EGFR gene copy number as a predictive biomarker for resistance to anti-EGFR monoclonal antibodies in metastatic colorectal cancer treatment: a meta-analysis
Wei-Dong Shen,Hong-Lin Chen,Peng-Fei Liu
1Department of Gastroenterology,the Affiliated Jiangyin Hospital of Nantong University,Shoushan Road 163#,Jiangyin 214400,China; 2Nantong University,Qixiu Road 19#,Nantong 226019,China.
Introduction
Colorectal cancer is the third most common cause of cancer,which ranked second to third of overall cancer incidence and mortality (1,2).Over the past decade,new biologic therapies beyond the old standard-of-care,biomodulated fluorouracil (5-FU),have become available for the treatment of metastatic colorectal cancer (mCRC) (3).The epidermal growth factor receptor (EGFR),represents an important target for cancer treatment because its activation stimulates key processes involved in tumor growth and progression,including proliferation,angiogenesis,invasion,and metastasis (4).Although EGFR tyrosine kinase inhibitors(TKIs) have shown little activity in mCRC treatment,the EGFR inhibitors monoclonal antibodies (MoAbs) have already shown the therapeutic effectiveness.Cetuximab,the first anti-EGFR MoAb,is a chimeric mouse-human monoclonal antibody that has been approved has clinically significant activity when given alone or in combination with chemotherapy in mCRC patients (5,6).Panitumumab,a fully human monoclonal antibody,has also shown efficacy as monotherapy in chemotherapy-refractory patients with mCRC (7).However,only 10-20% of mCRC patients are response and clinically benefited from anti-EGFR MoAbs(5,7).Response rate is an important predictor of survival in trials of mCRC (8).It is important to identify those who are more likely to respond and make the treatment more personalized.
Active KRAS mutations,in signaling pathways downstream of the EGFR,have been accepted to be a major predictive marker of resistance to EGFR targeted cetuximab- or panitumumab-based therapies.The presence of KRAS mutations was significantly associated with an absence of response to anti-EGFR MoAbs for mCRC patients [sensitivity=0.47 (0.43-0.52); specificity=0.93 (0.83-0.97); +LR=6.82; -LR=0.57].But the low sensitivity and relatively high -LR of KRAS mutations for determining non-responsiveness clearly shows that additional mechanisms of resistance to EGFR inhibitors exist (9).Other biomarkers from signaling pathways downstream of the EGFR have been investigated,and it is found that mutation of BRAF or PIK3CA or loss of PTEN expression may be also associated with resistance to EGFR-targeted cetuximab- or panitumumab-based therapies (10,11).
Some other biomarkers except the EGFR signaling pathways downstream also have been investigated.In 2005,EGFR gene copy number (GCN) was fi rst found have the association with clinical response to anti-EGFR treatment.Eight of nine patients with objective responses who were assessable by fluorescencein situhybridization (FISH)had an increased EGFR copy number.By contrast,one of 21 non-responders had an increased EGFR copy number(P<0.0001 for respondersvs.non-responders,Fisher’s exact test) (12).It has been confirmed by subsequent research(13,14).The positive predictive values for GCN were 40.0-48.3%,and the negative predictive values were 81.0% and 86.5% (14).But these results were all came from small sample size studies,and it has not been systematic reviewed.
In this article,we performed a meta-analysis to summarize the scientific evidence for the association between EGFR GCN and tumor response in mCRC patients treated with anti-EGFR MoAbs,and assess the sensitivity and specificity for predicting complete or partial response to anti-EGFR MoAbs.We also present the results of meta-analysis evaluating the relationship between increased EGFR GCN and survival for patients with mCRC receiving cetuximabor panitumumab-based therapies as secondary outcomes.
Methods
Database and literature search
Systematic computerized searches of the PubMed,EMBase and Cochrane Library were performed until 30 January 2013.The following search terms were used: “mCRC”,“metastatic colon cancer”,“metastatic rectal cancer”,“mCRC”,“EGFR”,“MoAbs”,“cetuximab”,“Erbitux”,“panitumumab”,“Vectibix”,“amplification” and “GCN”.The whole search strategies were listed in the appendix.We also looked at posters from the annual meetings of the American Society of Clinical Oncology (ASCO) (http://www.asco.org/ASCOv2/Meetings) and the European Society for Medical Oncology (ESMO) (http://www.esmo.org) in the past ten years.The references of all relevant studies were also manually reviewed to supplement our searches.Only studies published in English were included.
Study selection
The relevant clinical trials were manually selected carefully based on the following criteria: (I) investigated patients with mCRC who treated with anti-EGFR MoAbs; (II)EGFR GCN was tested by FISH or chromogenicin situhybridization (CISH) in all or part of the patients in the studies; and (III) reported or allowed the calculation of odd ratio (OR) with corresponding 95% confidence intervals (95% CIs) comparing objective response rate(ORR) stratified by EGFR GCN,reported or allowed the calculation of hazard ratios (HRs) with 95% CIs comparing progression-free survival (PFS) and overall survival(OS) stratified by EGFR GCN.When the same patient population was used in several papers,only the most recent studies were included in the meta-analysis.We excluded case reports and case series.
Assessment of study quality and data extraction
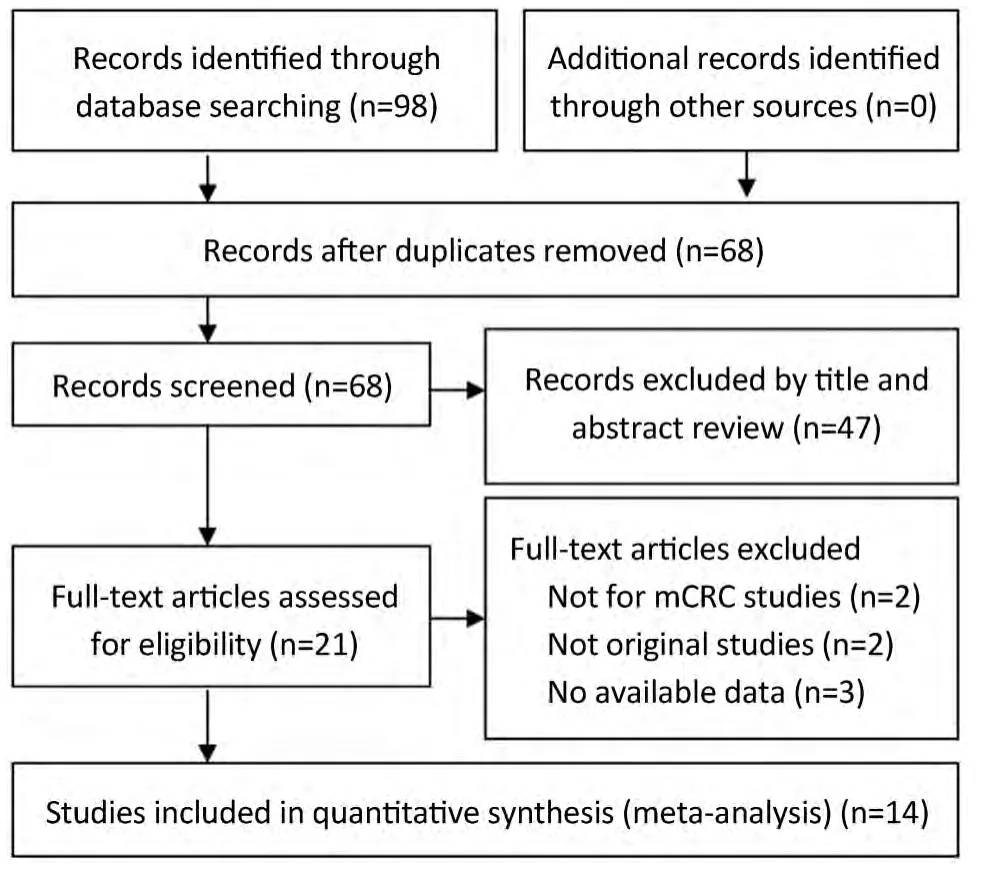
Figure 1 Flow diagram showing selection of studies.
Because there is no validated instrument to measure study quality for predictive marker studies in an observational setting,we adapted the Newcastle-Ottawa Scale and the frame work suggested by Wells (15).The Newcastle-Ottawa Scale (NOS) contains eight items,categorized into three dimensions including Selection [4],Comparability [1],and Exposure [3].A high-quality study can be awarded a maximum of one star for each numbered item within the Selection and Exposure categories.A maximum of two stars can be given for Comparability.The NOS ranges between zero up to nine stars.The following data were abstracted onto standardized forms: (I) basic information from papers such as fi rst author,publication year,country;(II) characteristics of patients such as age and gender; (III)information of treatment such as type of MoAbs (cetuximab or panitumumab); (IV) information of the outcome impact factors such as detection method,response criteria,GCN cutoff,and KRAS; and (V) information of outcome such as ORR,PFS and OS.Study quality assessment and data extraction were carried out independently by two reviewers.Disagreements were resolved by discussion between the two reviewers.
Statistical analysis
For the meta-analysis,ORR was defined as the primary outcome and PFS and OS as secondary outcomes.
For the primary outcome,the association between ORR and EGFR GCN was expressed as pooled OR.Overall effects were determined using the Z test.Predictive value was accessed by pooled sensitivity,pooled specificity and summary receiver operator characteristic (SROC).The area under the curve (AUC) and an index Q*are useful summaries of the curve (16).
For the secondary outcome,the association between PFS and OS and EGFR GCN was expressed as pooled hazard ratio (HR).The methods to combine time-toevent outcomes were summarized by the log HR and its variance (17,18).If the individual trials didn’t provide sufficient data,we extracted the data from the Kaplan-Meier survival curves by previously reported method (19)and the HR calculations spreadsheet (Additional file 1 of the paper,http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1920534/?tool=pubmed#S1).The survival curves were read by Engauge Digitizer version 4.1 (free software downloaded from http://digitizer.sourceforge.net).
For the primary outcome (ORR),we also did subgroup analyses.It were performed to evaluate the effect by ethnicity (Asian or Europe),MoAbs (cetuximab or panitumumab),EGFR GCN detection method (FISH or CISH),and response criteria [Response Evaluation Criteria in Solid Tumors (RECIST) or WHO].
Statistical heterogeneity was explored by χ2and inconsistency (I2) statistics; an I2value of 50 percent or more represented substantial heterogeneity (20).In the absence of heterogeneity,studies were pooled using a fi xedeffect model.If heterogeneity was observed,a randomeffects model was used.An estimate of potential publication bias for primary outcome was carried out by the Egger regression test and Begg adjusted rank correlation test.
The meta-analyses for pooled OR and pooled HR were performed with Stata software 12.0 (Stata Corp.,College Station,Texas).And the meta-analyses for pooled sensitivity,specificity and SROC were performed with Meta DiSc 1.4(By Joseph Lau) (21).
Results
Eligible studies
We identified 14 studies (12-14,22-32) that met our inclusion criteria for meta-analysis.The detailed steps of our literature search are shown inFigure 1.A total of 1,021 patients were used in the pooled analyses.Of the 14 studies,sample sizes ranged from 22 to 173.Seven of these studies were conducted in Italy (12,13,22,25,27,29,32),3 in France (28,30,31),and the rest of 4 conducted in Finland (23),China (24),Spain (26),and Belgium (14),respectively.The patients of 11 studies (14,22,24-32) received cetuximab treatment,while the patients of 2 studies (12,23) received cetuximab or panitumumab treatment,and only the patients of 1 study (13) received panitumumab treatment.RECIST were used as the response criteria in 13 studies (12-14,22-25,27-32),only a study (26) used criteria of WHO.A total of 10 studies (12-14,24-26,28-30,32) used FISH to detect EFGR GCN,3 studies (22,23,31) used CISH,and the rest one study (27) used FISH and CISH simultaneously.Three studies (22,27,30) provided KRAS wild-type patients and 11 studies (12-14,23-26,28,29,31,32) were in patients unselected by KRAS mutation status,while form these 11 studies,4 studies (12,14,23,32) provided KRAS wild-type patients data.The quality rating of the included studies ranged from 6 to 8 stars on the scale of 9.Table 1shows the main characteristics of the 14 included studies,andTable 2shows the outcome results of the studies by GCN cutoff.
Main results of overall response rate
The pooled ORR was 65.2% (167/256) in patients with high EFGR GCN,while in patients with low EFGR GCN,the pooled ORR was 12.2% (44/361).There was no heterogeneity in the studies (I2=0.0%,P=0.584).The pooled OR was 6.905 (95% CI: 4.489-10.620; Z=8.79,P=0.000)by fi xed-effect model (Figure 2),and 6.301 (95% CI: 4.023-9.870; Z=8.04,P=0.000) by random-effects model.The Begg’s test (Z=1.37,P=0.171) and the Egger’s test (t=2.29,P=0.041) suggested there was significant publication bias.
While in wild-type KRAS mCRC patients,the pooled ORR was 83.9% (78/93) in patients with high EFGR GCN,while in patients with low EFGR GCN,the pooled ORR was 14.3% (20/140).There was no heterogeneity in the studies (I2=0.0%,P=0.952).The pooled OR was 8.133(95% CI: 4.316-15.326; Z=6.48,P=0.000) by fixed-effect model (Figure 3),and 7.955 (95% CI: 4.211-15.027; Z=6.39,P=0.000) by random-effects model.The Begg’s test (Z=0.15,P=1.000) and the Egger’s test (t=1.14,P=0.307) suggested no publication bias.
Predictive value for overall response
The pooled sensitivity for increased EFGR GCN to predict ORR was 0.79 (95% CI: 0.73-0.84).The pooled specificity was 0.59 (95% CI: 0.55-0.62).The overall weighted AUC was 0.7632±0.0309,and the overall accuracy (Q*) was 0.7044±0.0259 (Figure 4A).
While wild-type KRAS mCRC patients,the pooled sensitivity was 0.80 (95% CI: 0.70-0.87),the pooled specificity was 0.60 (95% CI: 0.53-0.66),the overall weighted AUC was 0.7899±0.0454,and the overall accuracy(Q*) was 0.7271±0.0391 (Figure 4B).
Main results of progression-free survival (PFS)
The median of PFS ranged from 2.5 to 8.8 months.There was medium heterogeneity in the studies (I2=48.3%,P=0.060).The pooled HR was 0.557 (95% CI: 0.382-0.732;Z=6.26,P=0.000) by random-effects (Figure 5).It shows high EFGR GCN had benefit effect on PFS when treated with cetuximab or panitumumab in mCRC patients.
Main results of overall survival (OS)
The median of OS ranged from 4.8 to 21.2 months.There was no heterogeneity in the studies (I2=0.0%,P=0.852).The pooled HR was 0.579 (95% CI: 0.422-0.737; Z=7.21,P=0.000) fi xed-effect model (Figure 6).It shows high EFGR GCN had benefit effect on OS when treated with cetuximab or panitumumab in mCRC patients.
Subgroup analyses for overall response rate
The results of subgroup analysis are presented inTable 3.In ethnicity subgroup,increased EGFR GCN was statistically significantly associated with increased ORR in studies of Europe populations (HR=6.905; 95% CI: 4.489-10.620) and Asian populations (HR=6.787; 95% CI: 4.374-10.531).In MoAbs subgroup,increased EGFR GCN was statistically significantly associated with increased ORR in studies of cetuximab treated patients (HR=6.296; 95% CI: 3.990-9.935) and panitumumab treated patients (HR=34.517;95% CI: 1.826-652.370).In detection method subgroup,increased EGFR GCN was also statistically significantly associated with increased ORR in studies of CISH tested patients (HR=6.887; 95% CI: 2.756-17.231) and FISH tested patients (HR=6.910; 95% CI: 4.247-11.244).In response criteria subgroup,increased EGFR GCN was statistically significantly associated with increased ORR in studies used RECIST (HR=7.033; 95% CI: 4.522-10.940)but not used WHO criteria (HR=4.313; 95% CI: 0.606-30.669),which maybe result in small sample size of the one included study.The plots of subgroup analysis were listed in the appendix (Figures S1-S4).
Discussion

?
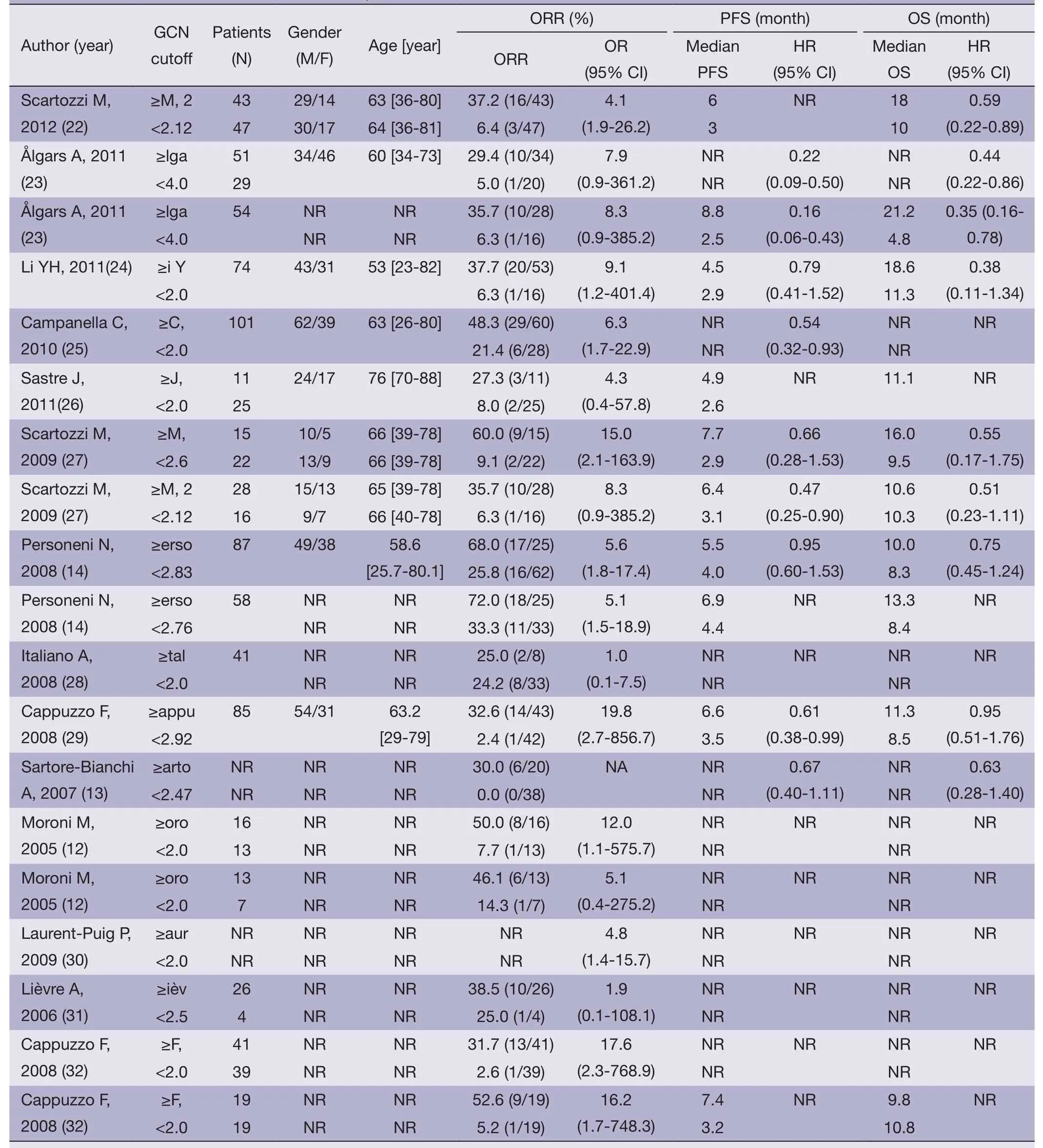
Table 2 Outcome results of the included studies by GCN cutoff

Figure 2 Meta-analysis of overall response rate in metastatic colorectal cancer (mCRC) patients treated with monoclonal antibodies (MoAbs)stratified by epidermal growth factor receptor (EGFR) gene copy number (GCN).
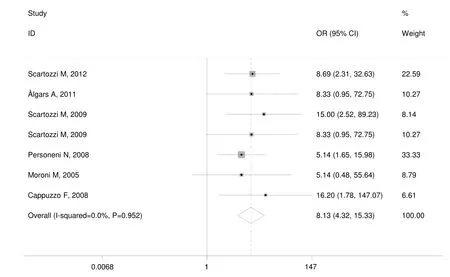
Figure 3 Meta-analysis of overall response rate in wild-type KRAS metastatic colorectal cancer (mCRC) patients treated with monoclonal antibodies (MoAbs) stratified by epidermal growth factor receptor (EGFR) gene copy number (GCN).
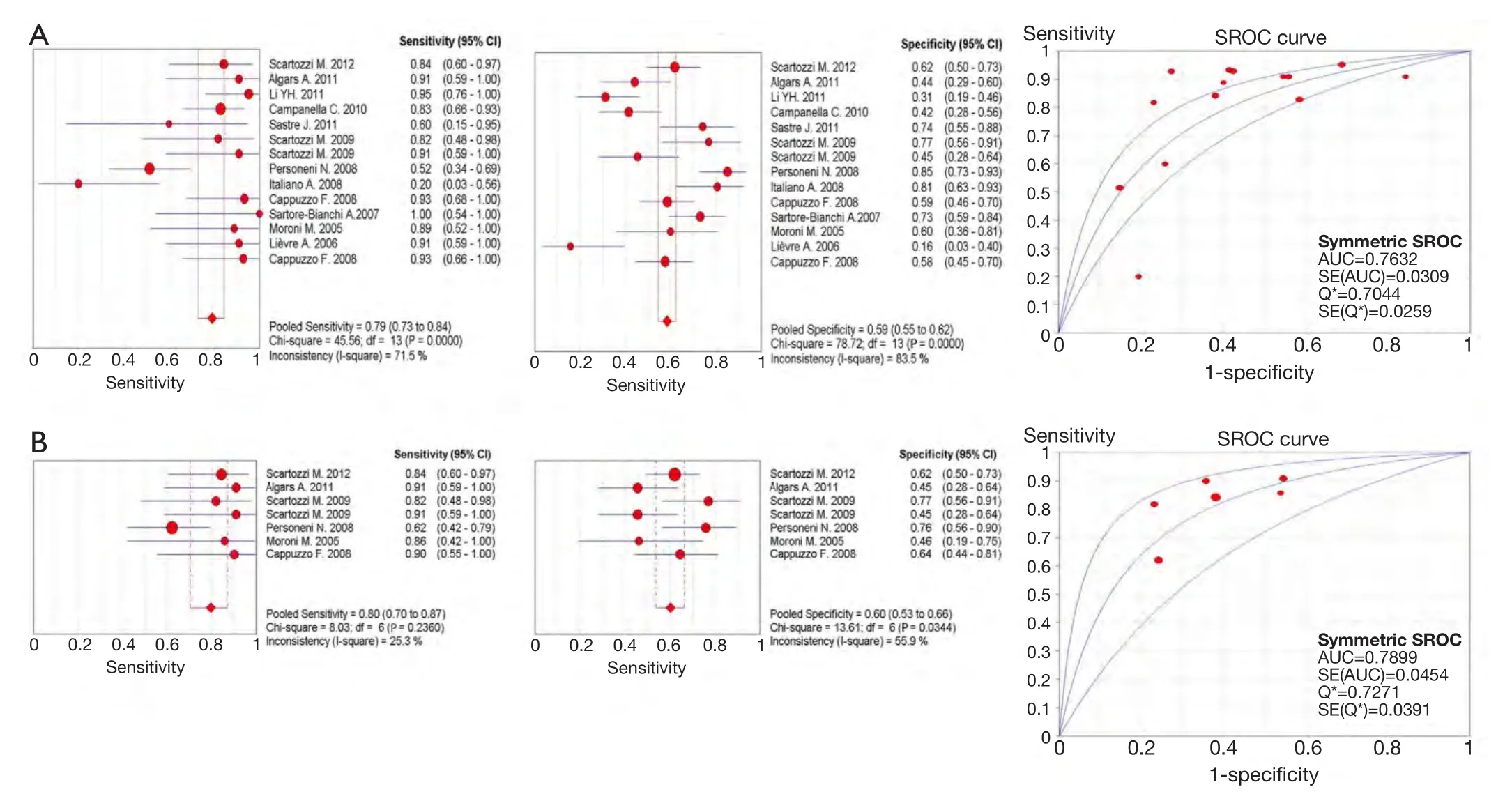
Figure 4 Meta-analysis of predictive value for overall response rate by increased epidermal growth factor receptor (EGFR) gene copy number (GCN).The upper half of the picture shows the predictive value in mixed KRAS (wild type and mutated) metastatic colorectal cancer (mCRC) patients; the lower half of the picture shows the predictive value in wild-type KRAS mCRC patients.
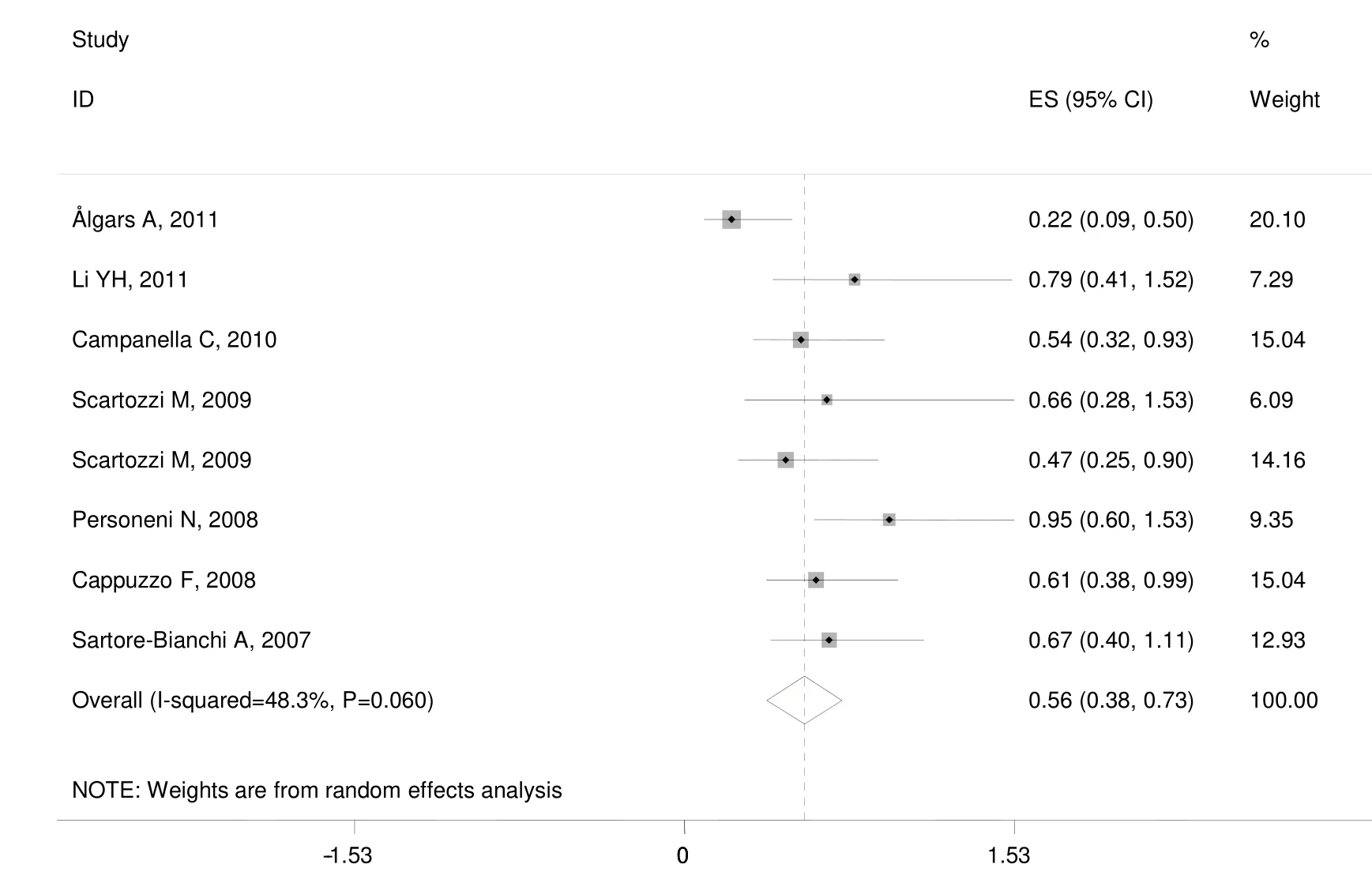
Figure 5 Meta-analysis of progression-free survival (PFS) in metastatic colorectal cancer (mCRC) patients treated with monoclonal antibodies (MoAbs) stratified by epidermal growth factor receptor (EGFR) gene copy number (GCN).
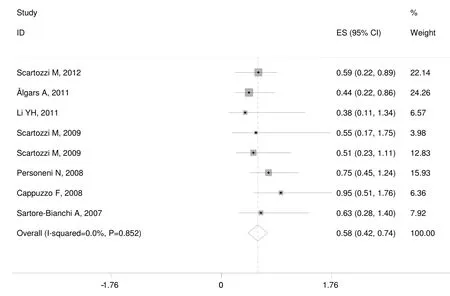
Figure 6 Meta-analysis of overall survival (OS) in metastatic colorectal cancer (mCRC) patients treated with monoclonal antibodies (MoAbs)stratified by epidermal growth factor receptor (EGFR) gene copy number (GCN).
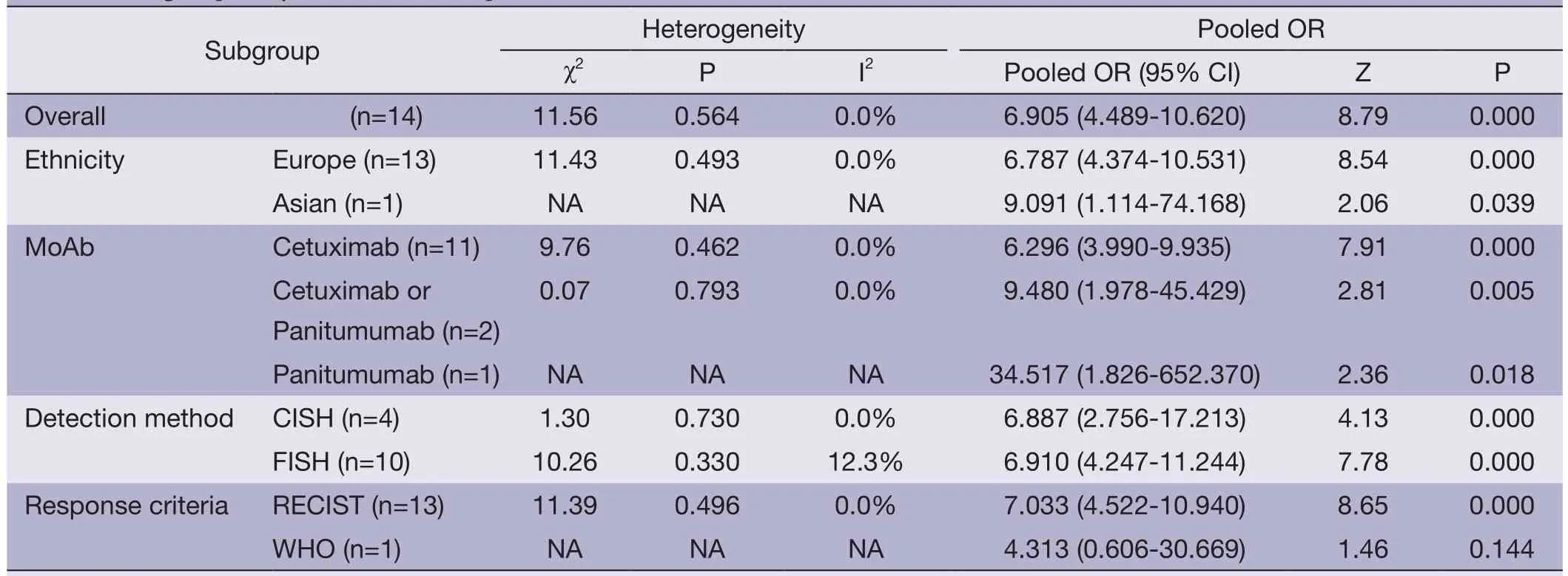
Table 3 Subgroup analyses for overall response rate
The EGFR gene is located on the short arm of chromosome 7 (7p21),which is commonly overrepresented or amplified in mCRC (33).A published meta-analysis showed,in nonsmall-cell lung cancer patients,increased EGFR GCN is associated with a moderate OS benefit and a substantial PFS benefit from EGFR TKIs treatment (34).Our metaanalysis showed mCRC patients with an increased EFGR GCN exhibit higher response rates to cetuximab- or panitumumab-based therapies,with the pooled OR 6.905(95% CI: 4.489-10.620).We also found increased EFGR GCN has predictive value for overall response treated with MoAbs for mCRC patients [the pooled sensitivity was 0.79(0.73-0.84) and the pooled specificity was 0.59 (0.55-0.62)].This result identified additional genetic determinants of resistance to EGFR-targeted cetuximab- or panitumumabbased therapies for further improving selection of patients;and this result also can explain rare cases of patients carrying KRAS-mutated tumors who have been reported to respond to either cetuximab or panitumumab.While a fair predicative test shows better than average accuracy,and has an AUC above 0.5; to demonstrate excellent accuracy,the AUC should be in the region of 0.97 or above; an AUC of 0.93 to 0.96 is very good; 0.75 to 0.92 is good; less than 0.75 can still be reasonable,but the test has obvious deficiencies in its diagnostic accuracy (35).The AUC of SROC is 0.7632±0.0309 in this meta-analysis.The predicative value is not good.In KRAS mutations predicating resistance to anti-EGFR MoAbs for mCRC patients,the sensitivity was low [0.47 (0.43-0.52)],while -LR was relatively high(-LR=0.57) (8).It showed KRAS and EGFR GCN all cannot independently predicate resistance to anti-EGFR MoAbs for mCRC.Combining KRAS with EGFR GCN,may be possible to improve the prediction performance for further improving selection of patients.This requires some other clinical trials to confirm.
While some studies found EGFR status has no relationship with response to MoAbs for mCRC patients.BOND trial showed EGFR faint stain,weak or moderate stain,strong stain in cetuximab and irinotecan group [high response group,overall response rate 22.9% (17.5-29.1%)]was 20.8%,24.7%,and 22.7%,respectively; in cetuximab group [low response group,overall response rate 10.8%(5.7-18.1%)] was 4.8%,12.7%,and 11.8% respectively,and no significant difference was found (P=0.64) (5).Chung KY also found in 16 chemotherapy-refractory,EGFR-negative colorectal cancer patients who received cetuximab,4 major objective responses were seen (response rate,25%; 95%CI: 4-46%) (36).These trails detected EGFR expression all by immunohistochemistry (IHC).Many technical reasons have been advocated for the lack of association between EGFR detection by IHC and response to EGFR-targeted treatment.These reasons include disparity between the form or epitope of EGFR detected by IHC and that targeted by anti-EGFR MoAbs,as well as issues related to processing and handling of tumor tissue samples,such as prolonged storage; IHC is also a semi-quantitative method that lacks a standardized scoring system and is subject to inter-observer variation (37).In our meta-analysis,the included studies were used FISH or CISH to detect EGFR GCN,and showed increased EFGR GCN exhibit higher response rates to cetuximab- or panitumumab-based therapies.EFGR GCN also can be detected by real-time quantitative polymerase chain reaction (qPCR).But EGFR qPCR was not predictive of response to treatment,disease control,PFS or OS (12,38).The reason may be tumor DNA dilution by DNA from normal cells during DNA extraction.
Active KRAS mutations have been accepted to be a major predictive marker of resistance to EGFR targeted cetuximab- or panitumumab-based therapies in mCRC patients (8,39,40).We also did EFGR GCN meta-analysis based on KRAS.The meta-analysis showed in wild-type mCRC patients,increased EFGR GCN was associated with better response,with the pooled OR 8.133 (95% CI:4.316-15.326); for predictive value,the pooled sensitivity was 0.80 (95% CI: 0.70-0.87),the pooled specificity was 0.60 (95% CI: 0.53-0.66),and the overall weighted AUC was 0.7899±0.0454.Mutation of PIK3CA or BRAF,or loss of PTEN expression also associated with resistance to EGFR-targeted cetuximab- or panitumumab-based therapies.The published meta-analysis showed: the RR for ORR by PIK3CA mutations was 0.59 (95% CI: 0.36-0.96)and statistically significant (P=0.034) in KRAS wild-type patients,and 0.75 (95% CI: 0.44-1.28) in unselected KRAS mutation status (41); the pooled RR for ORR by BRAF mutations was 0.14 (95% CI: 0.04-0.53; P=0.004) in KRAS wild-type patients,and 0.86 (95% CI: 0.57-1.30; P=0.48) in unselected KRAS mutation status (42); the pooled RR for ORR by loss of PTEN expression was 0.413 (95% CI: 0.177-0.965) in unselected KRAS mutation status (43).PIK3CA,BRAF,and PTEN are three biomarkers of EGFR signaling pathways downstream.They share a common characteristic that they are useful predictive biomarkers in KRAS wildtype patients.But the EFGR GCN showed is a useful predictive biomarker not only in KRAS wild-type patients,but also in unselected KRAS mutation status patients in our meta-analysis,which indicated the EFGR GCN is another important predictive biomarker for ORR to anti-EGFR MoAbs for mCRC patients,except for KRAS.
Treatment response in patients with mCRC receiving cetuximab- or panitumumab-based therapies is closely related to its prognosis.Mutation of KRAS,PIK3CA,or BRAF,or loss of PTEN expression has been found to affect prognosis.The meta-analysis for KRAS showed a significant PFS benefit for cetuximab-based therapy among mCRC patients with wild KRAS tumor (HR=0.64; 95%CI: 0.42-0.97; P=0.04),and no benefit for patients with mutated KRAS (HR=1.21; 95% CI: 0.92-1.59; P=0.17).It also showed significant OS benefit in the wild KRAS(HR=0.72; 95% CI: 0.56-0.93; P=0.01),and no benefit for patients with mutated KRAS (HR=1.06; 95% CI: 0.90 to 1.25; P=0.47) (44).The meta-analysis for PIK3CA exon 20 mutations was statistically significantly associated with shorter PFS (HR=2.52; 95% CI: 1.33-4.78; P=0.013) and OS (HR=3.29; 95% CI: 1.60-6.74; P=0.006) in KRAS wildtype mCRC (41).The meta-analysis for PTEN showed improved PFS (HR=0.466; 95 % CI: 0.292-0.640) and OS(HR=0.689; 95% CI: 0.482-0.896) in patients with normal PTEN expression over loss of PTEN expression (43).This meta-analysis also found mCRC patients with increased EGFR GCN are more likely to have better PFS and OS when treated with cetuximab or panitumumab.
There are some limitations in this meta-analysis.First,significant publication bias was found in these metaanalysis for the primary outcome.We only searched through English-language reports,and thus it may have missed studies in our literature review.Second,some of the included studies didn’t provide sufficient data of timeto-event outcomes for meta-analysis directly.We used Engauge Digitizer to extract data from survival curves,instead of using individual patient data meta-analysis.This may be result in basis.The limitations are needed to be taken into consideration when interpreting the fi ndings.
Despite of these limitations,our meta-analysis provided evidence that EGFR GCN represents a predictive biomarker for tumor response in mCRC patients treated with MoAbs.mCRC patients with increased EGFR GCN are more likely to have a better response,PFS and OS when treated with cetuximab or panitumumab.However,EGFR GCN only has the medium predictive value for overall response treated with MoAbs.Further research should potentially focus on comprehensive integrated analysis of the entire oncogenic pathway triggered by EGFR to enhance the prediction ability of the markers used individually.
Acknowledgements
Disclosure:The authors declare no conflict of interest.
1.Siegel R,Naishadham D,Jemal A.Cancer statistics,2012.CA Cancer J Clin 2012;62:10-29.
2.Jemal A,Bray F,Center MM,et al.Global cancer statistics.CA Cancer J Clin 2011;61:69-90.
3.Grothey A,Marshall JL.Optimizing palliative treatment of metastatic colorectal cancer in the era of biologic therapy.Oncology (Williston Park) 2007;21:553-64.
4.Baselga J,Arteaga CL.Critical update and emerging trends in epidermal growth factor receptor targeting in cancer.J Clin Oncol 2005;23:2445-59.
5.Cunningham D,Humblet Y,Siena S,et al.Cetuximab monotherapy and cetuximab plus irinotecan in irinotecanrefractory metastatic colorectal cancer.N Engl J Med 2004;351:337-45.
6.Van Cutsem E,K?hne CH,Hitre E,et al.Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer.N Engl J Med 2009;360:1408-17.
7.Van Cutsem E,Peeters M,Siena S,et al.Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer.J Clin Oncol 2007;25:1658-64.
8.Johnson KR,Ringland C,Stokes BJ,et al.Response rate or time to progression as predictors of survival in trials of metastatic colorectal cancer or non-small-cell lung cancer:a meta-analysis.Lancet Oncol 2006;7:741-6.
9.Linardou H,Dahabreh IJ,Kanaloupiti D,et al.Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-smallcell lung cancer and metastatic colorectal cancer.Lancet Oncol 2008;9:962-72.
10.Siena S,Sartore-Bianchi A,Di Nicolantonio F,et al.Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer.J Natl Cancer Inst 2009;101:1308-24.
11.Bardelli A,Siena S.Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer.J Clin Oncol 2010;28:1254-61.
12.Moroni M,Veronese S,Benvenuti S,et al.Gene copy number for epidermal growth factor receptor (EGFR)and clinical response to antiEGFR treatment in colorectal cancer: a cohort study.Lancet Oncol 2005;6:279-86.
13.Sartore-Bianchi A,Moroni M,Veronese S,et al.Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab.J Clin Oncol 2007;25:3238-45.
14.Personeni N,Fieuws S,Piessevaux H,et al.Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study.Clin Cancer Res 2008;14:5869-76.
15.Wells GA,Shea B,O’Connell D,et al.The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analysis.Ottawa Health Research Institute.Avaiable online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.Accessed 30 January 2013.
16.Walter SD.Properties of the summary receiver operating characteristic (sROC) curve for diagnostic test data.Stat Med 2002;21:1237-56.
17.Parmar MK,Torri V,Stewart L.Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints.Stat Med 1998;17:2815-34.
18.Williamson PR,Smith TC,Hutton JL,et al.Aggregate data meta-analysis with time-to-event outcome.Stat Med 2002;21:3337-51.
19.Tierney JF,Stewart LA,Ghersi D,et al.Practical methods for incorporating summary time-to-event data into metaanalysis.Trials 2007;8:16.
20.Higgins JPT,Thompson SG,Deek JJ,et al.Measuring inconsistency in meta-analyses.BMJ 2003;327:557-60.
21.Zamora J,Abraira V,Muriel A,et al.Meta-Disc: a software for meta analysis of test accuracy data.BMC Med Res Methodol 2006;6:31-47.
22.Scartozzi M,Giampieri R,Maccaroni E,et al.Analysis of HER-3,insulin growth factor-1,nuclear factor-kB and epidermal growth factor receptor gene copy number in the prediction of clinical outcome for K-RAS wild-type colorectal cancer patients receiving irinotecan-cetuximab.Ann Oncol 2012;23:1706-12.
23.?lgars A,Lintunen M,Carpén O,et al.EGFR gene copy number assessment from areas with highest EGFR expression predicts response to anti-EGFR therapy in colorectal cancer.Br J Cancer 2011;105:255-62.
24.Li YH,Wang F,Shen L,et al.EGFR fluorescence in situ hybridization pattern of chromosome 7 disomy predicts resistance to cetuximab in KRAS wild-type metastatic colorectal cancer patients.Clin Cancer Res 2011;17:382-90.
25.Campanella C,Mottolese M,Cianciulli A,et al.Epidermal growth factor receptor Gene copy number in 101 advanced colorectal cancer patients treated with chemotherapy plus cetuximab.J Transl Med 2010;16;8:36.
26.Sastre J,Aranda E,Grávalos C,et al.First-line singleagent cetuximab in elderly patients with metastatic colorectal cancer.A phase II clinical and molecular study of the Spanish group for digestive tumor therapy (TTD).Crit Rev Oncol Hematol 2011;77:78-84.
27.Scartozzi M,Bearzi I,Mandolesi A,et al.Epidermal Growth Factor Receptor (EGFR) gene copy number(GCN) correlates with clinical activity of irinotecancetuximab in K-RAS wild-type colorectal cancer: a fluorescence in situ (FISH) and chromogenic in situ hybridization (CISH) analysis.BMC Cancer 2009;9:303.
28.Italiano A,Follana P,Caroli FX,et al.Cetuximab shows activity in colorectal cancer patients with tumors for which FISH analysis does not detect an increase in EGFR gene copy number.Ann Surg Oncol 2008;15:649-54.
29.Cappuzzo F,Finocchiaro G,Rossi E,et al.EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients.Ann Oncol 2008;19:717-23.
30.Laurent-Puig P,Cayre A,Manceau G,et al.Analysis of PTEN,BRAF,and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer.J Clin Oncol 2009;27:5924-30.
31.Lièvre A,Bachet JB,Le Corre D,et al.KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer.Cancer Res 2006;66:3992-5.
32.Cappuzzo F,Varella-Garcia M,Finocchiaro G,et al.Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients.Br J Cancer 2008;99:83-9.
33.Spano JP,Lagorce C,Atlan D,et al.Impact of EGFR expression on colorectal cancer patient prognosis and survival.Ann Oncol 2005;16:102-8.
34.Dahabreh IJ,Linardou H,Kosmidis P,et al.EGFR gene copy number as a predictive biomarker for patients receiving tyrosine kinase inhibitor treatment: a systematic review and meta-analysis in non-small-cell lung cancer.Ann Oncol 2011;22:545-52.
35.Jones CM,Athanasiou T.Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests.Ann Thorac Surg 2005;79:16-20.
36.Chung KY,Shia J,Kemeny NE,et al.Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry.J Clin Oncol 2005;23:1803-10.
37.Shia J,Klimstra DS,Li AR,et al.Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: an immunohistochemical and chromogenic in situ hybridization study.Mod Pathol 2005;18:1350-6.
38.Lenz HJ,Van Cutsem E,Khambata-Ford S,et al.Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan,oxaliplatin,and fluoropyrimidines.J Clin Oncol 2006;24:4914-21.
39.Karapetis CS,Khambata-Ford S,Jonker DJ,et al.K-ras mutations and benefit from cetuximab in advanced colorectal cancer.N Engl J Med 2008;359:1757-65.
40.Khambata-Ford S,Garrett CR,Meropol NJ,et al.Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab.J Clin Oncol 2007;25:3230-7.
41.Mao C,Yang ZY,Hu XF,et al.PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis.Ann Oncol 2012;23:1518-25.
42.Mao C,Liao RY,Qiu LX,et al.BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis.Mol Biol Rep 2011;38:2219-23.
43.Wang ZH,Gao QY,Fang JY.Loss of PTEN expression as a predictor of resistance to anti-EGFR monoclonal therapy in metastatic colorectal cancer: evidence from retrospective studies.Cancer Chemother Pharmacol 2012;69:1647-55.
44.Ibrahim EM,Zekri JM,Bin Sadiq BM.Cetuximab-based therapy for metastatic colorectal cancer: a meta-analysis of the effect of K-ras mutations.Int J Colorectal Dis 2010;25:713-21.
 Chinese Journal of Cancer Research2014年1期
Chinese Journal of Cancer Research2014年1期
- Chinese Journal of Cancer Research的其它文章
- Build infrastructure in publishing scientific journals to benefit medical scientists
- A comparative study between Embosphere?and conventional transcatheter arterial chemoembolization for treatment of unresectable liver metastasis from GIST
- Prognostic value of post-treatment18F-FDG PET/CT for advanced head and neck cancer after combined intra-arterial chemotherapy and radiotherapy
- MiR-182 is up-regulated and targeting Cebpa in hepatocellular carcinoma
- Intraperitoneal chemotherapy and its evolving role in management of gastric cancer with peritoneal metastases
- Radiofrequency ablation or microwave ablation combined with transcatheter arterial chemoembolization in treatment of hepatocellular carcinoma by comparing with radiofrequency ablation alone
