilliermondii ATCC 6260]. 6: Endo-1,4-β-glucanase [Pichia stipitis CBS 6054].
The conserved motifs were used to design the degenerate primers to clone the partial gene encoding the CMCase in A. pullulans 98. In this case, amino acid sequences of the extracellular CMCases from different species of eukaryotic microorganisms were downloadedfrom GenBank (http://www.ncbi.nlm.nih.gov) and aligned. The PCR-generated fragments were sequenced. Then, an inverse PCR was performed to clone the DNA fragment of the gene encoding the CMCase. Analysis of the sequence by BLAST program indicated that the fragment (954 bp) of the putative CMCase gene was isolated as the amino acid sequence deduced from the fragment (954 bp) of the putative CMCase gene (EU978473) had high similarities to those of CMCase genes from N. fischeri NRRL 181 (XP_001258152), P. stipitis CBS 6054 (XP_001387349), P. nodorum SN15 (XP_001800923), A. terreus NIH 2624
(XP_001216992), D. hansenii CBS 767 (XP_461827) and P. guilliermondii ATCC 6260 (XP_461827) (Fig.7). Analysis of the protein deduced from the gene cloned in this study at http://motif.genome.jp/showed that the protein had the conserved domain of cellulase superfamily (glucosyl hydrolase family 5) from 90 to 328 amino acids (Fig.8), and the N-terminal amino acid sequence M-AP-H-A-E-P-Q-S-Q-T-T-E-Q-T-S-S-G-Q-F deduced from the cloned gene was completely in agreement with the N-terminal amino acid sequence of the purified CMCase (subsection 3.4).
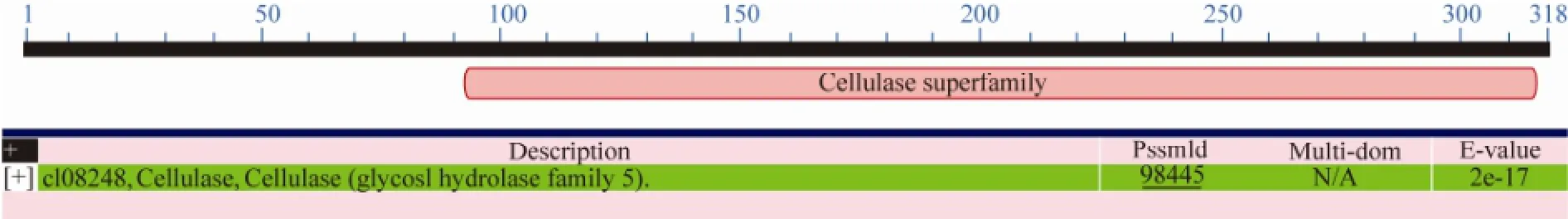
Fig.8 The conserved domain of the protein deduced from the cloned gene in this study. 90–318 amino acids deduced from the cloned gene are the same conserved domain as cellulase (glycosyl hydrolase family 5).
4 Discussion
The results (Figs.7 and 8) showed that the CMCase deduced from the cloned gene in this study was a member of the glycosyl hydrolase family 5. The family includes cellulase (EC 3.2.1.4); endo-1,4-β-xylanase (CMCase) (EC 3.2.1.8); cellulose 1,4-β-cellobiosidase (EC 3.2.1.91); xyloglucan-specific endo-β-1,4-glucanase (EC 3.2.1.151) and others (http://www.cazy.org/), which can hydrolyze cellulose and xylan. The N-terminal amino acid sequence of the purified enzyme with CMCase activity was M-AP-H-A-E-P-Q-S-Q-T-T-E-Q-T-S-S-G-Q-F (subsection 3.4). The results in Figs.7 and 8 showed that the N-terminal amino acid sequence deduced from the cloned CMCase gene in this yeast contained the same N-terminal amino acid sequence of the purified enzyme, suggesting that the purified enzyme was indeed a CMCase and was encoded by the cloned CMCase gene in this yeast. The sequence of the deduced CMCase exhibited 52% similarity with that of CMCase from Phaeosphaeria nodorum SN15. After comparing with the known N-terminal amino acid residues of CMCases, no CMCase with the same corresponding N-terminal amino acid sequence was observed (Fig.7). These results may imply that the CMCase from this marine yeast strain 98 was a novel enzyme.
Under the optimal conditions, maximum production of the CMCase of 4.51 U mg-1protein by A. pullulans 98 was obtained (Table 1). Because A. pullulans is widely distributed in different marine environments (Li et al., 2007) and can produce different kinds of extracellular enzymes, including cellulase (Zhang et al., 2007; Chi et al., 2007), it may play a role in degradation of polymers including cellulose in marine environments.
The relative molecular mass of the purified CMCase produced by A. pullulans 98 was found to be 67.0 kDa as indicated by a single band on the SDS-PAGE gel (Fig.1). Therefore, it may be concluded that the CMCase from the marine yeast strain was a monomer of 67.0 kDa protein. It has been reported that the molecular masses of an endoβ-1, 4-glucanase purified from Aspergillus niger strain IF031125, the extracellular endoglucanases, designated as RCE1 and RCE2, from Rhizopus oryzae, the CMCase IIIa from Favolus arcularius, the heterologously expressed CMCase encoded by CMCase 1 gene in the yeast Cryptococcus flavus and FI-CMCases from Aspergillus aculeatus were 40.0, 41.0, 61.0, 28.0, 34.0 and 24.0 kDa, respectively ( Murashima et al., 2002; Akiba et al., 1995; Enokibara et al., 1992; Hatano et al., 1994; Minamiguchi et al., 1995). These results indicated that the molecular mass of the CMCase from the marine yeast strain 98 was higher than that of the CMCase from filamentous fungi and other yeasts.
The enzyme activity produced by A. pullulans 98 was the highest at 40℃ and the enzyme was stable up to 30℃(Fig.2). It has been reported that the optimum temperature of the extracellular endoglucanases, designated as RCE1 and RCE2, from R. oryzae was 55℃ (Murashima et al., 2002), while the optimal temperature of the FI-CMCases from A. aculeatus was 50℃ (Minamiguchi et al., 1995). The optimal temperature of the endo-glucanase 3 and endo-glucanase 4 from Humicola grisea, and englucanase I from Scopulariopsis brevicaulis was 60℃, 75℃ and 60℃, respectively (Murashima et al., 2002). These results indicated that the optimal temperature of the CMCase from the marine yeast strain 98 was much lower than that of the CMCase from any other fungi. This may be related to the marine environment where the yeast was isolated. These characteristics of the CMCase may be of interest in biotechnology and textile industry as the CMCase activity could be easily inactivated at higher temperature after use.
The maximum enzyme activity produced by A. pullulans 98 was observed at pH 5.6, and the activity of the enzyme was stable in the range of pH 5.0 to pH 6.0 (Fig.3). It has been reported that the purified endo-β-1,4-glucanase from A. niger strain IF031125, has the optimal pH 6.0 and is stable in the pH range of 5.0-10.0 (Akiba et al., 1995). The optimal pH of FI-CMCases from A. aculeatus is 5.0 (Minamiguchi et al., 1995), and the enzyme is stable in the pH range of 2.0–9.0. For the extracellular endoglucanases, designated as RCE1 and RCE2 from R. oryzae, the optimum pH is between 5.0 and 6.0 (Enokibara et al., 1992). These results demonstrated that the CMCase isolated from fungi and yeasts worked the best at acidic environment.
It has been reported the purified endo-β-1,4-glucanase from A. niger strain IF031125 was inhibited by Cu2+(Akiba et al., 1995). The CMCase, designated as RCE1 and RCE2, from R. oryzae was inhibited by Cu2+and Zn2+, and in particular, Cu2+extremely inhibited this activity (Murashima et al., 2002). However, in our research, Cu2+could increase the activity of the purified CMCase (Fig.4). These results indicated that some biochemical properties of the CMCase from the marine yeast strain 98 were not the same as those of the CMCase from fungi.
The apparent Kmand Vmaxvalues of the CMCase produced by A. pullulans strain 98 for CMC were 4.7 mg mL-1and 0.6 μmol L-1min-1(mg protein)-1, respectively (Fig.5). It has been reported that the apparent Kmvalue of the CMCase from Favolus arcularius against CMC was 0.28 mg mL-1(Enokibara et al., 1992). These results revealed that the CMCase from the marine yeast strain 98 displayed low affinity for CMC.
The CMCase produced by A. pullulans strain 98 was found to be an endo-1, 4-β-D-glucanase (Fig.6). Both RCE1 and RCE2 from R. oryzae hydrolyzed the insoluble cellooligosaccharide (G33) to G2 (disaccharides), G3 (trisaccharides), and G4 (tetrasaccharides). G6 (hexosaccharides) was hydrolyzed to G2 and G4. G5 (pentosaccharides) was hydrolyzed to G2 and G3. G4 was weakly hydrolyzed to G2 by both the enzymes. Both the enzymes were found not to act on G2 or G3 (Murashima et al., 2002). This meant that like the CMCase from R. oryzae and the CMCase from the marine yeast strain 98 could not act on cellobiose.
Heterologous expression of the cloned gene in Bacillus subtilis or Saccharomyces cerevisiae and the biochemical characteristics of the recombinant CMCase, such as the CMC digestion capacity, are being investigated in this laboratory.
5 Conclusions
The molecular weight of the CMCase purified from A. pullulans 98 was 67.0 kDa. The optimal pH and temperature of the enzyme were 5.6 and 40℃, respectively. The enzyme was activated by Na+, Mg2+, Ca2+, K+, Fe2+and Cu2+. Only oligosaccharides with different sizes were released from CMC after the hydrolysis by the CMCase. The protein deduced from the putative gene encoding CMCase from A. pullulans 98 consisted of the conserved domain of cellulase superfamily (glucosyl hydrolase family 5), and the N-terminal amino acid sequence of the purified CMCase had the same corresponding sequence deduced from the cloned CMCase gene in this yeast.
Acknowledgement
The authors would like to thank Qingdao Municipal Science and Technology Commission, Qingdao, China for providing financial support to this work (06-2-2-22-jch).
Adams, A., Gottschling, D. E., Kaiser, C. A., and Stearns, T., 1998. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, New York, 84-88.
Akiba, S., Kimura, Y., Yamamoto, K., and Kumagai, H., 1995. Purification and characterization of a protease-resistant cellulase from Aspergillus niger. Journal of Fermentation and Bioengineering, 79: 25-30.
Bradford, M. M., 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72: 248-253.
Chi, Z. M., Ma, C., Wang, P., and Li, H. F., 2007. Optimization of medium and cultivation conditions for alkaline protease production by the marine yeast Aureobasidium pullulans. Bioresource Technology, 98: 534-538.
Chi, Z. M., and Liu, Z. Q., 2006. Marine yeasts and their applications in mariculture. Journal of Ocean University of China, 5 (3): 243-247.
Chi, Z. M., Yan, K. R., Gao, L. M., Li, J., and Wang, X. H., 2008. Diversity of marine yeasts with high protein content and evaluation of their nutritive compositions. Journal of Marine Biology Association UK, 88: 1347-1352.
Deshpande, M. S., Rale, V. B., and Lynch, J. M., 1992. Aureobasidium pullulans in applied microbiology: A status report. Enzyme and Microbial Technology, 14: 514-527.
Enokibara, S., Mori, N., and Kitamoto, Y., 1992. Purification and some properties of a carboxymethyl cellulase from Favolus arcularius. Journal of Fermentation Bioengineering, 73: 230-232.
Federici, F., 1982. Extracellular enzymatic activities in Aureobasidium pullulans. Mycology, 74: 738-743.
Fonseca, G. G., Heinzle, E., Wittmann, C., and Gombert, A. K., 2008. The yeast Kluyveromyces marxianus and its biotechnological potential. Applied Microbiology and Biotechnology, 79: 339-354.
George, V., and Diwan, A. M., 1983. Simultaneous staining of proteins during polyacrylamide gel electrophoresis in acidic gels by countermigration of Coomassie brilliantblue R-250. Analytical Biochemistry, 132: 481-483.
Gong, F., Sheng, J., Chi, Z. M., and Li, J., 2007. Inulinase production by a marine yeast Pichia guilliermondii and inulin hydrolysis by the crude inulinase. Journal of Industrial Microbiology and Biotechnology, 34: 179-185.
Hatano, T., Komura, T., Cui, Z., Mochizuki, D., Miyakawa, T., and Matsuzaki, H., 1994. Expression of the carboxymethylcellulase gene, CMC1, from Cryptococcus flavus in Saccharomyces cerevisiae. Journal of Fermentation Bioengineering, 77: 235-238.
Ikeda, Y., Park, E. Y., and Okida, N., 2006. Bioconversion of waste office paper to gluconic acid in a turbine blade reactor by the filamentous fungus Aspergillus niger. BioresourceTechnology, 97: 1030-1035.
Kim, K. C., Yoo, S. S., Oh, Y. A., and Kim, S. J., 2003. Isolation and characteristics of Trichoderma harzianum FJ1 producing cellulases and xylanase. Journal of Microbiology and Biotechnology, 13: 1-8.
Kudanga, T., and Mwenje, E., 2005. Extracellular cellulase production by tropical isolates of Aureobasidium pullulans. Microbiology, 51: 773-776.
Laemmli, U. K., 1970. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature, 227: 680-685.
Li, H. F., Chi, Z. M., Duan, X. H., Wang, L., and Sheng, J., 2007. Glucoamylase production by the marine yeast Aureobasidium pullulans N13d and hydrolysis of potato starch granules by the enzyme. Process Biochemistry, 42: 462-465.
Liu, G. L., Li, Y., Zhou, H. X., Chi, Z. M., and Madzak, C., 2012. Over-expression of a bacterial chitosanase gene in Yarrowia lipolytica and chitosan hydrolysis by the recombinant chitosanase. Journal of Molecular Catalysis B: Enzymatic, 83: 100-107.
Liu, G. L., Li, Y., Chi, Z., and Chi, Z. M., 2011a. Purification and characterization of κ-carrageenase from the marine bacterium Pseudoalteromonas porphyrae for hydrolysis of κcarrageenan. Process Biochemistry, 46: 265-271.
Liu, G. L., Wang, D. S., Wang, L. F., Zhao, S. F., and Chi, Z. M., 2011b. Mig1 is involved in mycelia formation and expression of the genes encoding extracellular enzymes in Saccharomycopsis fibuligera A11. Fungal Genetics and Biology, 48: 904-913.
Liu, J. J., and Cao X. J., 2013. Biodegradation of micro-crystalline cellulose in pH-pH recyclable aqueous two-phase systems with water-soluble immobilized cellulase. Biochemical Engineering Journal, 79: 136-143.
Lo, C. M., Zhang, Q., Callow, N. V., and Ju, L. K., 2010. Cellulase production by continuous culture of Trichoderma reesei Rut C30 using acid hydrolysate prepared to retain more oligosaccharides for induction. Bioresource Technology, 101: 717-723.
Ma, C. L., Ni, X. M., Chi, Z. M., Ma, L. Y., and Gao, L. M., 2007. Purification and characterization of an alkaline protease from the marine yeast Aureobasidium pullulans for bioactive peptide production from different sources. Marine Biotechnology, 9: 343-351.
Ma, L. J., Li, C., Yang, Z. H., Jia, W. D., Zhang, D. Y., and Chen, S. L., 2013. Kinetic studies on batch cultivation of Trichoderma reesei and application to enhance cellulase production by fed-batch fermentation. Journal of Biotechnology, 166: 192-197.
Minamiguchi, K., Ooi, T., Kawaguchi, T., Okada, H., Murao, S., and Arai, M., 1995. Secretive expression of the Aspergillus aculeatus cellulase (FI-CMCase) by Saccharomyces cerevisiae. Journal of Fermentation Bioengineering, 79: 363-366.
Murashima, K., Nishimura, T., Nakamura, Y., Koga, J., Moriya, T., and Sumida, N., 2002. Purification and characterization of new endo-1,4-β-D-glucanases from Rhizopus oryzae. Enzyme and Microbial Technology, 30: 319-326.
Singhania, R. R., Sukumaran, R. K., Patel, A. K., Larroche, C., and Pandey, A., 2010. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microbial Technology, 46: 541-549.
Song, C, Liu, G. L., Xu, J. L., and Chi, Z. M., 2010. Purification and characterization of extracellular β-galactosidase from the psychrotolerant yeast Guehomyces pullulans 17-1 isolated from sea sediment in Antarctica. Process Biochemistry, 45: 954-960.
Spiro, R. G., 1966. Analysis of sugars found in glycoproteins. Methods in Enzymology, 8: 3-26.
Sugumaran, K. R., Gowthami, E., Swathi, B., Elakkiya, S., Srivastava, S. N., Ravikumar, R., Gowdhaman, D., and Ponnusami, V., 2013. Production of pullulan by Aureobasidium pullulans from Asian palm kernel: A novel substrate. Carbohydrate Polymers, 92: 697-703.
Tang, B., Pan, H. B., Tang, W. J., Zhang, Q. Q., Ding, L. X., and Zhang, F. Q., 2012. Fermentation and purification of cellulose from a novel strain Rhizopus stolonifer var. reflexus TP-02, Biomass and Bioenergy, 36: 366-372.
Zhang, L., and Chi, Z. M., 2007. Screening and identification of a cellulase producing marine yeast and optimization of medium and cultivation conditions for cellulase production. Journal of Ocean University of China, 37: 101-108.
Zhou, J., Wang, Y. H., Chu, J., Zhuang, Y. P., Zhang, S. L., and Yin, P., 2008. Identification and purification of the main components of cellulases from a mutant strain of Trichoderma viride T 100-14. Bioresource Technology, 99: 6826-6833.
(Edited by Qiu Yantao)
(Received January 8, 2014; revised March 3, 2014; accepted May 26, 2015)
J. Ocean Univ. China (Oceanic and Coastal Sea Research)
DOI 10.1007/s11802-015-2574-4
ISSN 1672-5182, 2015 14 (5): 913-921
http://www.ouc.edu.cn/xbywb/
E-mail:xbywb@ouc.edu.cn
* Corresponding author. Tel: 0086-532-82032266
E-mail: zhenming@sdu.edu.cn
 Journal of Ocean University of China2015年5期
Journal of Ocean University of China2015年5期
- Journal of Ocean University of China的其它文章
- The Mechanism of the Acclimation of Nannochloropsis oceanica to Freshwater Deduced from Its Transcriptome Profiles
- Effects of Dietary Stachyose on Growth Performance, Digestive Enzyme Activities and Intestinal Morphology of Juvenile Turbot (Scophthalmus maximus L)
- Pharmacokinetics and Biodegradation of Chitosan in Rats
- Pharmacokinetics and Biodegradation Performance of a Hydroxypropyl Chitosan Derivative
- Changes in Plasma Osmolality, Cortisol and Amino Acid Levels of Tongue Sole (Cynoglossus semilaevis) at Different Salinities
- Effects of Different Microbes on Fermenting Feed for Sea Cucumber (Apostichopus japonicus)
