Research Progress in Digging and Validation of miRNA Target Genes Using Experimental Methods
Luo Xiao, Bai Xi, Cai Hua, Ji Wei, Liu Xin, Tang Li-li, and Zhu Yan-ming
Plant Bioengineering Laboratory, Northeast Agricultural University, Harbin 150030, China
Introduction
miRNAs are small non-coding RNAs, usually consisting of 20-25 nucleotides, depending upon the organism (Bartel, 2004; Reinhart et al., 2002),which function as post-transcriptional regulators of gene expression. miRNAs were initially discovered in 1993, regulating developmental transitions in Caenorhabditis elegans (Lee et al., 1993), at the time,the first miRNA was thought to be an oddity rather than a general phenomenon. However, in 2000, almost seven years after the initial identification of lin-4,the second miRNA, let-7 (Reinhart et al., 2000), was discovered. Since then, a large number of miRNAs has been discovered and cloned in many species, including more than 1 700 plant miRNAs. miRNAs are usually assembled into ribonucleoprotein (RNP)complexes called micro-RNPs (miRNPs), or miRNA-induced silencing complexes (miRISCs), to bind the mRNA target (Filipowicz et al., 2008; Zhao et al., 2011).Within this complex, miRNAs can play essential roles in biological functions.
As a class of non-coding RNA, miRNAs function as sequence-specific negative regulators in posttranscriptional gene silencing, pairing with target mRNAs and inducing transcript degradation or translational repression, depending upon the degree of sequence complementarity. In plants, miRNAs have been found to be involved in regulating many important biological processes, such as development,metabolism, signal transduction, and stress resistance(Duan et al., 2006; Mallory and Vaucheret, 2006). In animals, miRNAs have been found to be involved in regulating tumor metastasis cell proliferation, apoptosis, differentiation, metabolism, and development(Chen and Yin, 2005; Hwang and Mendell, 2006).miRNA research has shown good value and applicability, and thus has become a focal point in life sciences. That said, the functions of many miRNAs are still unclear, while the number of known miRNA target genes is very limited. Therefore, the identif i cation of miRNA targets is critically important to the study of miRNA function.
Several algorithms are available to predict miRNA target genes (Du et al., 2010; Griffiths-Jones et al.,2008; Lewis et al., 2005; Pei et al., 2010), where target predictions are usually based on the most general feature of miRNA regulation, the recognition of sequence motifs complementary to the seed region(nucleotides 2-7 of the miRNA)in the 3' UTR of target mRNAs (Lewis and Shih, 2003). However, the consistency between the different available miRNA prediction algorithms is limited, and they all suffer from high rates of false positives (John et al., 2004;Sethupathy et al., 2006). Additionally, prediction algorithms analyses usually do not consider whether the miRNA and the mRNAs are expressed in the same cells, so they do not accurately reflect in vivo processes, even when predictions are accurate, this is only determined after experimental validation.Therefore, many researchers hope to use experimental approaches to find and validate miRNA targets directly. Meanwhile, algorithms for the prediction of miRNA targets still depend on data provided by biological experimentation, especially the highthroughput identification of target genes. Thus, the development of precise, and rapid, assays for identifying and verifying miRNA targets will play a significant role in the study of miRNA functions and the biological processes in which they are involved. Compared with the variety of available miRNA target gene prediction tools, there are far fewer available approaches for the experimental verification of miRNA target genes, where traditional validation methods include: mutagenesis studies (O'Donnell et al., 2005), gene-silencing techniques (Johnson et al.,2005; Poy et al., 2004), and classic genetic studies(Johnston and Hobert, 2003; Lee et al., 1993). In this review, we summarized current experimental approaches in miRNA identif i cation and validation.
Large-scale miRNA Target Finding
Analysis of mRNAs to fi nd miRNA targets
Animal miRNAs down-regulate target genes by imperfectly complementing the 3'untranslated regions(3'UTRs)of their target mRNAs. Although, translational regulation has been recently demonstrated as being widespread in plants (Brodersen et al., 2008),typically, plant miRNAs repress their targets through mRNA degradation, cleaving at nucleotide positions 10-11 relative to the 5'end of the complementary miRNA sequence (Llave et al., 2002b). So we can find miRNA targets from mRNA level, because their expression levels might change, or be higher than the expected protein levels (Chen et al., 2010).
Combining miRNA over-expression with gene expression chip
Plant miRNAs usually complement target mRNA completely, resulting in target mRNA degradation.Thus, combinations of miRNA over-expression and gene expression chip (miRNA-Chip)were widely used in large-scale searches for miRNA targets in plants,and briefly used in mammals too. The identification and validation of mRNA targets of the heart and skeletal muscle miR-1, as well as the brain tissuespecific miR-124, used this methodology. First,miRNA-1 and miR-124 over-expression constructs were transfected into HeLa cells, whereupon microarrays were used to find out which genes were significantly down-regulated (P<0.001). Bioinformatics analyses were then required to find out which of the down-regulated genes contained motifs
complementary to the miRNAs, which successfully identif i ed more than 100 target genes for each miRNA(Lim et al., 2005). Alternatively, quantitative RTPCR can also be used to monitor changes in mRNA levels after a miRNA has been introduced into a cell(Boissonneault et al., 2008). This method opened the door to find miRNA targets in mammalian cells and quickly became common practice. Finding miRNA target genes via an analysis of down-regulated genes has the advantage of reducing search range, and making prediction faster, and more accurate. However,this methodology is not without its flaws: (1)the data from a gene chip cannot tell the difference between direct and indirect interactions; (2)the data can only be used for the identification of mRNAs which are silenced by degradation, and is not suited for identifying miRNA targets whose expression is inhibited at translation.
Combining Co-IP with gene expression chip
miRNA-mediated target cleavage involves a transient association of Argonaute (Ago)family containing protein complexes with the 3'UTR of target mRNA,whereas miRNA-mediated translational inhibition depends on a stable physical association between the miRNP and the target mRNA. This raises the possibility of using these interactions to identify miRNA-target interactions that occur in vivo. In this manner, Easow and his colleagues (2007)analyzed miRNA target genes by introducing a FLAG tag,followed by an HA epitope tag, onto the N terminus of the Ago1 protein. Using antibodies for the protein tag, Easow et al. purified the mRNA associated miRNP complexes, and then used microarrays to identify the enriched transcripts using the method outlined in Fig. 1A. The regulation of these mRNAs by miRNAs was then validated, using firefly luciferase, showing that this approach can be used to identify new miRNA targets (Beitzinger et al.,2007). Easow and his colleagues (2007)indicated that in a total of 108 miR-1 targets, only 32 had six mer seed matches to positions 2-7 or 3-8 of the miRNA.Other targets did not contain 6-mer seed matches,where ultimately ~40% of the identif i ed mRNAs had not been predicted by the three most widely used prediction algorithms (miRanda, TargetScan, and PicTar). This degree of discrepancy suggests that the currently used miRNA target prediction algorithms may miss a significant number of miRNA targets.miRNAs and their targets may interact by means not previously observed, and thus computational analyses may fail to identify many miRNA targets.Their research also indicated that only a comprehensive validation of isolated mRNAs will allow for a conclusive comparison of bioinformatics derived and experimentally derived miRNA target identif i cation.
The immunoprecipitation (IP)of Ago proteins has also been combined with the over-expression of synthetic miRNAs (Hendrickson et al., 2008; Karginov et al., 2007; Landthaler et al., 2008). In this variation,FLAG-tagged Ago proteins were used, requiring a signif i cant modulation of the cells which might result in target genes that are not physiologically relevant.In another variation, Tan et al. (2009)described an approach called antimiRNA-RIP-Chip (antimiRNA strategy combined with Ribonucleoprotein Immunoprecipitation-gene Chip), where combining RIPChip with the inhibition of specific miRNAs,allowing for the large-scale identification of endogenous transcripts that are targeted by the downregulated miRNA. In their case, wild-type human Ago2 protein was directly immunoprecipitated from untreated cells, and the Ago2-associated mRNA transcripts were analyzed by microarray to identify the miRNA-targetome (whole miRNA regulated gene set)of specific cells. This strategy allows for the identification of miRNA targets in human tissue samples, or purified cell populations, in an unbiased and physiologically relevant manner.
In another variation, Argonaute-containing complexes are pulled down, and the associated mRNAs are then subjected to silico searches for the seed sequences of associated miRNAs. This biochemical approaches may limit the identification of targets to those containing a perfect or near-perfect seed match,which may or may not be the case in vivo (Bartel,2009; Orom et al., 2008). Biochemical assays based on the introduction of biotinylated synthetic miRNAs into cells, followed by a pull-down on streptavidin beads (Rom and Lund, 2007), have helped identify new miRNA targets (Hsu et al., 2009; Orom et al.,2008); however, the applicability of this technique remains limited. One-step pull-down procedures are generally prone to high levels of background (Nonne et al., 2010). Recently, a target identif i cation approach called TAP-Tar (Tandem affinity purification of miRNA target mRNAs)was developed, based on tandem aff i nity purif i cation, in which mRNA/miRNA complexes were sequentially pulled down, first via the Ago moiety, and then via the miRNA (Fig. 1B)(Nonne et al., 2010). This procedure greatly reduces the background noise, and provides an assay for the direct validation of the target mRNAs.
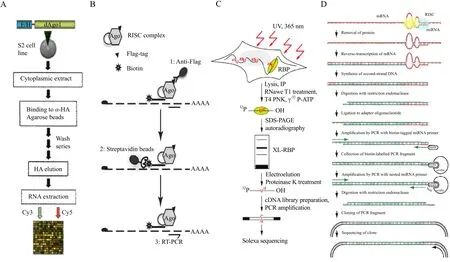
Fig. 1 Diagrammatic representation of recent experimental methods for fi nding miRNA targets
RIP-Chip identifies targets at the transcriptome level. However, its application is limited to the characterization of kinetically stable interactions,and only in rare cases allows for the identification of the RBP (RNA-binding proteins)recognition element (RRE)within the long target RNA. More direct RBP target site information can be obtained by crosslinking and IP (CLIP). CLIP uses ultraviolet light to cross-link Ago proteins to the associated RNA and miRNA, followed by the isolation of crosslinked RNA segments, and then cDNA sequencing.
HITS-CLIP (High-throughput Sequencing-Crosslinking and Immunoprecipitation)has been used in order to identify and sequence specific miRNA binding sites on targeted mRNAs (Chi et al., 2009).In one study, Ago protein complexes were immunoprecipitated and purified from mouse brains, whereupon associated RNAs were identif i ed by sequencing,and, in so doing, produced a transcriptome-wide interaction map for miR-124, as well as maps for the 20 most abundant miRNAs. HITS-CLIP provides a general platform for exploring the specificity and range of miRNA action in vivo, and identifies precise sequences by targeting relevant miRNA-mRNA interactions. The option of studying individual miRNAs with this approach would give even more insightful knowledge on target recognition properties,without having to guess at some of the interactions,or make assumptions regarding which miRNAs are binding the identif i ed target mRNA. However, CLIP is limited by ineff i cient RNA-protein crosslinking, where the location of the crosslink is not readily identif i able within the sequenced RNA fragments, making it difficult to separate UV-crosslinked RNA segments from background RNA fragments in the sample.Further development of this technique is ongoing in several laboratories. Recently, an improved method called PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation)was developed (Hafner et al., 2010a; 2010). In this method, photoreactive ribonucleoside analogs, such as 4-thiouridine (4SU)was incorporated into the transcripts of cultured cells expressing FLAG/HA-tagged RBP IGF2BP1. Cells were irradiated at UV 365 nm, and the cross-linked RNA-protein complexes were isolated by immunoprecipitation. The covalently bound RNA was partially digested with RNaseT1,and then radiolabeled. Separation of the radiolabeled RNPs by SDS-PAGE indicated that 4SU-containing RNA cross-linked most efficiently to IGF2BP1. This method also allowed for the identif i cation of the RBP binding sites by scoring for thymidine (T)to cytidine(C)transitions in the sequenced cDNA. Compared to conventional UV 254 nm cross-linking with unlabeled RNA, the photoactive nucleosides improved RNA recovery 100-to 1000-fold, using the same levels of radiation (Fig. 1C). The author also uncovered tens of thousands of binding sites for several important RBPs and RNPs, and was able to assess the regulatory impact of the target binding.
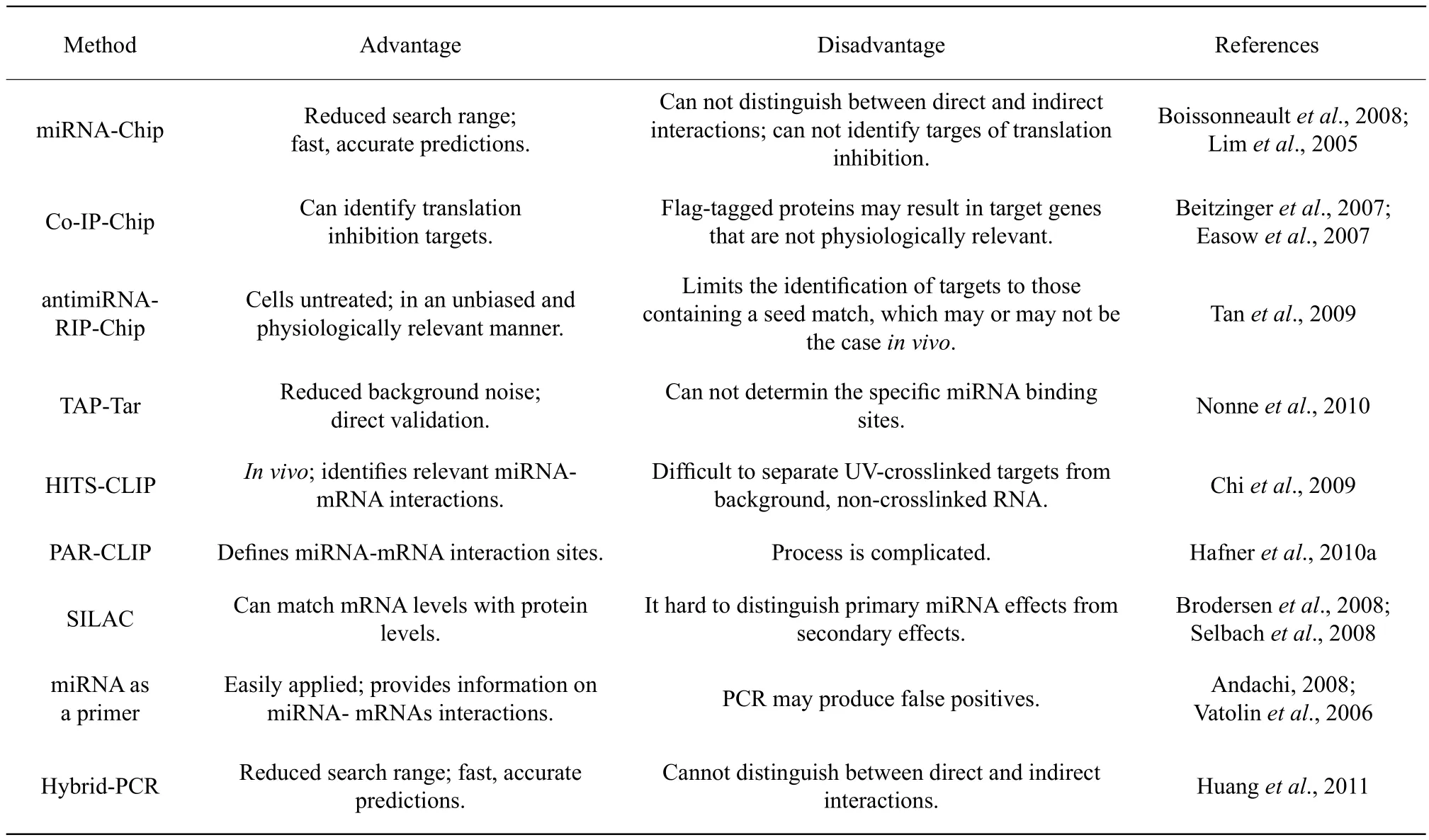
Table 1 Comparing experimental method for fi nding miRNA targets
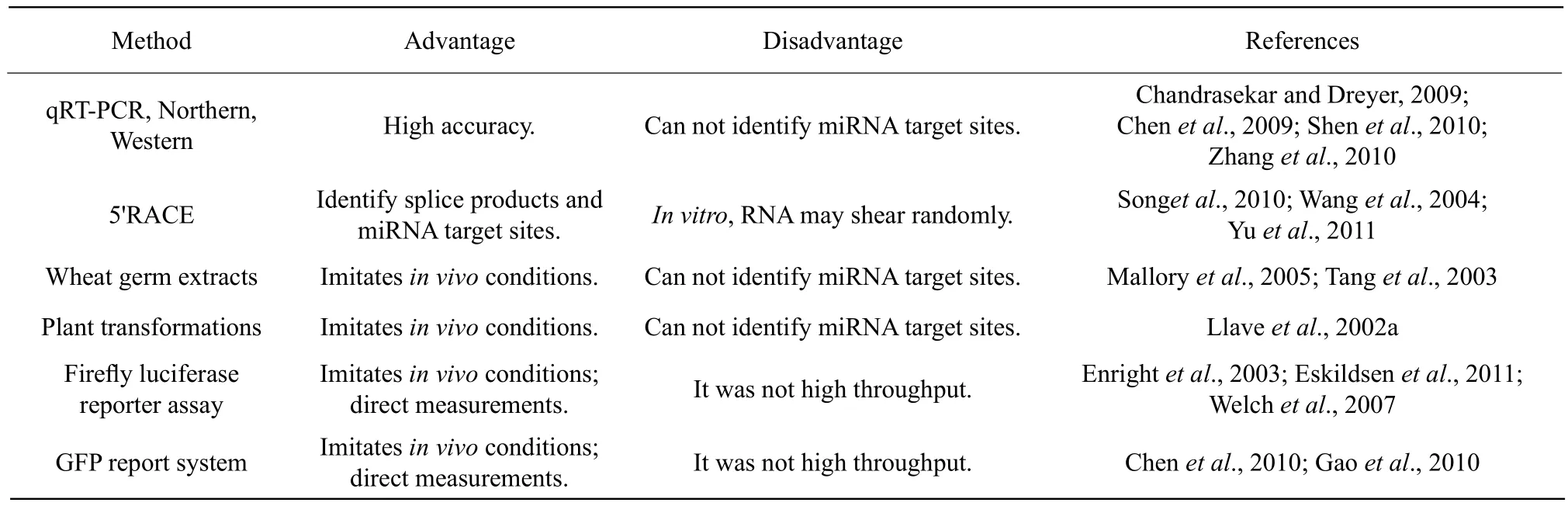
Table 2 Comparing experimental method for validation miRNA targets
Analysis of protein level to fi nd miRNA targets
Because some miRNAs mediate translational repression, which does not alter mRNA levels,problems arise when trying to detect miRNA target genes according to the changes in mRNA levels. Two groups established new ways of looking for miRNA target genes using proteomics techniques. They introduced mature double-stranded miRNA into HeLa cells, and applied a quantitative-mass-spectrometrybased approach called SILAC (stable isotope labelling with amino acids in cell culture)to investigate the influence of specific miRNAs on the levels of many proteins. After introducing the miRNA, HeLa cells were grown in media containing amino acids marked with a heavier stable isotope. By comparing the levels of light and heavy peptides, between untreated and miRNA treated cultures respectively, the researchers determine how the protein levels had changed for 2 000-5 000 proteins. Between the two teams, they found hundreds of proteins whose levels were changed after the introduction of one miRNA, which hadn't been ref l ected as changes in mRNA levels (Brodersen et al., 2008; Selbach et al., 2008).
Calin et al. (2008)identified miR15/16 targets by microarray and proteomics, systematically revealing the miR15/16 gene regulatory network. First, using a microarray, they identif i ed the genes whose expression levels changed, when miR15/16 was over-expressed.Then, using two-dimensional electrophoresis and mass spectrometry, they identified the genes whose protein levels changed. Another approach to verify a miRNA-target interaction would be to knockout the miRNA gene and examine the effects on protein levels(Baek et al., 2008). The introduction of proteomics to miRNA target finding has improved the detection rate of miRNA target genes, being a more direct test for miRNA targets. Nevertheless, mass spectrometry techniques carry some inherent fl aws, especially with regards to the difficulty identifying lowly expressed proteins, and the cost to perform.
PCR based methods
Polymerase Chain Reaction (PCR)is widely held as one of the most important experimental methods in molecular biology. Several PCR based techniques were established specif i cally to fi nd miRNA targets.
miRNA as a primer for cDNA synthesis
miRNA molecules bind to complementary or weakly complementary sites of target mRNAs (Doench and Sharp, 2004; Elbashir et al., 2001; Hutvagner and Zamore, 2002), Based on this, a method to detect miRNAs targeting specified genes was proposed by Vatolin et al (2006), in which an endogenous miRNA was used as a primer for cDNA synthesis on a target mRNA template. The cDNA fragment was ligated to an adapter oligonucleotide at the 5' terminus and amplif i ed by PCR with a primer from the adapter and a gene specific primer corresponding to the target mRNA. The 5' terminal analyses of cloned PCR fragments showed signature sequences that help to identify the miRNA. Another technique was developed to identify the miRNA targets, in the Andachi lab, where target genes of individual miRNAs are identif i ed by isolating cDNA clones of mRNAs (Fig.1D)(Andachi, 2008). In this version, the cytoplasmic extract was mixed with a detergent, to destabilize the proteins, and incubated in a reverse-transcription reaction buffer to synthesize a first-strand cDNA using the miRNA as a primer. They then removed the protein and leftover polynucleotides, and synthesized the second strand. The cDNA fragments were ligated to an adaptor oligonucleotide, and then amplified by PCR with one primer complementary to the adaptor and a second biotin-tagged miRNA-derived primer.Amplified fragments were then cloned into a vector and sequenced. The application of this technique to Caenorhabditis elegans miRNA lin-4 yielded many copies of lin-14, a known target of lin-4. In the case of C. elegans miRNA let-7, a new target gene responsible for the lethal phenotype in let-7 mutants had been identif i ed. These results demonstrated that this method was useful in identifying targets on the basis of base pairing with individual miRNAs.
In contrast to the previously described microarray approaches, this PCR based technique has the advantage of requiring minimal RNA, where the extract from a single cell is sufficient, making it easier to perform. This technique can also provide information on how miRNAs interact with mRNAs.
Currently, our research team is synthesising cDNAs from target mRNAs using endogenous miRNAs as reverse-transcription primers, as proposed by Vatolin et al. (2006)and Andachi (2008), where we are attempting to fi nd targets of Glycine soja's miRNA398(unpublished data).
Hybrid-PCR
In addition to base pair complementary, the stability of primer-template hybridizations is essential for successful PCR reactions. These requirements are also true for miRNA target recognition. With this in mind,Huang et al. (2011)established a novel screening approach named hybrid-PCR, which entails a seminested PCR using a hybrid-primer and outer/inner primers homologous to the oligo dT-adaptor primer.To begin, an oligo dT-adaptor was introduced onto the 5'-terminus of mRNA-derived cDNA during reverse transcription (Fig. 2A). miRNA specific hybridprimers were designed, and the reverse complement of the miRNA seed region was ligated to the 3' terminus of the hybrid primer. The specificity of a miRNA to a given mRNA would affect the hybridization of the hybrid-primer to the mRNA-derived cDNA. A low annealing temperature of 37℃ was applied in the fi rst round ampli fi cation, so as to make the hybrid-primer hybridize with the putative target. This was followed by a second round of PCR, with a higher annealing temperature of 55℃, for further amplification of sequences from putative target mRNAs. The amplification products were variable in length, and were subsequently cloned and sequenced (Fig. 2A). Using this method, 15 putative target mRNAs of human cytomegalovirus (HCMV)miR-UL112-1, including the previously confirmed HCMV IE72, were identifi ed. Moreover, the random validation of six candidate mRNAs, by luciferase reporter assays, con fi rmed that they were down-regulated by HCMV miR-UL112-1.Hybrid-PCR is thus an effective and rapid approach for screening for miRNA targets, with the advantage of simplicity, low cost, and the ease of implementation.
Validation of miRNA Target Genes
Accurate target gene validation has proven notoriously difficult, as apparent by the relatively few miRNA targets that have been validated thus far. There are not many methods for validating miRNA target genes, and none of the current methods can be applied in a highthroughput fashion. The most direct validation method involves the measurement of mRNA or protein levels in miRNA over-expression or knock out lines, using qRT-PCR (Chandrasekar and Dreyer, 2009; Zhang et al., 2010), northern blots (Chandrasekar and Dreyer,2009; Chen et al., 2009), or Western blots (Shen et al.,2010; Zhang et al., 2010). This method can directly identify miRNA target genes, but cannot identify miRNA target sites. 5' RACE (Rapid amplif i cation of the cDNA ends)directly analyzes the spliced products of miRNA-mRNA interactions and can be used to identify miRNA target sites and mRNA splice sites(Song et al., 2010; Wang et al., 2004; Yu et al., 2011).However, this technique is carried out in vitro, and errors may occur as RNA can experience shearing during the experiment. Wheat germ extracts contain endogenous miRNA-induced silencing complex(miRISCs). This RISC complex can direct the eff i cient cleavage of wild-type Arabidopsis mRNA, and has been used to study the splicing of mRNA in vitro(Mallory et al., 2005; Tang et al., 2003). One study demonstrated that miR160 directed ARF17 cleavage,as they were able to detect a product of the size expected from miR160-directed cleavage.
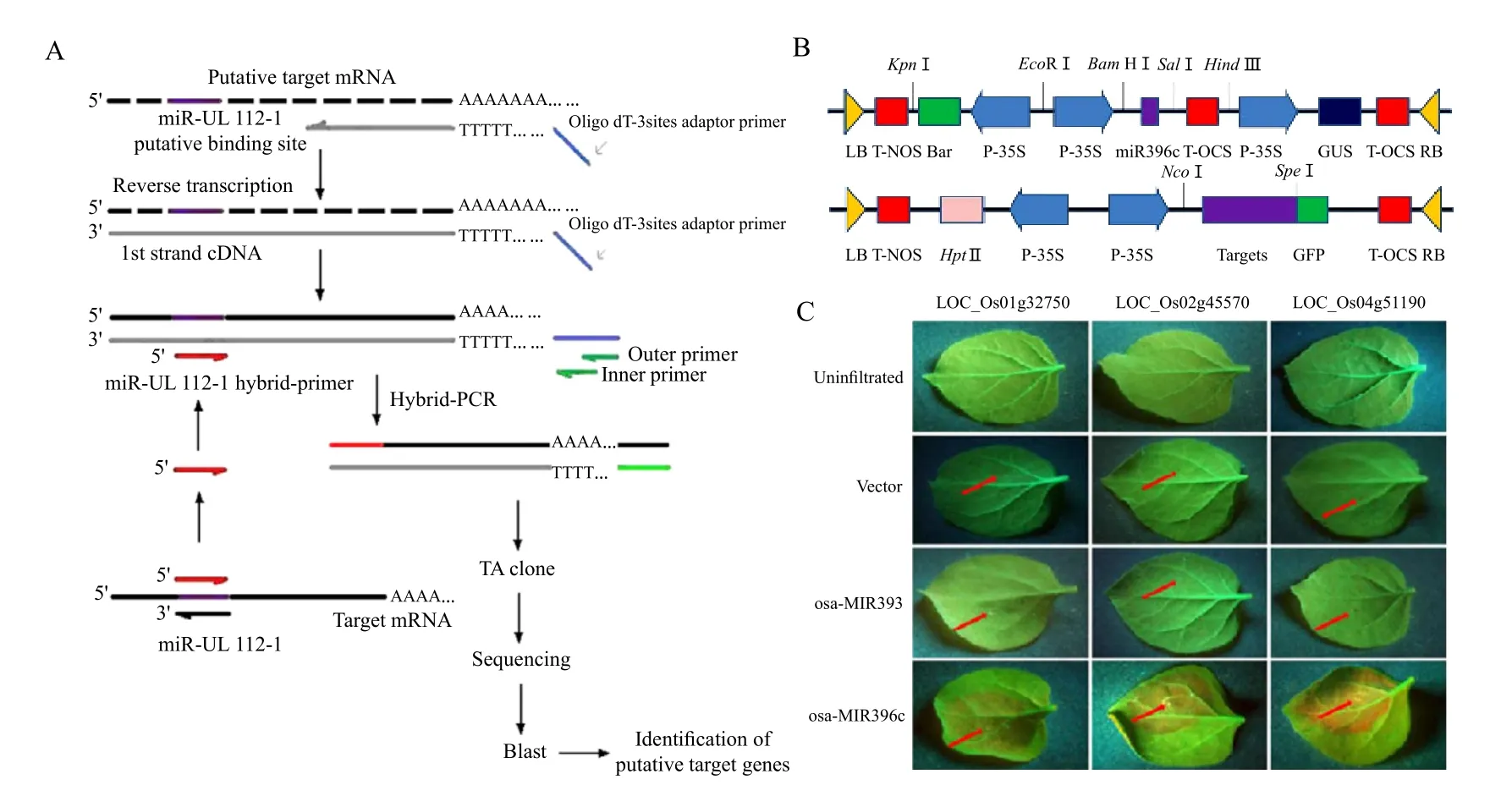
Fig. 2 Diagrammatic representation of hybrid-PCR and a GFP report system
An Agrobacterium-mediated delivery system (Agroinoculation)was developed to validate miRNA target genes, where miRNA and target mRNA were coexpressed in N. benthamiana leaf tissue, and the splice product was then detected with a northern blot. Using this method, Llave et al. (2002a)showed that miRNA 39 targeted SCL mRNAs.
At present, the most commonly used method for miRNA target validation is a luciferase reporter assay. The basic principle is that the luciferase gene,tagged with the miRNA target site, is transfected into cells expressing said miRNA, where changes in luminescence correspond to changes in transcript and protein levels, as conferred by miRNA silencing(Enright et al., 2003; Eskildsen et al., 2011; Welch et al., 2007). Liu and his colleagues (2008)used this method to study the function of the miR-16 family,where their studies showed that miR-16 family could regulate ccnd1.
GFP system is also used to detect miRNA targets,Chen et al. found that the MLK2 was directly regulated by miR-181b. They co-transfected HL-60 cells with miR-181b locked nucleic acid (LNA)and EGFP reporter vector containing either MLK23'UTR or MLK2 3'UTR carrying a mutational miR-181b binding site. The level of EGFP expression was significantly increased when endogenous miR-181b was blocked by miR-181b LNA. However, when the reporter vector containing a mutational miR-181b binding site was used, the increase in the level of EGFP was aborted (Chen et al., 2010). Our lab has also designed an in vivo assay, using leaf cells of Nicotiana benthamiana, to verify the interaction between miRNAs and their targets (Gao et al., 2010).Leaves normally appear red under ultraviolet light,while transgenic plants expressing GFP show up green under ultraviolet light (Brigneti et al., 1998).Putative target mRNAs were thus fused to the 5' end of GFP and introduced into N. benthamiana alongside a 35S promoter driven osa-MIR396c precursor. The area surrounding the infiltration points would turn red under a UV lamp due to degradation of the target-GFP fusion mRNA, thus allowing us to validate three osa-MIR396c targets (Fig. 2C). However, this method requires two plasmids, and, in order to get better results, we would occasionally need to develop fully transformed plants, which is a time-consuming process. Our research team is currently researching the use of protoplasts, in lieu of tobacco leaves, for miRNA target validation. We've constructed a vector which has three promoters to control expression of the miRNA, as well as RFP and GFP. GFP will be fused to the miRNA target site of putative mRNA targets,and RFP will be used as control to determine whether the vector has been introduced into the protoplasts.We can then see whether an interaction exists, which we can observe by the intensity of the GFP signal. We believe this method will be more direct and eff i cient.
Perspectives
miRNAs are post-transcriptional regulators of gene expression, and are involved in numerous cellular processes. Consequently, miRNAs are an important component of gene regulatory networks, and an improved understanding of miRNAs will further our knowledge of these networks. In recent years,there have been many studies concentrated on the discovery of the new miRNAs and the identification of their mRNA targets. Although researchers have identified many miRNAs, few miRNA targets have been identif i ed by experimentation. There is complex relationship between miRNAs and mRNAs, as a single miRNA can target multiple mRNAs and a single mRNA can be targeted by multiple miRNAs. However, most of the current methods for identifying regulatory miRNAs and their target mRNAs ignore this observed phenomenon and focus on miRNA-mRNA pairs.
Although current methods of miRNA target identif i cation contain some fl aws, the application and popularization of these methods has promoted the rapid expansion of miRNA targets as a fi eld research.Still, several key principles have emerged regarding the pattern of miRNA target recognition, and these have been applied to computationally predict targets of miRNA regulation (Bartel, 2009)
Currently, there is a demand for a more complete understanding of target gene-miRNA interactions, in an effort to fully map complete regulatory networks.Taken together, the study of miRNA target genes may be more complex than imagined. The development of novel bioinformatics techniques will undoubtedly lead to the prediction of numerous miRNAs and target genes. However, the challenge still remains to verify these predictions experimentally. In the future, we predict that miRNA target identification will focus more on high throughput, in vivo, identifications of target genes. The development and application of these new methods will no doubt result in the identif i cation and validation of many more miRNAs, and their target genes, expanding our understanding of the breadth and function of miRNA networks.
We are grateful to Mr Neil Hobson (University of Alberta, Canada)for the language editing assistance.
Andachi Y. 2008. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA, 14: 2440-2451.
Baek D, Villén J, Shin C, et al. 2008. The impact of microRNAs on protein output. Nature, 455(7209): 64-71.
Bartel D P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116: 281-297.
Bartel D P. 2009. MicroRNAs: target recognition and regulatory functions. Cell, 136: 215-233.
Beitzinger M, Peters L, Zhu J Y, et al. 2007. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol, 4: 76-84.
Boissonneault V, St-Gelais N, Plante I, et al. 2008. A polymerase chain reaction-based cloning strategy applicable to functional microRNA studies. Analytical Biochemistry, 381: 166-168.
Brigneti G, Voinnet O, Li W X, et al. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J, 17: 6739-6746.
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, et al. 2008.Widespread translational inhibition by plant miRNAs and siRNAs.Science, 320: 1185.
Calin GA, Cimmino A, Fabbri M, et al. 2008. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A, 105:5166-5171.
Chandrasekar V, Dreyer J L. 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci, 42:350-362.
Chen F, Yin J. 2005. Gene expression regulators -microRNAs. Chinese Science Bulletin, 50: 1281-1292.
Chen H, Chen Q, Fang M, Mi Y. 2010. microRNA-181b targets MLK2 in HL-60 cells. Sci China Life Sci, 53: 101-106.
Chen X, Guo X, Zhang H, et al. 2009. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene, 28: 1385-1392.
Chi SW, Zang J B, Mele A, et al. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature, 460: 479-486.
Doench J G, Sharp P A. 2004. Specif i city of microRNA target selection in translational repression. Genes Dev, 18: 504-511.
Du J F, Wu Y J, Fang X F, et al. Prediction of sorghum miRNAs and their targets with computational methods. Chinese Science Bulletin,55: 1263-1270.
Duan C, Wang C, Guo H. 2006. Regulation of microRNA on plant development and viral infection. Chinese Science Bulletin, 51:269-278.
Easow G, Teleman A A, Cohen S M. 2007. Isolation of microRNA targets by miRNP immunopurif i cation. RNA, 13: 1198.
Elbashir S M, Martinez J, Patkaniowska A, et al. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J, 20: 6877-6888.
Enright A J, John B, Gaul U, et al. 2003. MicroRNA targets in Drosophila. Genome Biol, 5: R1.
Eskildsen T, Taipaleenmaki H, Stenvang J, et al. 2011. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal)stem cells in vivo. Proc Natl Acad Sci USA, 108: 6139-6144.
Filipowicz W, Bhattacharyya S N, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics, 9: 102-114
Gao P, Bai X, Yang L, et al. 2010. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta, 231: 991-1001.
Griff i ths-Jones S, Saini H K, Van Dongen S, et al. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Research, 36: D154.
Hafner M, Landthaler M, Burger L, et al. 2010a. Transcriptome-wide identif i cation of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 141: 129-141.
Hafner M, Landthaler M, Burger L, et al. 2010. PAR-CliP - A method to identify transcriptome-wide the binding sites of RNA binding proteins. JoVE. 41. http://www.jove.com/details.php?id=2034, doi:10.3791/2034.
Hendrickson D G, Hogan D J, Herschlag D, et al. 2008. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance.PLoS One, 3: e2126.
Hsu R J, Yang H J, Tsai H J. 2009. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identif i cation of microRNA-target mRNAs. Nucleic Acids Research, 37: e77.
Huang Y, Qi Y, Ruan Q, et al. 2011. A rapid method to screen putative mRNA targets of any known microRNA. Virol J, 8: 8.
Hutvagner G, Zamore P D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297: 2056-2060.
Hwang H, Mendell J. 2006. MicroRNAs in cell proliferation, cell death,and tumorigenesis. British Journal of Cancer, 94: 776-780.
John B, Enright A J, Aravin A, et al. 2004. Human microRNA targets.PLoS biology, 2: e363.
Johnson S M, Grosshans H, Shingara J, et al. 2005. RAS is regulated by the let-7 microRNA family. Cell, 120: 635-647.
Johnston R J, Hobert O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature, 426: 845-849.Karginov F V, Conaco C, Xuan Z, et al. 2007. A biochemical approach to identifying microRNA targets. Proceedings of the National Academy of Sciences, 104: 19291.
Landthaler M, Gaidatzis D, Rothballer A, et al. 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA, 14: 2580.
Lee R C, Feinbaum R L, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75: 843-854.
Lewis B P, Burge C B, Bartel D P. 2005. Conserved seed pairing, often fl anked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120: 15-20.
Lewis B P, Shih I. 2003. Prediction of mammalian microRNA targets.Cell, 115: 787-798.
Lim L P, Lau N C, Garrett-Engele P, et al. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature, 433: 769-773.
Liu Q, Fu H, Sun F, et al. 2008. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res, 36:5391-5404.
Llave C, Xie Z, Kasschau K D, et al. 2002a. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science,297: 2053-2056.
Llave C, Xie Z, Kasschau K D, et al. 2002b. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science,297: 2053.
Mallory A C, Bartel D P, Bartel B. 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell Online, 17: 1360.
Mallory A C, Vaucheret H. 2006. Functions of microRNAs and related small RNAs in plants. Nature Genetics, 38: S31-S36.
Nonne N, Ameyar-Zazoua M, Souidi M, et al. 2010. Tandem affinity purif i cation of miRNA target mRNAs (TAP-Tar). Nucleic Acids Res,38: e20.
O'Donnell K A, Wentzel E A, Zeller K I, et al. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature, 435: 839-843.
Orom U A, Nielsen F C, Lund A H. 2008. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation.Molecular Cell, 30: 460-471.
Pei Y F, Wang Z M, Fei F, et al. 2010. Bioinformatics study indicates possible microRNA-regulated pathways in the differentiation of breast cancer. Chinese Science Bulletin, 55: 927-936.
Poy M N, Eliasson L, Krutzfeldt J, et al. 2004. A pancreatic isletspecif i c microRNA regulates insulin secretion. Nature, 432: 226-230.Reinhart B J, Slack F J, Basson M, et al. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans.Nature, 403: 901-906.
Reinhart B J, Weinstein E G, Rhoades M W, et al. 2002. MicroRNAs in plants. Genes & Development, 16: 1616.
Rom U A, Lund A H. 2007. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods, 43: 162-165.
Selbach M, Schwanhausser B, Thierfelder N, et al. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature, 455:58-63.
Sethupathy P, Megraw M, Hatzigeorgiou A G. 2006. A guide through present computational approaches for the identif i cation of mammalian microRNA targets. Nature Methods, 3: 881-886.
Shen Q, Cicinnati V R, Zhang X, et al. 2010. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer, 9: 227.
Song C, Fang J, Wang C, et al. 2010. MiR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata)microRNAs. PLoS One, 5: e10861.
Tan L P, Seinen E, Duns G, et al. 2009. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res, 37: e137.
Tang G, Reinhart B J, Bartel D P, et al. 2003. A biochemical framework for RNA silencing in plants. Genes Dev, 17: 49-63.
Vatolin S, Navaratne K, Weil R J. 2006. A novel method to detect functional microRNA targets. J Mol Biol, 358: 983-996.
Wang X J, Reyes J L, Chua N H, et al. 2004. Prediction and identif i cation of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol, 5: R65.
Welch C, Chen Y, Stallings R. 2007. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene, 26: 5017-5022.
Yu H, Song C, Jia Q, et al. 2011. Computational identification of microRNAs in apple expressed sequence tags and validation of their precise sequences by miR-RACE. Physiol Plant, 141: 56-70.
Zhang J G, Wang J J, Zhao F, et al. 2010. MicroRNA-21 (miR-21)represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta, 411: 846-852.Zhao LQ, Chen X, Cao Y. 2011. miRNA and nasopharyngeal carcinoma. Chinese Science Bulletin, 56: 722-728.
 Journal of Northeast Agricultural University(English Edition)2012年2期
Journal of Northeast Agricultural University(English Edition)2012年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- 3' Noncoding Region Construction of GHR Gene-luciferase Report Vector and Valuation
- Instruction to Authors
- Journal of Northeast Agricultural University (English Edition)
- Effects of Acute and Chronic Cold Stress on Antioxidant Function in Intestinal Tracts of Chickens
- Development of the SCNT Embryo from Butyrolactone I Prematured Bovine Oocytes
